Abstract
Recent many researchers focus on image segmentation methods due to the rapid development of artificial intelligence technology. Hereinto, pulse-coupled neural network (PCNN) has a great potential based on the properties of neuronal activities. This paper elaborates internal behaviors of the PCNN to exhibit its image segmentation abilities. There are three significant parts: dynamic properties, parameter setting and complex PCNN. Further, we systematically provide the related segmentation contents of the PCNN, and hope to help researchers to understand suitable segmentation applications of PCNN models. Many corresponding examples are also used to exhibit PCNN segmentation effects.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Image segmentation is regarded as one of the most important issues for image processing, and its main characteristic is that an image is divided into a certain number of regions according to the static and dynamic properties of the images. When image segmentation results are given by appropriate segmentation algorithms, we can further extract related features, and conduct the identification and classification of objects.
In the past decades, Eckhorn et al.’s [1,2,3,4] bio-inspired neural network based on cat visual cortex, can synchronously release pulses for similar neuron inputs. Johnson et al. [5,6,7,8,9,10] developed the above model and proposed a pulse coupled neural network (PCNN). Subsequently, Ranganath and Kinser et al. [11,12,13,14,15] presented modified PCNN models and further exploited image processing capacities of the PCNN.
PCNN has broad applications in image processing field, such as image fusion, image segmentation, image denoising, image enhancement, feature extraction. In recent years, PCNN have significant potentials for evolving image segmentation algorithms, due to its synchronous dynamic properties of the neuronal activity, including synchronous pulse release, capture behavior, nonlinear modulation and automatic wave. Further, in contrast to other prevalent segmentation methods, PCNN has low computational complexity and high segmentation accuracy, which becomes quite suitable for image segmentation. Thus, PCNN can obtain good segmentation application effects in natural images, medical images and other types of images.
In this paper, we provide basic and classical PCNN models to introduce fundamental properties of the PCNN, then explain internal segmentation behaviors of the PCNN according to three main aspects: dynamic properties, parameter setting and complex PCNN. We also give image segmentation applications of the PCNN as possible in subsequent section. The flowchart of the whole framework in this paper is shown as Fig. 1.
The rest of paper is organized as follows. Section 2 introduces the basic model and classical modified models of the PCNN. Section 3 elaborates the dynamic properties of the PCNN for image segmentation. Section 4 deduces main parameter setting methods of the prevalent PCNN models. Section 5 explains complex PCNN models by analyzing heterogeneous PCNN and multi-channel PCNN. Section 6 reviews image segmentation applications of the PCNN. Section 7 makes a conclusion for the paper.
2 The PCNN Models
2.1 Basic PCNN Model
A neuron is regarded as a basic unit for constructing neural networks, and is also an electrically excitable cell which can transmit and receive information via chemical and electrical signals. A typical neuron comprises a cell body, an axon and dendrites. Most neurons transmit and receive one particular type of signal by the axon and the dendrites, respectively. The majority of neurons belong to central nervous system that is simulated as artificial neural networks to analyze and solve practical problems.
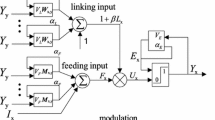
Different from traditional artificial neural networks, PCNN only has a single layer of laterally linked pulse coupled neurons, which main includes four crucial components: the dendritic tree, the membrane potential, the action potential and the dynamic threshold potential.
On the dendritic tree, the feeding synapses receive the external stimulus which is main input signal, and the action potential of the neighboring neurons. Moreover, the linking synapses are only associated with their adjacent neurons. The interaction of the above two synapses produces the membrane potential of the neuron, which is compared with the dynamic threshold potential to judge whether the action potential generates or not.
Lindblad and Kinser et al. [13] developed pulse-coupled neural network and presented its discrete model. The model in the position (i,j) for neuron Nij has five main parts: feeding input Fij[n], linking input Lij[n], internal activity Uij[n], dynamic threshold Eij[n] and pulse output Yij[n]. Hereinto, feeding input Fij[n] and linking input Lij[n] are given as follows:
In (1) and (2), e−αf and e−αl denote the exponential decay factors recording previous input states, VF and VL are the weighing factors modulating the action potentials of surrounding neurons. Additionally, Mijkl and Wijkl denote the feeding and linking synaptic weights, respectively. Sij is external feeding input stimulus which has a great influence for pulse-coupled synaptic modulation.
The decrease of numerical values for the first terms in (1) and (2) is with the increase of iteration times, moreover, corresponding values of the second terms in the above equations are always larger than zero owing to neighboring neurons firing. The third term in (1) denotes an external input stimulus with a nonzero positive number. Obviously, if the decay values of the first term in (1) surpass the sum of the second and third terms, the values of the feeding input Fij[n] will be smaller than the previous iteration Fij[n − ]. Similarly, Lij[n] and Lij[n − 1] can also acquire a comparison result after comparing with the values of two input terms in (2).
According to the computational mechanisms of pulse-coupled neural network, the non-linear modulation between feeding and linking inputs produces internal activity Uij[n] to deduce the coupling result of the membrane potential. The corresponding equation is given by:
where β denotes the linking strength, which directly determines the contribution of the linking input Lij[n] in internal activity Uij[n]. The above formula shows the modulatory coupling to the neuronal inputs. Obviously, the feeding input Fij[n] plays a most significant role in coupling modulating because of its weighing assignment, while the linking input Lij[n] has a secondary influence from neighboring neurons. Based on the comparison result between internal activity Uij[n] and dynamic threshold Eij[n], the neuronal firing condition and the dynamic threshold Eij[n] are described as follows:
In (4), if internal activity Uij[n] of a neuron is more than its dynamic threshold Eij[n], it will instantaneously fire and generate an output pulse (Yij[n] = 1); otherwise, the neuron will continue to keep quiet (Yij[n] = 0) until the above firing condition is ultimately satisfied.
In (5), αe is an exponential decay factor. The smaller its value, the more obviously dynamic threshold is affected from previous iteration. VE is the amplitude of dynamic threshold which generates an obvious influence only after previous neuron firing. The structure of basic PCNN model is shown in Fig. 2.
2.2 Classical PCNN-Modified Models
For image segmentation, there are four classical modified PCNN models: the intersecting cortical model (ICM) [14], the region growing PCNN model (RG-PCNN) [16], the spiking cortical model (SCM) [17], Simplified PCNN model (SPCNN) [18]. Most modified or simplified PCNN models derive from the above models because of high segmentation accuracy and low computational complexity.
2.2.1 Icm
Ekblad et al. [14] presented the ICM to extract image features without obvious boundaries. This PCNN structure simplifies the feeding and linking inputs, and remains the characteristics of the basic PCNN. Corresponding equations are given as
where f and g are decay constants of internal activity Fij[n] and dynamic threshold Eij[n], respectively. h is the amplitude of the dynamic threshold.
