Abstract
Trehalose plays an important role in mediating stress responses in plants, and trehalose-6-phosphate synthases and trehalose-6-phosphate phosphatases are essential for trehalose biosynthesis. Here, we address the function of rice (Oryza sativa) OsTPP3. We analyzed the expression of OsTPP3 in different tissues and stress conditions, and generated OsTPP3-overexpressing rice plants. These plants showed a higher tolerance to simulated drought conditions (10% PEG treatment) than wild-type (WT) plants. Reverse-transcription quantitative PCR analysis indicated that transcript levels of genes related to stress responses and abscisic acid biosynthesis were significantly higher in the OsTPP3 overexpressors than in WT plants. These results highlight the importance of OsTPP3 in conferring drought tolerance in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The non-reducing disaccharide trehalose and its intermediate product, trehalose-6-phosphate (T6P), play important roles in the regulation of metabolism and stress (Paul et al. 2008). In trehalose biosynthesis, trehalose-6-phosphate synthases (TPSs) catalyze the synthesis of T6P, and trehalose-6-phosphate phosphatases (TPPs) dephosphorylate T6P to produce trehalose (Paul 2007; Paul et al. 2008). Genome analyses have identified variable numbers of TPS and TPP genes in different plant species. For example, rice (Oryza sativa) has 11 TPS genes clustered into two subfamilies (Zang et al. 2011). Winter wheat (Triticum aestivum) has 12 TPS genes (Xie et al. 2015), and different types of cotton have different numbers of these two genes, with 15, 14, and 24 TPS genes in Gossypium raimondii (group D), G. arboreum L. (group A), and G. hirsutum L. (group AD), respectively (Mu et al. 2016). This diversity and expansion of these two gene families likely reflects the many roles TPS and TPP proteins play in plant growth and development, as well as stress tolerance.
In growth and development, inflorescence branching is a major yield-related trait in grain crops and is controlled by the developmental fate of axillary shoot meristems (Ward and Leyser 2004). In maize, RAMOSA3 (RA3) encodes a trehalose-6-phosphate phosphatase that is expressed in discrete domains subtending axillary inflorescence meristems; RA3 regulates inflorescence branching by modifying a sugar signal that moves into axillary meristems (Satoh-Nagasawa et al. 2006). Sucrose levels decrease and trehalose levels increase in salt-treated flowering maize, thereby providing a resource for studying primary metabolic pathways in C4 plants (Henry et al. 2015). In garden peas (Pisum sativum L.), T6P regulates bud dormancy, activation, and growth through sucrose (Fichtner et al. 2017). In Arabidopsis thaliana, loss of TPS1 function causes delayed embryonic development and disrupts vegetative growth by affecting sugar metabolism and abscisic acid (ABA) biosynthesis (Gómez et al. 2010).
Genes associated with trehalose biosynthesis also function in responses to abiotic stress (Garg et al. 2002), and manipulation of these genes can affect key agronomic traits. For example, in maize (Zea mays), TPP overexpression significantly increases yield under drought and normal conditions (Nuccio et al. 2015). In rice, OsTPP7 promotes seed germination under anaerobic conditions, which allows rice farmers to shift from transplanting seedlings to direct seeding, thus reducing labor and increasing yield (Kretzschmar et al. 2015). In rice, OsTPP1 expression is regulated by stress and OsTPP1-overexpressing plants exhibit increased resistance to salt and low temperature, with increased expression of stress response genes (Ge et al. 2008). In rice, the mitogen-activated protein kinase (MAPK) OsMAPK3 phosphorylates the helix–loop–helix (HLH) transcription factor OsbHLH002 to increase OsTPP1 expression, which in turn increases trehalose levels (Zhang et al. 2017). OsTPP2 expression is regulated by stress factors including low temperature, drought, and salinity stress, as well as ABA treatment (Shima et al. 2007). Overexpression of OsTPS1 increases tolerance to cold, salt, drought, and other abiotic stresses in transgenic rice (OsTPS1; Li et al. 2011a). Resistance to drought, salt, and low temperature can be improved through heterologous expression of Escherichia coli TPS and TPP in rice (Jang et al. 2003).
