Abstract
Purpose of Review
Asthma is a common inflammatory airway disease, which affects more than 300 million people worldwide. Although conventional drugs are effective for most of the patients with mild-to-moderate asthma, they are less effective for patients with difficult-to-treat or severe asthma. Identification of asthma endotypes and biomarkers will lead to more precise approaches to treat asthma.
Recent Findings
Asthma subphenotypes and endotypes have been described based on clinical variables and sputum granulocytes. A recent asthma endotype study has been summarized based on the combination of T2 (FeNO) and non-T2 (IL-6) biomarkers. Discovery of potential biomarkers for asthma has been discussed in the context of omics approaches. Current biologic drugs for asthma have been summarized, and the future direction of precise treatment of asthma has been suggested.
Summary
This review provides a concise overview of the current state of subphenotypes, endotypes, biomarkers, omics approaches, and biologic drugs in asthma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Asthma is a common respiratory disease characterized by chronic airway inflammation, history of respiratory symptoms, and variable expiratory airflow limitation, which affects more than 300 million people worldwide [1]. The main conventional drugs used for asthma control are inhaled corticosteroids (ICS), short-acting β2-agonist (SABA), long-acting β2-agonist (LABA), and leukotriene receptor antagonist (LTRA). Although these drugs are effective for most of the patients with mild-to-moderate asthma, they are less effective or even have side effects for patients with difficult-to-treat (~ 17% of asthma) or severe asthma (~ 4% of asthma) [1]. Identification of asthma endotypes linking pathophysiological pathways and molecular mechanisms to asthma clinical phenotypes and incorporating systems biology or omics findings should lead to more precise approaches to categorize and treat asthma.
Subphenotypes and Endotypes
Asthma is currently considered a heterogeneous syndrome rather than a single Th2 cytokine-mediated allergic disease. Asthma can be clinically categorized based on age onset of asthma, atopic status, obesity, and asthma severity. An unsupervised hierarchical cluster analysis has been performed in 726 subjects with asthma from the NHLBI-sponsored Severe Asthma Research Program (SARP) using 34 clinical variables covering a broad spectrum of measurements of asthma [2•]. Five distinct asthma subphenotypes have been identified in SARP, which can be described as early-onset atopic mild, moderate, and severe asthma, late-onset non-atopic obese asthma, and late-onset non-atopic COPD-like asthma [2•]. Although cluster analysis can identify more homogeneous subphenotypes of asthma, the identified subphenotypes are not always associated with definite pathophysiological pathways or treatment response. That’s why identification of asthma endotypes (subphenotypes with clear pathophysiological mechanism) is an essential step of precision medicine for asthma.
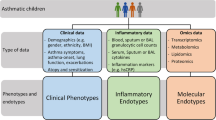
Combination of epidemiological, clinical, and pathophysiological characteristics with biomarkers, genetics, and treatment response may define endotypes of asthma. Stratified by sputum granulocytes (eosinophils and neutrophils), subjects with asthma can be categorized into four groups: mixed granulocytic, eosinophilic, neutrophilic, and paucigranulocytic asthma. Eosinophilic and neutrophilic asthma present type 2 (T2) and non-type 2 (non-T2) asthma, respectively. Paucigranulocytic asthma may present well-controlled asthma or asthma with low airway inflammation and is driven by structural changes of the airway or other mechanisms [3]. Subjects with mixed granulocytic asthma have the lowest lung function and highest health care use and asthma symptoms in SARP [4]. Cluster analysis in SARP using both clinical variables and sputum granulocytes shows that subjects with increased sputum neutrophils are associated with more severe asthma [5]. Sputum granulocytes are good biomarkers for airway inflammation; however, induced sputum is mainly a research-based approach and difficult for routine clinical use. Instead, measurement of fractional exhaled nitric oxide (FeNO) and serum IL-6 levels is non-invasive and may reflect airway inflammation process. Subjects with higher IL-6 levels tend to be more obese with higher blood neutrophil counts and have more severe asthma and asthma exacerbations [6••]. Subjects with higher FeNO levels tend to have T2 asthma with eosinophilic inflammation and have more severe asthma, asthma exacerbations, and greater improvement after systemic corticosteroid administration [6••]. Combination of T2 and non-T2 biomarkers may identify distinct asthma endotypes: high T2 & high non-T2, high T2 & low non-T2, low T2 & high non-T2, and low T2 & low non-T2 asthma (see Fig. 1).