2.2.2 RG-pcnn
Stewart et al. [16] proposed the RG-PCNN to achieve region growing based on seed points, and avoided over-segmentation or under-segmentation of pixel regions. This is the first time that the PCNN combines basic region growing algorithm to segment interesting regions. The model is formulated as
In (9)–(12), Fij[n] is feeding input corresponding the pixel intensity at the n-th iteration. Tij[n] is firing time matrix which records neuronal firing states at each iteration. d is an inhibition term to limit the linking input and improve the smoothness of the segmented image. The parameters ω and \(\varOmega\) are fixed thresholdings at each iteration.
2.2.3 SCM
Referring to the literature of [7], the SCM is reasonably presented for image processing [17]. its equations are given by
In (13)–(15), f and g denote exponential decay factors. VL and VE are the amplitudes of internal activity and dynamic threshold, respectively.β is the linking strength. Obviously, the SCM retains main characteristics of the basic PCNN, and reduces setting parameters from seven to five. Additionally, the negative time matrix of the SCM can associate human subjective perception with objective input stimulus of the PCNN.
2.2.4 SPCNN
Based on Zhan et al.′s. SCM model, Chen et al. [18] proposed a SPCNN model with an automatic parameter setting method to segment assigned objects. The setting parameters of the former are obtained from empirical image attribution values, while those of the latter can be automatically determined according to static and dynamic properties of the PCNN. Hereinto, the setting parameters of SPCNN are formulated as
Subsequently, two segmentation examples of the SPCNN taken from Berkeley Segmentation Dataset [19], are given as shown in Fig. 3. Obviously, the SPCNN can generates good segmentation results with high contrast as shown in Fig. 3a–e, and bad results with low contrast as shown in Fig. 3f–j, due to automatic setting method of the above parameters.
The segmentation process of the SPCNN from Berkeley Segmentation Dataset: Images in the first column represent natural gray images; images in the second, third and fourth column represent the segmentation results of the SPCNN with iteration times 1–3; images in the fifth column represent the final segmentation results of the SPCNN with iteration times 3–6 (blue regions, cyan regions, red regions and yellow regions denote the third, fourth, fifth and sixth iteration results, respectively). (Color figure online)
3 Dynamic Properties
3.1 Feeding Input F
The feeding input Fij[n] of a neuron in (1) always includes three terms, and its expression can be rewritten as
Hereinto, the first term F1 denotes previous feeding input state based on an exponential decay factor e−αf. The second term F2 shows action potential outputs of the neighboring neurons with the weighing factor VF. The third term F3 is an external input stimulus Sij. A neuron changes its own previous state by adjusting the parameter e−αf in F1, and is always affected by its neighboring neurons according to F2. The neuron also receives external stimuli based on F3. Since F1 and F2 have similar input expressions and practical influences in contrast to traditional linking input in (2), simplified feeding inputs tend to be defined as F3, such as Refs. [20, 21], and its mathematical equation is expressed as
The input stimulus Sij is always defined as image attribution values, such as pixel intensities and pixel gradient values. In Addition, there are still little literatures that the feeding input is composed of last two items in (22), like Cheng et al. [22]. The literature adopted a simplified feeding input to strongly associate central neurons with neighboring neurons to detect the existence of the cracks. The simplified feeding input is given as
In (24), the first and second items actually derive from F2 and F3 in (22). Referring to [22], subsequent feeding input of basic PCNN can be divided into any combination of F1, F2 and F3 to solve practical image segmentation problems.
3.2 Linking Input L
The linking input Lij[n] of a neuron in (2) has two terms, and its equation can be also simplified as
Hereinto, the first term L1 denotes previous linking input state from an exponential decay factor e−αl analogous to the above parameter e−αf in (1). The second term L2 gives action potential outputs of the neighboring neurons. However, the above two terms would be difficult to distinguish from previous two terms of the feeding input in (22). In order to build a clear corresponding relationship between the feeding and the linking inputs, most simplified PCNN models mainly retain the second term L2 of the linking input in (25). Further, the linking inputs mainly focus on real influences of neighboring firing neurons to exhibit the connectivity and the interoperability between neurons. Most simplified linking input is written as
Additionally, the simplified linking input Lij[n] can also increase a positive constant d as an inhibition term to improve the smoothness of neuronal firing regions, especially when the whole image has a smaller difference for pixel intensities [16]. The linking input Lij[n] is rewritten as
In (27), the distribution range of the parameter d is 1 ≥ d > 0. The larger the values of the parameter d, the smoother the segmented image obtains final segmenting region. Neighboring firing neurons can also directly affect the linking input. It is obvious that the modified expression of the linking input always depends on the inhibition term d and neighboring action potentials Ykl[n − 1].
3.3 Internal Activity U
The internal activity Uij[n] in (3) derives from the coupled results between the feeding input and the linking input with linking strength β. The increase of Uij[n] is with the increase of the above two inputs. In recent years, there are several significantly improved expressions of the internal activity from the SPCNN [18], the enhanced PCN [23], the pulse coupled neural filter [24].
In the SPCNN, the internal activity Uij[n]1 includes its previous neuron state and the coupled result in (3). Its mathematical expression is given as
where the parameter αf is the exponential decay factor based on the internal activity. Comparing with most simplified PCNN models, the internal activity Uij[n] of the SPCNN provides the neuronal modulation results more reasonably.
The enhanced PCN is a two-layer recurrent PCNN whose internal activity Uij(t)2 in each layer increases an inhibition term γLij(t) modulated by the linking strength γ. The internal activity terms of the enhanced PCN are shown as
In (29) and (30), the inhibition terms readjust the firing times of corresponding neurons than basic PCNN. What is more, neurons firing speed of two-layer network is faster than single-burst PCNN.
For the pulse coupled neural filter, traditional coupled modulation is changed for avoiding direct influence of zero-valued pixels, and new internal activity is given as follows:
3.4 Action Potential Y
For basic PCNN in (4), the generation of action potential Y derives from the comparisons of internal activity Uij[n] and dynamic threshold Eij[n]. If Uij[n] > Eij[n], neuron Nij outputs a pulse (Yij[n] = 1); otherwise, it does not fire (Yij[n] = 0). Additionally, output values of action potential Y can occasionally be set by logical operation rules [25]. Certainly, referring to the SCM model [17], action potential Y can also be described as
In (32), action potential Yij[n]1 adopts the sigmoid function with the parameter γ. This indicates that the action potential can generate various types of pulse outputs, which is analogous to traditional action potential Yij[n] in (4).
Besides the above sigmoid function, the action potential can also adopt the radial-basis function [26]. Its equation is described as
Additionally, The action potential can generate other output values except ‘0’ or ‘1’, such as [27], and its equation is given as
Where ξij(n) is initial output results based on the comparisons between internal activity and dynamic threshold. Gij denotes the difference between the parameter ξij(n) and neighboring thresholdings. Yij(n)3 and k are final action potential and the hierarchy parameter, respectively. The above action potential setting method merge more related image information based on spatial adjacency proximity into the expression of the action potential.