Although many studies on TPS and TPP have been conducted to date, investigations of OsTPP3 remain limited. This study addresses this gap by measuring OsTPP3 expression in various tissues and in response to different abiotic stressors. Additionally, we generated transgenic lines overexpressing OsTPP3 and show that these lines have increased tolerance to drought treatment. Our findings further indicate that the enhanced drought tolerance in transgenic rice resulted from changes in the expression of ABA biosynthetic and abiotic stress-related genes.
Materials and methods
Plant and other experimental materials
Rice (Oryza sativa L.) plants were grown in a paddy field at South China Agricultural University under natural conditions. Zhonghua 11 (ZH11) was used as the wild type. E. coli DH10B and Agrobacterium tumefaciens EHA105 were used for cloning and transformation experiments. pCAMBIA1380 was used as the binary vector for Agrobacterium-mediated transformation.
ABA treatments
To evaluate root length, seedling height, and germination rates, rice seeds were immersed in water and 3 or 6 μM ABA. Germination rates were determined 6 days after treatment. The root length and seedling height were directly measured 10 days after germination. Germination and seedling growth took place in a greenhouse with a 13/11-h day/night cycle (25/23 °C).
Stress treatments
Six-day-old seedlings were transferred into Kimura B nutrition solution. Two-week-old seedlings were subjected to abiotic stress treatments. For salt and simulated drought conditions, seedlings were treated with Kimura B solution supplemented with 10% PEG 6000 and were maintained in a greenhouse with a 13/11-h day/night cycle (25/23 °C). Seedlings were individually harvested at 0-, 2-, 4-, 6-, and 8-day intervals. For low- and high-temperature treatments, seedlings in Kimura B solution were subjected to 8/10 °C (day/night) or 42/37 °C (day/night) in the growth chamber (Canada Conviron PGV-36) with a 13/11-h day/night cycle. Seedlings were individually harvested at 0-, 3-, 12-, and 48-h intervals. The tissues were frozen in liquid nitrogen and stored at − 80 °C until RNA extraction.
To analyze drought tolerance of OsTPP3-overexpressing plants, WT and T2 homozygous seeds were germinated and transferred to Kimura B solution. The 2-week-old seedlings were then transferred into new Kimura B nutrition solution with 10% PEG 6000. The plants were photographed before treatment and at 6 and 10 days after drought treatment followed by 4 days of rewatering recovery.
Vector construction and genetic transformation
For overexpression of OsTPP3 (Os07g0624600, LOC_Os07g43160), the open reading frame was amplified from cDNA using the primers OETPPF (5’-aaagcttttgcttcggcttccgctgctc-3′) and OETPPR (5′-aaggatcccatgatcagttaccatccatgc-3′). After subcloning and sequencing, the correct gene was inserted into pCAMBIA 1380-Ubi (driven by the maize Ubiquitin promoter). The transformation was performed as previously described (Li et al. 2011b; Zhou et al. 2014).
DNA extraction and PCR detection
The DNA extraction was conducted as described previously (Zhou et al. 2014). The primers used in detection of hygromycin phosphotransferase (HPT) are as follows (HPTF, 5′-gaagtgcttgacattggggagt-3′; HPTR, 5′-agatgttggcgacctcgtatt-3′). The total volume of PCR reaction was 25 μL with 1 U Taq DNA polymerase (Takara, Dalian), 0.3 μM HPTF and HPTR, and 50 ng of genomic DNA. The wild-type plants were set as the negative control, and verified transgenic rice was set as the positive control. The PCR product was sent to Thermo Fisher Scientific for sequencing.