Biomarkers and Omics
Biomarkers are biological indicators of a specific pathophysiological process, which can be used to stratify patients, track disease status and progression, and indicate drug effects. A good biomarker should be quantifiable, reproducible, non-invasive, and cost-effective [7]. Identification of biomarkers for different asthma endotypes is another essential step of precision medicine for asthma. At the current stage, blood eosinophils, sputum eosinophils, FeNO, and total serum IgE have been commonly used as biomarkers for T2 asthma (see Fig. 1). These T2 asthma biomarkers are positively correlated to each other; however, the correlations are not perfect which may indicate more than one T2 asthma endotype [6••]. Furthermore, different cutoff criteria have been suggested for these T2 biomarkers. Combination of several T2 biomarkers may be applied to fine-tune T2 asthma endotypes. In contrast, biomarkers for non-T2 asthma are less well defined. Some potential non-T2 asthma biomarkers are sputum neutrophils, Th1 cytokines, Th17 cytokines, and inflammation biomarkers such as IL-6 (see Fig. 1). Importantly, non-T2 asthma is not homogeneous but composed of multiple endotypes, and thus, identification of non-T2 asthma biomarkers is urgently needed.
Omics aims to collect and quantify biological molecules at genome-wide scales, including genomics, epigenomics, transcriptomics, proteomics, metabolomics, and microbiomics. Early trials of using single nucleotide polymorphisms (SNPs) to predict asthma are not very successful due to the limited heritability explained from the small number of SNPs identified in genome-wide association studies (GWAS). For example, area under the ROC curve (AUC) for asthma susceptibility is only 0.58 using SNPs in GSDMB, IL33, IL1RL1, TSLP, IL13, and HLA-DRA [8]. With the large biobanks and meta-analyses, more than a hundred asthma susceptibility genes have been identified [9, 10]. More importantly, Incorporation of omics findings into asthma subphenotypes or endotypes may reveal new biomarkers. A recent GWAS has identified SNPs in GATA3, MUC5AC, and KIAA1109 associated with moderate-to-severe asthma [11]. Another GWAS has identified shared and distinct SNPs/genes for childhood-onset and adult-onset asthma [12]. Polygenic risk score (PRS) approaches which account for the variable yet cumulative effects of GWAS loci may help identify subjects with high risk for asthma or asthma endotypes. Epigenomics, transcriptomics, proteomics, metabolomics, and microbiomics studies have also been used to identify biomarkers for asthma [13]; however, interpretation needs to be more careful since the findings are time-sensitive and tissue-specific. Integration of multi-omics findings is necessary to identify causal and/or hub genes. The NHLBI-sponsored Trans-Omics for Precision Medicine (TOPMed) program, ~ 144,000 subjects with whole-genome sequence and subset with other omics data (RNAseq, epigenomics, and metabolomics), may help identify novel biomarkers for asthma (https://www.nhlbiwgs.org).
Precision Medicine for Asthma
Precision medicine is a clinical approach to treat patients according to genes, environment, and lifestyle of each individual. To utilize precision medicine approach, we must understand not only molecular and pathophysiological mechanisms for disease risk in patients, but also therapeutic response to drugs. Pharmacogenetic studies evaluate the drug effects on patients based on genetic differences between individuals. Many candidate gene and some genome-wide pharmacogenetic studies have been performed for conventional asthma drugs such as ICS, SABA, LABA, and LTRA; however, the findings are largely inconsistent or unconvincing mainly due to small sample size and lack of replication [14]. To solve this problem, post hoc analyses of existing drug trial cohorts, large meta-analyses of these cohorts, and prospective studies stratified by genotypes and asthma subphenotypes or endotypes are necessary.