3.5 Dynamic Threshold E
Dynamic threshold Eij[n] influenced by the exponential decay factor αe and the amplitude parameter VE, plays an important role for PCNN segmentation. In (4), the comparison results between internal activity Uij[n] and dynamic threshold Eij[n] can directly judge whether a neuron fire or not. This indicates that the calculation results of dynamic threshold Eij[n] obviously affect final firing results. The smaller the values of dynamic threshold Eij[n], the greater the number of firing neuron at the iteration is. Based on the dynamic threshold of basic PCNN, we will give several main improved equations and analyze their image segmentation characters.
Firstly, Gao et al. [20] adopted a regularized Heaviside function Hε including the cluster center m2[n] of the object to modify the dynamic threshold as
In (35), the dynamic threshold cannot form periodic oscillation because of the uses of the parameters Hε and m2[n]. Further, if the internal activity of a neuron is lower than m2[n], it will be prevented from firing.
Secondly, Cheng et al. [22] introduced a decay constant λ to control the changing rate of firing signal inputs and normalized pixel intensity Sij. The new dynamic threshold is described as
In (36), the dynamic threshold is gradually growing with the increase of the number of neighboring firing neurons before the new threshold satisfies the terminate condition of the whole cycle. Here, the setting method of the dynamic threshold focuses on the influences of neighboring pixels and normalized pixel intensity rather than dynamic threshold itself.
Thirdly, Xiang et al. [28] used five proper parameters to form a new dynamic threshold rather than the parameters αe and VE of basic PCNN. The dynamic threshold can be given by
where
In (37) and (38), N is the length of the attenuation step. T+ and K are the upper limit and the width of the variation range for dynamic thresholds, respectively. n denotes iteration number and M is maximum number of iterations. As the parameters T+, N, M and K are set to constant values, the change of dynamic threshold depends only on the parameter n. This setting method provides a new thinking that the dynamic threshold is determined by iteration times of the PCNN.
3.6 Feature Expression
3.6.1 Entropy Sequence
Ma et al. [29] proposed an automatic image segmentation method based on the maximum entropy to choose optimal segmentation result after n iterations. They also gave an entropy sequence to extract feature values of testing images. For entropy sequence, information entropy is firstly calculated at each iteration
where H(P) denotes the information entropy of a binary image. P1 and P0 are the occurring probabilities of ‘1’ and ‘0’, respectively. Secondly, all information entropies are combined into one sequence, which is regarded as an entropy sequence. It has invariant texture features in rotation, translation and scale. Significantly, most images always generate a unique entropy sequence by setting suitable PCNN parameters.
3.6.2 Time Sequence and Time Matrix
Time sequence is given by the sum of firing neurons at each iteration [16, 17]. Its expression is given as follow:
Obviously, time sequence can save neuronal firing information of the whole image, and further accomplish a feature conversion between multi-dimensional and one-dimensional information. Further, related mathematical definitions of time matrix are also given to determine its final expression [26]. Their corresponding equations are described as
In (41), the parameters g and θij(0) are the decay coefficient, and the amplitude of the dynamic threshold, respectively. As T 2ij [n] is an implicit function and cannot obtain the calculating result, we get reasonable time matrix by calculating the formula (42).
3.6.3 Neighboring Firing Matrix
We adopt a neighboring firing matrix to record the number of neighboring firing neurons at each iteration [30]. The mathematical equation of the firing matrix for four neighboring neurons is given as
Neighboring firing matrix based on human visual system (HVS) can improve the image description capacity of edge regions, which easily generate a more reasonable input stimulus for the PCNN.
3.6.4 Sub-intensity Ranges of Neurons Firing
The sub-intensity ranges exhibit corresponding pixel intensities of firing neurons at each iteration [18, 29]. Thus, it is significant to calculate and analyze the sub-intensity ranges of neurons firing for PCNN. Subsequently, we give two examples about the sub-intensity ranges of the SPCNN in Fig. 4a, b to clearly illustrate the change of pixel intensities at each iteration.
4 Parameter Setting
4.1 Input Stimulus
For traditional PCNN, input stimulus Sij is usually defined as a normalized pixel intensity, which directly affects computational results of the feeding inputs. Since more and more researchers pay attention to HVS, subsequent modified strategies are proposed to associate input stimuli of the PCNN with HVS. Based on Weber-Fechner law, Ref. [31] used a new input stimuli to simulate human vision perception, and the expression is defined by
where B and S 1ij denote objective and subjective pixel intensities, respectively. K and K0 denote two modulated constants. Referring to [31], Huang et al. [21] adopted a more reasonable external stimulus analogous to human eyes inputs
where K, C and r are three empirical constants. Tij records corresponding iteration times from firing neurons. The above formula builds the relationship between the time matrix Tij and subjective visual brightness. Ref. [30] also used the new expression of the input stimulus as follow:
where Soriij is a normalized pixel intensity in position (i,j) from an original image. S′denotes a normalized Otsu thresholding of the original image. Qij denotes the corresponding value of neighboring firing matrix Q in position (i,j). In (46), neighboring firing matrix and Otsu thresholding determine the input stimulus of modified PCNN model.
4.2 Exponential Decay Factor
4.2.1 Exponential Decay Factors α f and α l
For image segmentation, the exponential decay factors αf in (1) and αl in (2) are always set to empirical values under a complex scene. In order to satisfy desired requirements for identifying complex objects, several self-adaptive parameter setting methods, such as the SPCNN [18] and the PA-PCNN [32], are proposed.
In the SPCNN, the parameters αf and αl are merging into one parameter α 1f , the equation of which is given as
Based on the SPCNN, the parameter α 2f in the PA-PCNN adopts normalized Otsu thresholding S′ and its mathematical equation is written as
Comparing with the PA-PCNN, the parameter α 1f in the SPCNN is easy to generate more obviously exponent decay.
4.2.2 Exponential Decay Factor α e
Exponential decay factor αe has several expression approaches proposed by Refs. [18, 25, 31,32,33]. Chen et al. [18] adopted an adaptive-parameter expression and its equation is formulated by
In (49), S′ denotes the normalized Otsu thresholding of the whole image. What is more, Zhou et al. [25] used the cluster mean m2(n) from the fired region at previous iteration to build the relationship between the parameters m2(n) and α 2e
In addition, the parameter α 3e in Wei et al. [31] and Helmy et al. [33] adopted the average gray level μ and an adjustable constant C as follows:
Based on the above equations, Ref. [32] also provided a simplified expression of the parameter α 4e as
4.3 Input Amplitudes
4.3.1 Feeding Input Amplitude V F and Linking Input Amplitude V L
The parameters VF and VL in basic PCNN denote the amplitudes of the sum of output values from neighboring neurons at previous iteration for the feeding input and the linking input, respectively. In most prevalent modified models, the above two parameters are often eliminated to reduce computational complexity. Meanwhile, several important PCNN models only retain the parameter VL, which is empirically set to 1 [17, 18].