RNA extraction, cDNA synthesis, RT-PCR, and RT-qPCR
RNA was isolated using the RNA extraction kit with TRIzol reagent (Invitrogen, USA) and quantified with a DU730 spectrophotometer (Beckman Coulter, Germany). cDNA was synthesized with a 5 × iScript RT supermix kit (Takara, Dalian) using about 2 µg of total RNA. RT-qPCR was performed with SYBR premix Ex Taq II (Takara, Dalian) in a total volume of 20 μL on the Bio-Rad CFX 96 following the manufacturer’s protocol. Data were normalized to the internal rice UBIQUITIN (UBI) gene, and relative quantification was used for data analysis. RT-PCR was performed with the Takara Ex Taq Hot Start Version kit following the manufacturer’s manual. The primers used are as follows: (TPPF, 5′-caaggagatcgtcgtgttcctcg-3′; TPPR, 5′-atgcacctcccgctcacgatcg-3′; UBIF, 5′-aaccagctgaggcccaaga-3′; UBIR, 5′-acgattgatttaaccagtccatga-3′).
Primers for OsNCED1, OsNCED2, OsNCED3, and OsNCED4 were described by Zhu et al. (2009), and primers for OsRAB21, OsDREB2a, OsMYB2, OsPP2C49, OsPP2C48, OsPP2C6, OsRAB21, OsbZIP23, OsProt, and OsRab1 were described by Hong et al. (2016).
Statistical analysis
The statistical significance of each parameter was determined using t tests. P < 0.001, P < 0.01 and P < 0.05 were considered significant differences and labeled ***, **, and *, respectively.
Results
OsTPP3 expression was induced in response to abiotic stress
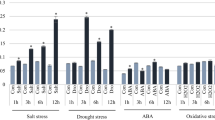
A query of the public database (https://www.ncbi.nlm.nih.gov) showed that the OsTPP3 gene is 1518 bp in length and encodes a 366-amino-acid protein. To study the expression of this gene, we used reverse-transcription quantitative PCR (RT-qPCR) to analyze the transcript levels of OsTPP3 in different tissues. We found that OsTPP3 is expressed in the roots, stems, stem nodes, seedlings, young panicles, leaves, and immature seeds, with expression levels in stem nodes being the highest (Fig. 1a).
Transcript analysis of OsTPP3 in different tissues and abiotic stresses. Relative expression of OsTPP3 was analyzed by RT-qPCR. a Expression pattern in different tissues. b Expression pattern of OsTPP3 in 2-week-old rice seedlings exposed to low temperature (8/10 °C) or high temperature (42/37 °C). Samples were collected at 0, 3, and 12 h. c Expression pattern of OsTPP3 in 2-week-old rice seedlings exposed to 10% PEG 6000 treatment. Samples were collected at 0, 2, 4, 6, and 8 days. RT-qPCR data were normalized using the rice UBI gene and are shown relative to 0 h. Error bars represent the mean ± SD of three biological replicates
Several studies have shown that expression of the TPP gene family is affected by stress conditions. Therefore, we measured the transcript level of OsTPP3 in high-temperature, low-temperature, and drought stress conditions. The high- and low-temperature treatments induced OsTPP3 expression, with low temperature producing a large response at 3 h after treatment. Under high temperature, OsTPP3 expression increased at 3 and 12 h and decreased relative to its previous levels at 12 h (Fig. 1b). During the 10% PEG treatment simulating drought, the expression levels of OsTPP3 were also increased at 2, 4, 6, and 8 days (Fig. 1c). These results indicate that OsTPP3 expression responds to various abiotic stresses.
Overexpression of OsTPP3 does not affect normal plant development
To investigate the function of OsTPP3 in rice, we generated 10 transgenic rice overexpressor lines containing OsTPP3 driven by the Ubiquitin promoter. Wild-type (WT) and transgenic plants did not show any obvious phenotypic differences when grown in normal conditions (Fig. 2a). In addition, PCR detection of the hygromycin phosphotransferase (HPT) selectable marker gene in transgenic lines revealed a 500-bp band present in overexpression lines (OE), but not in the WT, indicating that the construct was successfully inserted into the rice genome (Fig. 2b).