Biologic drugs are products from living organisms with known molecular mechanisms based on biotechnology methods. Currently, there are five FDA-approved biologic drugs for asthma treatment: omalizumab (anti-IgE antibody), mepolizumab and reslizumab (anti-IL-5 antibody), benralizumab (anti-IL-5Rα antibody), and dupilumab (anti-IL-4Rα antibody). These biologic drugs are recommended for patients with severe T2 asthma as add-on drugs when asthma symptoms are uncontrolled by high doses of ICS. Biologic drugs for non-T2 asthma are still in clinical trials including anti-inflammation, anti-IL-17, and anti-CXCR2 drugs [15]. Combination of anti-T2 and anti-non-T2 drugs may be required for high T2 & high non-T2 asthma; anti-T2 and anti-non-T2 drugs may be effective for high T2 & low non-T2 and low T2 & high non-T2 asthma, respectively; and other biologic drugs may be needed for low T2 & low non-T2 asthma (see Fig. 1). In the future clinical trials, collection of omics data and stratification of patients by endotypes should be taken into account at the stage of experimental design. The NHLBI-sponsored Precision Interventions for Severe asthma and/or Exacerbation-Prone Asthma Network (PrecISE), a clinical study from over 30 locations across the USA, may forward precision medicine treatment for severe asthma (https://preciseasthma.rog/preciseweb/).
Conclusions
In conclusion, it is time to re-analyze available clinical data stratified by genotypes and endotypes, to identify novel biomarkers using systems biology approaches on omics data, and to incorporate subphenotypes/endotypes and biomarkers/omics into future experimental design and clinical trials. Identification of asthma endotypes and biomarkers will lead to more precise approaches to treat asthma.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention (GINA). [accessed July 1, 2019; updated 2019]. Available at https://ginasthma.org
• Moore WC, Meyers DA, Wenzel SE, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181:315–23 This is a study showing heterogeneous clinical asthma subphenotypes.
Tliba O, Panettieri RA Jr. Paucigranulocytic asthma: uncoupling of airway obstruction from inflammation. J Allergy Clin Immunol. 2019;143:1287–94.
Hastie AT, Moore WC, Meyers DA, Vestal PL, Li H, Peters SP, et al. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J Allergy Clin Immunol. 2010;125:1028–36.
Moore WC, Hastie AT, Li X, Li H, Busse WW, Jarjour NN, et al. Sputum neutrophil counts are associated with more severe asthma phenotypes using cluster analysis. J Allergy Clin Immunol. 2014;133:1557–63.
•• Li X, Hastie AT, Peters MC, et al. Investigation of serum IL-6 and IL6R levels as biomarkers for asthma severity and asthma exacerbations. Am J Respir Crit Care Med. 2019;199:A2687 This is a study showing asthma endotypes based on T2 (FeNO or blood eosinophils) and non-T2 (IL-6) biomarkers.
Tiotiu A. Biomarkers in asthma: state of the art. Asthma Res Pract. 2018;4:10.
Li X, Ampleford EJ, Howard TD, Moore WC, Torgerson DG, Li H, et al. Genome-wide association studies of asthma indicate opposite immunopathogenesis direction from autoimmune diseases. J Allergy Clin Immunol. 2012;130:861–8.
Demenais F, Margaritte-Jeannin P, Barnes KC, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018;50:42–53.
Zhu Z, Lee PH, Chaffin MD, Chung W, Loh PR, Lu Q, et al. A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat Genet. 2018;50:857–64.
Shrine N, Portelli MA, John C, Soler Artigas M, Bennett N, Hall R, et al. Moderate-to-severe asthma in individuals of European ancestry: a genome-wide association study. Lancet Respir Med. 2019;7:20–34.
Pividori M, Schoettler N, Nicolae DL, Ober C, Im HK. Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma: genome-wide and transcriptome-wide studies. Lancet Respir Med. 2019;7:509–22.
Kan M, Shumyatcher M, Himes BE. Using omics approaches to understand pulmonary diseases. Respir Res. 2017;18:149.
Kersten ET, Koppelman GH. Pharmacogenetics of asthma: toward precision medicine. Curr Opin Pulm Med. 2017;23:12–20.
Agache I, Akdis CA. Precision medicine and phenotypes, endotypes, genotypes, regiotypes, and theratypes of allergic diseases. J Clin Invest. 2019;130:1493–503.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The author declares that he has no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by the author.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Asthma
Rights and permissions
About this article
Cite this article
Li, X. Hot Topic: Precision Medicine for Asthma—Has the Time Come?. Curr Allergy Asthma Rep 19, 45 (2019). https://doi.org/10.1007/s11882-019-0881-3
Published:
DOI: https://doi.org/10.1007/s11882-019-0881-3