4.3.2 Dynamic Threshold Amplitude V E
The parameter VE is reasonably evolved from the experience data to the adaptive results. Significantly, several adaptive setting methods, such as the SPCNN and the MSPCNN, should be introduced due to their practical roles on image segmentation [18, 30].
For the SPCNN, the parameter V 1E is set to an adaptive value by reasonable formula derivation as follows:
Referring to the SPCNN, the parameter V 2E in the MSPCNN decreases the number of the setting parameters and its formula is written as
In (54), the parameter S′ denotes normalized Otsu thresholding and the parameter S′8 is an offset value.
4.4 Linking Strength
Kuntimad and Ranganath calculated the range of the linking strength β, and built the relationship between the linking strength and the linking input by acquiring neuronal capture ranges [34]. If the intensity range of object regions is [I4, I3], and the intensity range of background regions is [I2, I1] (I4 > I2 > I3 > I1), the capture results of the whole image can be given by
In (55)–(57), Lmin1(T1) and Lmin2(T2) are minimum linking inputs of the object and the background, respectively. According to the above three formulae, the range of the parameter β can be given as
Refs. [20, 23] also adopted the above method to determine the range of the parameter β. Refs. [16, 35, 36] gave the parameters βini and Δβ to redefine β. Hereinto, the parameter βini contains the minimum distance diffmin and the neuronal pulsing seed input inputseed. The parameter Δβ is a constant. Obviously, the value of β is gradually added by the parameter Δβ at each iteration until the statistical termination is met. Refs. [37, 38] set the value of the parameter β by calculating the distance between the neighboring pixels and the central pixel for a fixed region. Ref. [39] proposed the setting method of the parameter β, and can evidently improve neuron firing speed. Ref. [40] supplied a new expression based on the mean and variance of the pixel intensity. Ref. [22] gave a 3 × 3 matrix to obtain the linking strength β, which can decrease the influences of the bed pixels.
Besides the above semi-automatic setting method through calculating the empirical values and the constant values, several automatic parameter setting methods of the linking strength β are also presented, such as the SPCNN and the PA-PCNN. The SPCNN gives the expression of the parameter β1 as
Based on simplified parameter β1, the parameter β2 in the PA-PCNN is given as
In (59) and (60), Smax denotes the maximum normalized intensity of the whole image. S′ denotes the normalized Otsu thresholding.
4.5 Synaptic Weight
The parameters Mijkl and Wijkl in basic PCNN represent synaptic weights that are the sum of neighboring neuron outputs. Ranganath et al. [12] firstly adopted the inverse of the Euclidean distance to describe Mijkl and Wijkl as follows:
In (61), synaptic weights for neuron Nij in position (i, j) have many linking values by calculating the distances between neurons. Hereinto, the Euclidean distances of four and eight neighboring neurons are shown in Fig. 5.
References [28, 39, 41] adjusted synaptic weights based on the exponential function. Hereinto, Ref. [39] used a synaptic weight Mijkl with Gaussian distribution and its mathematical equation is defined as
where the parameter C is the normalized coefficient, and the parameter represents the smoothness of neighboring regions. For most modified models, the parameter Mijkl is removed and the parameter Wijkl is retained to reduce parameters number. Refs. [18, 30] expressed the synaptic weights by the matrices with constant values as follows:
Besides the constant values, the synaptic weight Wijkl also contains fixed parameter values such as [42].
5 Complex PCNN
5.1 Heterogeneous PCNN
Ref. [21] presented a heterogeneous PCNN model that is more suitable for human to observe. Based on the above PCNN, Ref. [43] proposed a heterogeneous SPCNN, including SPCNN 1, SPCNN 2 and SPCNN 3 for image segmentation. There are three types of the SPCNN models to produce the heterogeneous structure of the SPCNN. The above SPCNN models have different internal activities, linking strengths and synaptic weights, while they have same other setting parameters. They can be also connected via the linking parameter L12 linking the SPCNN 1 and the SPCNN 2, and the linking parameter L23 linking the SPCNN 2 and the SPCNN 3. Hereinto, the changes of internal activities directly influence final segmentation results, and three types of internal activities are given by
In (65)–(67), the heterogeneous SPCNN can acquire three different values for the internal activities at each iteration, meanwhile, it also gives the comparison results between the dynamic thresholds and the above internal activities to produce three different outputs as follows:
In (68)–(70), a1, a2 and a3 are final output values of SPCNN 1, SPCNN 2 and SPCNN 3, respectively. Their parameter values are set to 0.3, 0.5 and 0.2 (the sum of the parameter values is 1). The above three outputs Y1ij[n], Y2ij[n] and Y3ij[n] can be combined to one output Yij[n], corresponding values of which have ‘0’, ‘0.2’, ‘0.3’, ‘0.5’, ‘0.7’, ‘0.8’ and ‘1’ rather than ‘0’ or ‘1’. This indicates that the heterogeneous SPCNN has more clearly segmentation levels, which easily generate more reasonable segmentation results. The structure flowchart of heterogeneous SPCNN is shown in Fig. 6. The segmentation results of the heterogeneous PCNN from two examples of Berkeley Segmentation Dataset are given in Fig. 7.
5.2 Multi-channel PCNN
For color image segmentation, single-channel PCNN is difficult to provide reasonable segmentation results, because of corresponding color spaces of normalized Red Green Blue (RGB). Therefore, multi-channel PCNN with appropriate setting parameters has a significant meaning for further conducting color image segmentation.
Ref. [44] gave a reasonable segmentation strategy and identified objects in complex real-world scenes. Significantly, the achievement of multi-channel PCNN mainly depends on normalized expressions of different channels in color spaces. The resulting images based on RGB color spaces are shown as follows:
In (71)–(74), R, G and B denote pixel intensities of Red, Green and Blue in color space channels, respectively. r, g and b are the normalized expressions of different channels in RGB color models. O1, O2, O3, O4 and O5 denote five channels of transformed color images. For multi-channel PCNN, we first analyze transformed color channels, and then adopt modified PCNN models to acquire the result of each channel and integrate all the above results into final results for achieving the segmentation goal of multi-channel PCNN.
6 Related Applications of PCNN for Image Segmentation
6.1 Natural Image Segmentation
Stewart et al. [16] proposed a seeded region growing (SRG) algorithm that uses the inhibition term d to control the amplitude of the linking input in the PCNN and designs the fixed threshold Tx[t] to take the place of the dynamic threshold. Refs. [45,46,47] gave the termination conditions and the increment step Δβ of the linking strength to deduce fine segmentation steps of the modified region growing algorithms. Xu et al. [35] proposed a color region growing PCNN (CRG-PCNN) model, converting RGB color space into LAB and assessing the color distance between the neurons of feeding inputs. Zhou et al. [48] presented an extended PCNN based on a decision tree, which builds a direct relation between the adjustable parameters and the image characteristics. Zhao et al. [49]. designed a gradient-coupled spiking cortex model for further smoothing the pixels of same regions and enhancing the pixels of the contours from different regions.