Phenotypic comparison of ZH11 and OsTPP3-overexpressing rice plants. a Appearance of WT and OsTPP3 overexpressor plants at ripening stage. b Detection of the HPT selectable marker. cOsTPP3 expression measured by RT-PCR. UBIQUITIN was used as a loading control. WT ZH11, OE1.1 and OE2.2 OsTPPT3 overexpression lines, M molecular marker DL2000, N negative control, P positive control
To verify the expression levels of OsTPP3 in the transgenic lines, RT-PCR was performed, which revealed that expression levels of OsTPP3 in the transgenic lines OE1.1 and OE2.2 were considerably higher than in WT (Fig. 2c), indicating that the incorporated OsTPP3 gene was successfully overexpressed.
OsTPP3-overexpressing rice plants show increased drought tolerance
To investigate drought tolerance in OsTPP3-overexpressing plants, we used osmotic stress to simulate drought stress. To this end, we treated WT and transgenic plants with 10% PEG 15 days after germination. Before treatment, no significant phenotypic differences between the overexpressing plants and the WT were detected (Fig. 3a). At 6 days of PEG treatment, the leaves of the WT were all curled (Fig. 3b). Although older leaves in the overexpressor lines were curled, the young leaves appeared normal (Fig. 3b). When the drought treatment was extended to 10 days, followed by a 4-day recovery period in fresh water, a few new leaves emerged from the WT and exhibited normal green color, whereas most leaves from the transgenic plants recovered to normal, with only the tips of some old leaves remaining withered (Fig. 3c). These results suggest that OsTPP3 overexpression enhances drought tolerance and recovery compared to WT plants.
Drought tolerance of WT and OsTPP3-overexpressing plants. Seeds were germinated in water and transferred to Kimura B nutrition solution. Two-week-old seedlings were treated in Kimura B nutrition solution with 10% PEG 6000. Plants before treatment (a), 6 days after drought treatment (b), and 10 days after drought treatment followed by 4 days of rewatering recovery (c) are shown. WT, wild type; OE1.1, OsTPP3 overexpression line. Three independent experiments were performed
OsTPP3-overexpressing rice lines show increased sensitivity to ABA
The plant hormone ABA is involved in the response to drought. To test the sensitivity of OsTPP3-overexpressing plants to ABA, we treated germinated seeds with different concentrations of ABA and measured plant height, root length, and seed germination, all of which are affected by ABA. WT plants did not show any significant differences in height between untreated and 3 μM ABA-treated plants, whereas the overexpressor lines exhibited significant differences between untreated and 3 μM ABA-treated plants (Fig. 4a).
Effect of ABA on root length, seedling height, and germination rate. The seedling height (a) and root length (b) were measured on 10-day-old plants after ABA treatment (n = 40). c Germination rates after immersion in water solution for 6 days (n > 200). The seeds and seedlings were cultured in hydroponic solution with 0, 3, or 6 μM ABA. *P < 0.05; **P < 0.01; ***P < 0.001 (t test)
Furthermore, root lengths were more strongly affected in the OsTPP3 overexpressor lines compared to WT. With 6 μM ABA treatment, the root length of the WT seedlings measured 6.08 ± 1.25 cm, which was significantly shorter than that of untreated plants, at 7.35 ± 0.98 cm (P = 0.015). Difference between 6 μΜ ABA-treated OsTPP3 overexpressor line OE1.1 and untreated plants was highly significant (P = 3.29 × 10−8) (Fig. 4b). Root lengths of the 6 μM ABA-treated OsTPP3-overexpressing line OE1.1 were 4.98 ± 0.95 cm, whereas in untreated plants, root length measured 7.86 ± 0.91 cm (Fig. 4b). A significant difference was also observed in another overexpressor line, OE2.2 (P = 3.11 × 10−8) (Fig. 4b). Root lengths of 6 μM ABA-treated OE2.2 were 4.27 ± 1.26 cm, whereas in untreated plants, root length measured 7.22 ± 1.01 cm (Fig. 4b).