Xiao et al. [50] modified the expression of dynamic thresholds for PCNN, and used fuzzy mutual information as the criterion of optimal segmentation results. Nie et al. [51, 52] merged Unit-Linking PCNN, maximum Shannon entropy rule, minimum cross-entropy rule and pre-processing strategies into image segmentation schemes. Ma et al. [53] gave the evaluation criterion of maximum information-entropy to find suitable iteration times for PCNN to obtain reasonable segmentation results. Zhou et al. [39] changed the expressions of the synaptic weights, the linking strength and the dynamic threshold to improve the automatic segmentation control ability of the PCNN. Zhan et al. [54] showed a fast-linking SCM model that can achieve the optimal thresholding selection and make better homogeneous objects as soon as possible. Li et al. [55] found a parameter optimal method of simplified PCNN based on immune algorithm for adjusting setting parameters automatically. Jiao et al. [56, 57] combined the SPCNN and the gbest led gravitational search (GLGSA) into a novel image segmentation method that is applied to 23 standard benchmark function. Guo et al. [58] first defined adaptive-semi-local feature contrast as the input stimulus of the original images, and then introduce saliency motivated improved simplified pulse coupled neural network based on HVS to locate interest regions.
There are still other modified algorithm based the PCNN, which are suitable for image segmentation, such as, oscillatory correlation PCNN [59], automatic design PCNN [60], clustering threshold PCNN [61], simplified parameters PCNN [62], unsupervised texture multi-PCNN [63], genetic PCNN [64], bidirectional search PCNN [65] and sine–cosine oscillation heterogeneous PCNN [66].
6.2 Medical Image Segmentation
For medical images, the lesions tend to be determined by appropriate segmentation steps, together including pre-processing, regions segmentation and post-processing. Hereinto, most modified PCNN models has a good potential for finding the locations of the lesions rapidly and accurately for different types of diseases.
Micro-calcification detection methods in digitized mammograms are separately proposed, including contourlet transform and simplified pulse coupled neural network, to extract micro-calcification clusters [67, 68]. PCNN-based level set also supplies a reasonable segmentation strategy with PCNN coarse segmentation and level set refined segmentation [69]. A PCNN-based segmentation algorithm in breast MR images can identify corresponding interest regions and detect the boundary of the breast regions [70]. An evolutionary PCNN is proposed to segment and detect breast lesions in ultrasound images [42]. Basic PCNN model is adopted to improve segmentation accuracy rate of potential masses based on digitized mammograms [71]. Moreover, MSPCNN [30] and PA-PCNN [32], simplifying adaptive parameters than traditional PCNN, are proposed to achieve initial segmentation of mass regions for digitized mammograms. Hereinto, the segmentation results of the MSPCNN based on Digital Database for Screening Mammography (DDSM) and Mammographic Image Analysis Society (MIAS) database [72, 73], are shown in Fig. 8.
Besides breast images, several modified PCNN is gradually used to segment the lesions of other common diseases. A biologically-inspired spiking neural network with median filter can reasonably detect the boundary of the prostate in ultrasound images [74]. A modified SPCNN obtains a reliable stone segmentation result in the ultrasound images of the gallbladder [75]. A spatial pulse coupled neural network with the statistical expectation maximization is proposed in brain MR images [76]. A PCNN-based segmentation algorithm can obtain the segmentation results of rat brain volumes in MR images [77]. A segmentation method combining the PCNN and Selective Binary and Gaussian Filtering Regularized Level Set (GFRLS) is adopted to segment and classify teeth based on anatomical structure in MicroCT slices [27]. A modified PCNN derived from Zhan et al.’s [78]. SCM model acquires nuclei in reflectance confocal images of epithelial tissue. A self-adaptive PCNN model with any colony optimization (ACO) obtains reasonable segmentation results of the lesion in MR medical images [79]. A Tandem Pulse Coupled Neural Network (TPCNN) with Contrast Limited Adaptive Histogram Equalization (CLAHE) and Deep Learning Based Support Vector Machine (DLBSVM) is proposed to acquire segmentation results of retinal blood vessels in ophthalmologic diabetic retinopathy images [80]. A memristive pulse coupled neural network (M-PCNN) based on hardware implementation, is applied to medical image edge extraction [81].
Other PCNN segmentation methods of medical images also provide reasonable segmentation frameworks, such as, the SCM-motivated enhanced CV [82, 83], the SPCNN-based segmentation approaches [84,85,86,87], the particle-swarm optimization PCNN [88].
6.3 Image Segmentation Based on HVS
Image segmentation algorithms based on HVS for PCNN can achieve desired segmentation goal, where the segmentation results are much closer to human vision classification than other prevalent algorithms. Huang et al. [21, 89] introduced the Web–Fechner law to build the relationship between objective image brightness and subjective human perception. Its equation is given as
Meanwhile, PCNN time matrix was also given to associate adjustable PCNN parameters with image brightness values as follows:
In (75), K and r denote setting constants. S and I represent objective image brightness values and subjective perception values, respectively. In (76), Tij[n] is time matrix of PCNN at the nth iteration. c denotes a constant. VE and αE denote the amplitudes of dynamic threshold and the exponential decay factor, respectively. According to the above two equations, final expression of subjective human perception based on PCNN can be written as
Based on (77), an objective expression with neighboring firing matrix based on HVS is described to provide a new input stimulus of the feeding input [30]. Its final equation is given in (46). Corresponding experimental results show segmentation performances analogous to human visual perception. The flowchart is given in Fig. 9.
6.4 Other Applications
PCNN also has broad applications in other aspects based on image segmentation, such as satellite image analysis [33, 90,91,92,93], visual saliency detection [58, 94, 95], plant recognition [28, 53, 96,97,98], iris feature extraction [99, 100], infrared human segmentation [101], palmprint verification [102], crack detection [22], power line detection [24], catenary fault detection [37], fabric defection [103], constrained ZIP code segmentation [104], vehicle recognition [38], aquatic feeding detection [105], heterogeneous material segmentation [106], fingerprint orientation field estimation [107], character recognition [108], feature extraction and object detection based on color images [44, 109,110,111]. Other segmentation applications can also be elaborated by the corresponding reviews [112,113,114,115].
7 Conclusion
PCNN plays important roles for image segmentation. In this paper, we give a comprehensive review to analyze PCNN segmentation properties and show related applications. There are three main steps. We first provide basic PCNN model and classical modified PCNN models, then introduce dynamic properties, parameter setting and multi-channel PCNN. In addition, we also provide main image segmentation applications of the PCNN. In the future, the PCNN merging semantic segmentation and deep learning will probably generate new image segmentation frameworks and bring a rapid progress than state-of-the-art algorithms.
References
Eckhorn R, Reitbock HJ, Arndt M, Dicke P (1989) A neural network for feature linking via synchronous activity: results from cat visual cortex and from simulations. In: Cotterill RMJ (ed) Models of brain function. Cambridge University Press, Cambridge
Eckhorn R, Reitboeck HJ, Arndt M, Dicke P (1990) Feature linking via synchronization among distributed assemblies: simulations of results from cat visual cortex. Neural Comput 2:293–307
Reitboeck HJ, Eckhorn R, Arndt M, Dicke P (1990) A model for feature linking via correlated neural activity. Springer, Berlin, pp 112–125
Eckhorn R (1999) Neural mechanisms of scene segmentation: recordings from the visual cortex suggest basic circuits for linking field models. IEEE Trans Neural Netw 10:464–479
Johnson JL, Ritter D (1993) Observation of periodic waves in a pulse-coupled neural network. Opt Lett 18:1253–1255
Johnson JL (1994) Pulse-coupled neural network, adaptive computing: mathematics, electronics, and optics, pp 47–76
Johnson JL (1994) Pulse-coupled neural nets: translation, rotation, scale, distortion, and intensity signal invariance for images. Appl Opt 33:6239
Johnson JL, Padgett ML (1999) PCNN models and applications. IEEE Trans Neural Netw 10:480
Padgett ML, Johnson JL (1997) Pulse coupled neural networks (PCNN) and wavelets: biosensor applications. In: Proceedings of international conference on neural networks, pp 2507–2512
Johnson JL, Padgett ML, Omidvar O (1999) Guest editorial overview of pulse coupled neural network (PCNN) special issue. IEEE Trans Neural Netw 10:461–463
Ranganath HS, Kuntimad G, Johnson JL (1995) Pulse coupled neural networks for image processing. In: Proceedings IEEE Southeastcon 95 visualize the future, pp 37–43
Ranganath HS, Kuntimad G (1996) Iterative segmentation using pulse-coupled neural networks. In: Applications and science of artificial neural networks II, International Society for Optics and Photonics, pp 543–555
Lindblad T, Kinser JM, Taylor J (2005) Image processing using pulse-coupled neural networks. Springer, Berlin
Ekblad U, Kinser JM, Atmer J, Zetterlund N (2004) The intersecting cortical model in image processing. Nuclear Instrum Methods Phys Res Sect A Accel Spectrom Detect Assoc Equip 525:392–396
Kinser JM (1996) Simplified pulse-coupled neural network. In: Applications and science of artificial neural networks II, International Society for Optics and Photonics, pp 563–568
Stewart RD, Fermin I, Opper M (2002) Region growing with pulse-coupled neural networks: an alternative to seeded region growing. IEEE Trans Neural Netw 13:1557–1562
Zhan K, Zhang H, Ma Y (2009) New spiking cortical model for invariant texture retrieval and image processing. IEEE Trans Neural Netw 20:1980–1986
Chen Y, Park S-K, Ma Y, Rajeshkanna A (2011) A new automatic parameter setting method of a simplified PCNN for image segmentation. IEEE Trans Neural Netw 22:880–892
Martin D, Fowlkes C, Tal D, Malik J (2001) A database of human segmented natural images and its application to evaluating segmentation algorithms and measuring ecological statistics. In: ICCV Vancouver
Gao C, Zhou D, Guo Y (2013) Automatic iterative algorithm for image segmentation using a modified pulse-coupled neural network. Neurocomputing 119:332–338
Huang Y, Ma Y, Li S, Zhan K (2016) Application of heterogeneous pulse coupled neural network in image quantization. J Electron Imaging 25:061603
Cheng Y, Tian L, Yin C, Huang X, Cao J, Bai L, Cheng Y, Tian L, Yin C, Huang X (2018) Research on crack detection applications of improved PCNN algorithm in MOI nondestructive test method. Neurocomputing 277:249–259
Ranganath HS, Bhatnagar A (2018) Image segmentation using two-layer pulse coupled neural network with inhibitory linking field. GSTF J Comput (JoC) 2018:1
Li Z, Liu Y, Walker R, Hayward R, Zhang J (2010) Towards automatic power line detection for a UAV surveillance system using pulse coupled neural filter and an improved Hough transform. Mach Vis Appl 21:677–686
Zhou D, Zhou H, Gao C, Guo Y (2016) Simplified parameters model of PCNN and its application to image segmentation. Pattern Anal Appl 19:939–951
Zhan K, Shi J, Wang H, Xie Y, Li Q (2017) Computational mechanisms of pulse-coupled neural networks: a comprehensive review. Arch Comput Methods Eng 24:533–588
Wang L, Li S, Chen R, Liu S-Y, Chen J-C (2016) An automatic segmentation and classification framework based on PCNN model for single tooth in MicroCT images. PLoS ONE 11:e0157694
Xiang R (2018) Image segmentation for whole tomato plant recognition at night. Comput Electron Agric 154:434–442
Ma Y, Dai R, Li L (2002) Automated image segmentation using pulse coupled neural networks and images entropy. J China Inst Commun 23:46–50
Lian J, Yang Z, Sun W, Guo Y, Zheng L, Li J, Shi B, Ma Y (2019) An image segmentation method of a modified SPCNN based on human visual system in medical images. Neurocomputing 333:292–306
Wei S, Hong Q, Hou M (2011) Automatic image segmentation based on PCNN with adaptive threshold time constant. Neurocomputing 74:1485–1491
Lian J, Shi B, Li M, Nan Z, Ma Y (2017) An automatic segmentation method of a parameter-adaptive PCNN for medical images. Int J Comput Assist Radiol Surg 12:1511–1519
Helmy AK, El-Taweel GS (2016) Image segmentation scheme based on SOM–PCNN in frequency domain. Appl Soft Comput 40:405–415
Kuntimad G, Ranganath HS (1999) Perfect image segmentation using pulse coupled neural networks. IEEE Trans Neural Netw 10:591–598
Xu G, Li X, Lei B, Lv K (2018) Unsupervised color image segmentation with color-alone feature using region growing pulse coupled neural network. Neurocomputing 306:1–16
Yonekawa M, Kurokawa H (2009) An automatic parameter adjustment method of pulse coupled neural network for image segmentation. In: International conference on artificial neural networks, Springer, pp 834–843
Wu C, Liu Z, Jiang H (2018) Catenary image segmentation using the simplified PCNN with adaptive parameters. Optik 157:914–923
Yang N, Chen H, Li Y, Hao X (2012) Coupled parameter optimization of PCNN model and vehicle image segmentation. J Transp Syst Eng Inf Technol 12:48–54
Zhou D, Gao C, Guo Y (2014) A coarse-to-fine strategy for iterative segmentation using simplified pulse-coupled neural network. Soft Comput 18:557–570
Bi Y, Qiu T, Li X, Guo Y (2004) Automatic image segmentation based on a simplified pulse coupled neural network. In: International symposium on neural networks, Springer, pp 405–410