In addition, we measured the effects of ABA on seed germination. When we treated seeds with 6 μM ABA, we observed no change in germination rates in WT plants, whereas seed germination in the two OsTPP3-overexpressing lines was significantly reduced (Fig. 4c). These results indicate that OsTPP3 overexpression increased sensitivity to ABA compared to WT plants.
ABA biosynthesis-related genes are upregulated in OsTPP3-overexpressing plants
To explore the reasons for the increased drought tolerance observed in OsTPP3-overexpressing plants, we measured the expression levels of several ABA biosynthesis-related genes using RT-qPCR. Expression levels of rice 9-cis-epoxycarotenoid dioxygenase (OsNCED) genes OsNCED1, OsNCED2, OsNCED3, and OsNCED4 in the two overexpressor lines were higher than in WT plants (Fig. 5). Of these genes, OsNCED2 expression was the most affected, increasing 30-fold compared to WT plants (Fig. 5b). OsNCED3 expression also increased 10-fold compared to WT levels (Fig. 5c). Expression of OsNCED1 and OsNCED4 was also significantly upregulated in the overexpressor lines relative to WT but to a lesser extent (Fig. 5a, d). These findings suggest that the increased expression of ABA biosynthesis-related genes may alter the ABA content in OsTPP3-overexpressing plants, which might explain the increased drought resistance in these plants.
Changes in expression levels of stress-responsive genes in transgenic plants
To examine whether OsTPP3 overexpression affects expression of drought-tolerance-related genes, 10 genes known to be involved in the drought response were analyzed by RT-qPCR. Expression levels of these 10 genes (OsRAB21, OsDREB2a, OsMYB2, OsPP2C49, OsPP2C48, OsPP2C6, OsRAB21, OsbZIP23, OsProt, and OsRab1) all significantly increased (Fig. 6). In the two OsTPP3-overexpressing lines, OsPP2C6 expression increased by 6.85- and 8.29-fold, respectively; OsProt expression increased by 6.72- and 5.64-fold, respectively; and OsRAB16 increased by 12.47- and 12.29-fold, respectively (Fig. 6). These findings indicate that these drought response genes increased the drought tolerance observed in the OsTPP3-overexpressing lines.
Discussion
Since the discovery of trehalose, its function as a sugar signaling molecule has been extensively investigated (Ramon and Rolland 2007; Smeekens et al. 2010). Trehalose can increase the tolerance of plants to drought, salt, low temperature, and other abiotic stress conditions. For instance, OsTPP1 overexpression improves resistance to different stresses (Ge et al. 2008) and increases trehalose levels to improve the resistance of transgenic rice to chilling (Zhang et al. 2017). In another example, OsTPP7 overexpression increases germination of seeds under submergence and anaerobic conditions (Kretzschmar et al. 2015). In this study, no significant phenotypic changes were observed in the OsTPP3-overexpressing rice variety ZH11. Drought simulated by PEG treatment showed that overexpressing plants were more tolerant than WT, which is consistent with previously reported functions of TPP family genes in rice (Ge et al. 2008; Kretzschmar et al. 2015; Shima et al. 2007; Zhang et al. 2017). Researchers have conducted functional studies on TPP and TPS gene families in rice, wheat, and maize that indicate that these genes are intimately associated with growth and development under stress conditions (OsTPS1; Li et al. 2011a; Nuccio et al. 2015; Xie et al. 2015).
ABA is a central regulator of abiotic stress responses in plants. Here, we show that expression levels of genes known to play a role in ABA biosynthesis (OsNCED1, OsNCED2, OsNCED3, and OsNCED4) are significantly higher in plants overexpressing OsTPP3 than in WT. Among these genes, expression levels of OsNCED2 and OsNCED3 were much higher than the WT plants, which may increase ABA contents and lead to an increase in drought tolerance compared to WT plants. In addition, the expression levels of various genes involved in drought responses were significantly higher in OsTPP3-overexpressing plants, particularly OsPP2C6 in the ABA signal transduction pathway (Han et al. 2017), the ABA marker gene OsRAB16 (Hong et al. 2009), and OsProt, which is responsible for proline transport (Igarashi et al. 2000). These findings suggest that enhanced drought resistance in OsTPP3-overexpressing plants may be due to an increase in ABA content and transport, and may be associated with higher levels of proline and other antioxidant substances.