Chacon-Murguia MI, Ramirez-Quintana JA (2018) Bio-inspired architecture for static object segmentation in time varying background models from video sequences. Neurocomputing 275:1846–1860
Gómez W, Pereira W, Infantosi AFC (2016) Evolutionary pulse-coupled neural network for segmenting breast lesions on ultrasonography. Neurocomputing 175:877–887
Yang Z, Lian J, Li S, Guo Y, Qi Y, Ma Y (2018) Heterogeneous SPCNN and its application in image segmentation. Neurocomputing 285:196–203
Chen Y, Ma Y, Kim DH, Park S-K (2015) Region-based object recognition by color segmentation using a simplified PCNN. IEEE Trans Neural Netw Learn Syst 26:1682–1697
Lu Y, Miao J, Duan L, Qiao Y, Jia R (2008) A new approach to image segmentation based on simplified region growing PCNN. Appl Math Comput 205:807–814
Chen N, Qian ZB, Zhao SX, Fan JS (2007) Region growing based on pulse-coupled neural network. In: 2007 International conference on machine learning and cybernetics, IEEE, pp 2832–2836
Ma T, Zhan K, Wang Z (2010) Applications of pulse-coupled neural networks. Springer, Berlin
Zhou D, Shao Y (2017) Region growing for image segmentation using an extended PCNN model. IET Image Process 12:729–737
Zhao R, Ma Y (2012) A region segmentation method for region-oriented image compression. Neurocomputing 85:45–52
Xiao Z, Shi J, Chang Q (2009) Automatic image segmentation algorithm based on PCNN and fuzzy mutual information. In: 2009 Ninth IEEE international conference on computer and information technology, IEEE, pp 241–245
Nie R, Zhou D, Zhao D (2008) Image segmentation new methods using unit-linking PCNN and image’s entropy. J Syst Simul 20:222–227
Nie R, Cao J, Zhou D, Qian W (2019) Analysis of pulse period for passive neuron in pulse coupled neural network. Math Comput Simul 155:277–289
Ma Y, Dai R, Li L, Wei L (2002) Image segmentation of embryonic plant cell using pulse-coupled neural networks. Chin Sci Bull 47:169–173
Zhan K, Shi J, Li Q, Teng J, Wang M (2015) Image segmentation using fast linking SCM. In: 2015 International joint conference on neural networks (IJCNN), IEEE, pp 1–8
Li J, Zou B, Ding L, Gao X (2013) Image segmentation with PCNN model and immune algorithm. J Comput 8:2429–2437
Jiao K, Pan Z (2019) A novel method for image segmentation based on simplified pulse coupled neural network and Gbest led gravitational search algorithm. In: IEEE access
Jiao K, Xu P, Zhao S (2018) A novel automatic parameter setting method of PCNN for image segmentation. In: 2018 IEEE 3rd international conference on signal and image processing (ICSIP), IEEE, pp 265–270
Guo Y, Yang Z, Ma Y, Lian J, Zhu L (2018) Saliency motivated improved simplified PCNN model for object segmentation. Neurocomputing 275:2179–2190
Wang D, Terman D (1997) Image segmentation based on oscillatory correlation. Neural Comput 9:805–836
Berg H, Olsson R, Lindblad T, Chilo J (2008) Automatic design of pulse coupled neurons for image segmentation. Neurocomputing 71:1980–1993
Li H, Guo L, Yu P, Chen J, Tang Y (2016) Image segmentation based on iterative self-organizing data clustering threshold of PCNN. In: 2016 2nd International conference on cloud computing and internet of things (CCIOT), IEEE, pp 73–77
Huang Y, Shuang W (2008) Image segmentation using pulse coupled neural networks. In: International conference on multimedia and information technology
Wang M, Han G, Tu Y, Chen G, Gao Y (2008) Unsupervised texture Image segmentation based on Gabor wavelet and multi-PCNN. In: 2008 Second international symposium on intelligent information technology application, IEEE, pp 376–381
Ma Y, Qi C (2006) Study of automated PCNN system based on genetic algorithm. J Syst Simul 18:722–725
Yang L, Lei K (2010) A new algorithm of image segmentation based on bidirectional search pulse-coupled neural network. In: 2010 International conference on computational aspects of social networks, IEEE, pp 101–104
Yang Z, Lian J, Li S, Guo Y, Ma Y (2019) A study of sine–cosine oscillation heterogeneous PCNN for image quantization. Soft Comput 2019:1–12
Yang Z, Dong M, Guo Y, Gao X, Wang K, Shi B, Ma Y (2016) A new method of micro-calcifications detection in digitized mammograms based on improved simplified PCNN. Neurocomputing 218:79–90
Guo Y, Dong M, Yang Z, Gao X, Wang K, Luo C, Ma Y, Zhang J (2016) A new method of detecting micro-calcification clusters in mammograms using contourlet transform and non-linking simplified PCNN. Comput Methods Progr Biomed 130:31–45
Xie W, Li Y, Ma Y (2016) PCNN-based level set method of automatic mammographic image segmentation. Optik Int J Light Electron Opt 127:1644–1650
Hassanien AE, Kim T-H (2012) Breast cancer MRI diagnosis approach using support vector machine and pulse coupled neural networks. J Appl Log 10:277–284
Ali JM, Hassanien AE (2006) PCNN for detection of masses in digital mammogram. Neural Netw World 16:129
Heath M, Bowyer K, Kopans D, Moore R, Kegelmeyer WP (2000) The digital database for screening mammography. In: Proceedings of the 5th international workshop on digital mammography Medical Physics Publishing, pp 212–218
Suckling P (1994) The mammographic image analysis society digital mammogram database. In: Digital Mammo, 375–386
Hassanien AE, Al-Qaheri H, El-Dahshan E-SA (2011) Prostate boundary detection in ultrasound images using biologically-inspired spiking neural network. Appl Soft Comput 11:2035–2041
Lian J, Ma Y, Ma Y, Shi B, Liu J, Yang Z, Guo Y (2017) Automatic gallbladder and gallstone regions segmentation in ultrasound image. Int J Comput Assist Radiol Surg 12:553–568
Fu J, Chen C, Chai J, Wong ST, Li I (2010) Image segmentation by EM-based adaptive pulse coupled neural networks in brain magnetic resonance imaging. Comput Medl Imaging Gr 34:308–320
Murugavel M, Sullivan JM Jr (2009) Automatic cropping of MRI rat brain volumes using pulse coupled neural networks. Neuroimage 45:845–854
Harris MA, Van AN, Malik BH, Jabbour JM, Maitland KC (2015) A pulse coupled neural network segmentation algorithm for reflectance confocal images of epithelial tissue. PLoS ONE 10:e0122368
Xu X, Liang T, Wang G, Wang M, Wang X (2017) Self-adaptive PCNN based on the ACO algorithm and its application on medical image segmentation. Intell Autom Soft Comput 23:303–310
Jebaseeli TJ, Durai CAD, Peter JD (2019) Segmentation of retinal blood vessels from ophthalmologic diabetic retinopathy images. Comput Electr Eng 73:245–258
Zhu S, Wang L, Duan S (2017) Memristive pulse coupled neural network with applications in medical image processing. Neurocomputing 227:149–157
Guo Y, Gao X, Yang Z, Lian J, Du S, Zhang H, Ma Y (2018) SCM-motivated enhanced CV model for mass segmentation from coarse-to-fine in digital mammography. Multimed Tools Appl 77:24333–24352
Gao X, Wang K, Guo Y, Yang Z, Ma Y (2015) Mass segmentation in Mammograms based on the combination of the spiking cortical model (SCM) and the improved CV Model. In: International symposium on visual computing, Springer, pp 664–671
Ma Y, Wang L, Ma Y, Dong M, Du S, Sun X (2016) An SPCNN-GVF-based approach for the automatic segmentation of left ventricle in cardiac cine MR images. Int J Comput Assist Radiol Surg 11:1951–1964
Ma Y, Wang D, Ma Y, Lei R, Wang K (2017) Novel approach for automatic segmentation of LV endocardium via SPCNN. In: Eighth international conference on graphic and image processing (ICGIP 2016), International Society for Optics and Photonics, pp 1022519
Wang K, Ma Y, Lei R, Yang Z, Ma Y (2017) Automatic right ventricle segmentation in cardiac MRI via anisotropic diffusion and SPCNN. In: Eighth international conference on graphic and image processing (ICGIP 2016), International Society for Optics and Photonics, pp 1022527
Guo Y, Wang X, Yang Z, Wang D, Ma Y (2016) Improved saliency detection for abnormalities in mammograms. In: 2016 International conference on computational science and computational intelligence (CSCI), IEEE, pp 786–791
Hage IS, Hamade RF (2013) Segmentation of histology slides of cortical bone using pulse coupled neural networks optimized by particle-swarm optimization. Comput Med Imaging Gr 37:466–474
Huang Y, Ma Y, Li S (2015) A new method for image quantization based on adaptive region related heterogeneous PCNN. In: International symposium on neural networks, Springer, pp 269–278
Li H, Jin X, Yang N, Yang Z (2015) The recognition of landed aircrafts based on PCNN model and affine moment invariants. Pattern Recognit Lett 51:23–29
Waldemark K, Lindblad T, Bečanović V, Guillen JL, Klingner PL (2000) Patterns from the sky: satellite image analysis using pulse coupled neural networks for pre-processing, segmentation and edge detection. Pattern Recognit Lett 21:227–237
Karvonen JA (2004) Baltic sea ice SAR segmentation and classification using modified pulse-coupled neural networks. IEEE Trans Geosci Remote Sens 42:1566–1574
Del Frate F, Latini D, Pratola C, Palazzo F (2013) PCNN for automatic segmentation and information extraction from X-band SAR imagery. Int J Image Data Fusion 4:75–88
Wang B, Wan L, Li Y (2016) Saliency motivated pulse coupled neural network for underwater laser image segmentation. J Shanghai Jiaotong Univ (Sci) 21:289–296
Guo Y, Luo C, Ma Y (2017) Object detection system based on multimodel saliency maps. J Electron Imaging 26:023022
Wang Z, Sun X, Zhang Y, Ying Z, Ma Y (2016) Leaf recognition based on PCNN. Neural Comput Appl 27:899–908
Guo X, Zhang M, Dai Y (2018) Image of plant disease segmentation model based on pulse coupled neural network with Shuffle Frog Leap Algorithm. In: 2018 14th International conference on computational intelligence and security (CIS), IEEE, pp 169–173
Wang Z, Li H, Zhu Y, Xu T (2017) Review of plant identification based on image processing. Arch Comput Methods Eng 24:637–654
Xu G, Zhang Z, Ma Y (2008) An image segmentation based method for iris feature extraction. J China Univ Posts Telecommun 96:101–117
Wang Z, Ma Y, Xu G (2006) A novel method of iris feature extraction based on the ICM. In: 2006 IEEE international conference on information acquisition IEEE, pp 814–818
He F, Guo Y, Gao C (2019) A parameter estimation method of the simple PCNN model for infrared human segmentation. Opt Laser Technol 110:114–119
Wang X, Lei L, Wang M (2012) Palmprint verification based on 2D–Gabor wavelet and pulse-coupled neural network. Knowl Based Syst 27:451–455
Shi M, Jiang S, Wang H, Xu B (2009) A simplified pulse-coupled neural network for adaptive segmentation of fabric defects. Mach Vis Appl 20:131–138
Shang L, Yi Z, Ji L (2009) Constrained ZIP code segmentation by a PCNN-based thinning algorithm. Neurocomputing 72:1755–1762
Ruan C, Zhao D, Chen X, Jia W, Liu X (2016) Aquatic image segmentation method based on hs-PCNN for automatic operation boat in crab farming. J Comput Theor Nanosci 13:7366–7374
Skourikhine AN, Prasad L, Schlei BR (2000) Neural network for image segmentation. In: Proceedings of SPIE: the international society for optical engineering, vol 4120, pp 28–35
Ji L, Yi Z, Shang L (2008) An improved pulse coupled neural network for image processing. Neural Comput Appl 17:255–263
Gu X, Zhang L, Yu D (2005) General design approach to unit-linking PCNN for image processing. In: Proceedings. 2005 IEEE international joint conference on neural networks, IEEE, pp 1836–1841
Gu X (2008) Feature extraction using unit-linking pulse coupled neural network and its applications. Neural Process Lett 27:25–41
Mohammed MM, Badr A, Abdelhalim M (2015) Image classification and retrieval using optimized pulse-coupled neural network. Expert Syst Appl 42:4927–4936
Mureşan RC (2003) Pattern recognition using pulse-coupled neural networks and discrete Fourier transforms. Neurocomputing 51:487–493
Zhan K, Shi J, Wang H, Xie Y, Li Q (2017) Computational mechanisms of pulse-coupled neural networks: a comprehensive review. Arch Comput Methods Eng 24:573–588
Yang Z, Lian J, Guo Y, Li S, Wang D, Sun W, Ma Y (2018) An overview of PCNN model’s development and its application in image processing. Arch Comput Methods Eng 2018:1–15
Wang Z, Ma Y, Cheng F, Yang L (2010) Review of pulse-coupled neural networks. Image Vis Comput 28:5–13
Subashini MM, Sahoo SK (2014) Pulse coupled neural networks and its applications. Expert Syst Appl 41:3965–3974
Acknowledgments
The authors thank all the reviewers for their valuable comments, which further improved the quality of the paper. This study was funded National Natural Science Foundation of China (Grant Nos. 61175012, 61962034 and 61861024), Natural Science Foundation of Gansu Province of China (Grant Nos. 148RJZA044 and 18JR3RA288) and Youth Foundation of Lanzhou Jiaotong University of China (Grant No. 2014005).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lian, J., Yang, Z., Liu, J. et al. An Overview of Image Segmentation Based on Pulse-Coupled Neural Network. Arch Computat Methods Eng 28, 387–403 (2021). https://doi.org/10.1007/s11831-019-09381-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11831-019-09381-5