In previous reports, the expression level of OsTPP1 and OsTPP2 were induced by exogenous ABA. Both of them confer stress tolerance in rice (Ge et al. 2008; Shima et al. 2007). Sweet potato IbMIPS1 (Zhai et al. 2016), cotton GhTPS11 (Wang et al. 2016), were induced too under stress treatment. Oropetium thomaeum under exogenous ABA stress application can increase trehalose accumulation (Zhang et al. 2018b). ABA biosynthesis and signaling genes were generally up-regulated and trehalose synthesis-related genes were up-regulated in tea plant (Camellia sinensis) (Liu et al. 2016), in switchgrass (Panicum virgatum L.) (Zhang et al. 2018a), and in IbMIPS1 overexpression of sweet potato (Zhai et al. 2016). These results suggest ABA and trehalose have some connection when plants encounter stress. One trehalose gene, OsTRE1, its overexpression of transgenic plants showed remarkable increases in trehalase activity, while had no morphological alterations or growth defects, except enhanced salt tolerance (Islam et al. 2019). It is similar with the results in our study. Arabidopsis TPS1 gene product plays an essential role in regulating the growth of vegetative, as well as embryogenic tissue in a mechanism involving ABA and metalose metabolism (Avonce et al. 2004; Gómez et al. 2010). OsTPS8 may regulate suberin deposition and trehalose in rice through ABA signaling to confer salinity tolerance (Vishal et al. 2019). All these studies suggest that trehalose and ABA have intimate connection in stress tolerance and growth development. These data will serve as a valuable resource for stress tolerance breading through genetic engineering.
TPP and TPS gene families have important functions in plants (Ramon and Rolland 2007), and research on these genes may reveal possible downstream applications to improve abiotic stress tolerance in plants. When rice TREHALOSE-6-PHOSPHATE PHOSPHATASE1 (OsTPP1) was overexpressed in developing maize ears using a floral promoter, kernel set and harvest index increased (Nuccio et al. 2015). Field data from several sites over multiple seasons showed that the activity of OsTPP1 improved yields from 9 to 49% under non-drought or mild drought conditions and from 31 to 123% under more severe drought conditions, relative to non-transgenic controls (Nuccio et al. 2015). In another example, transgenic rice expressing a fusion of TPS and TPP from E. coli that produces a bifunctional enzyme showed an increase in trehalose levels in leaves and seeds, which resulted in improved tolerance to drought, salt, and cold (Jang et al. 2003). These examples suggest that trehalose protects against abiotic stress and increases yield in extreme environmental conditions. In this study, the phenotype of OsTPP3-overexpressing plants appeared wild type under normal conditions but showed improved tolerance under drought conditions. This finding underscores the importance of further studies of OsTPP3 and the TPP and TPS gene families to develop molecular tools that might benefit future agricultural applications.
References
Avonce N, Leyman B, Mascorro-Gallardo JO, Van Dijck P, Thevelein JM, Iturriaga G (2004) The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol 136:3649–3659
Fichtner F, Barbier FF, Feil R, Watanabe M, Annunziata MG, Chabikwa TG, Hofgen R, Stitt M, Beveridge CA, Lunn JE (2017) Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). Plant J 92:611–623
Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu R (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99:15898–15903
Ge L, Chao D, Shi M, Zhu M, Gao J, Lin H (2008) Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 228:191–201
Gómez LD, Gilday A, Feil R, Lunn JE, Graham IA (2010) AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J 64:1–13
Han S, Min MK, Lee SY, Lim CW, Bhatnagar N, Lee Y, Shin D, Chung KY, Lee SC, Kim BG, Lee S (2017) Modulation of ABA signaling by altering VxGΦL motif of PP2Cs in Oryza sativa. Mol Plant 10:1190–1205
Henry C, Bledsoe SW, Griffiths CA, Kollman A, Paul MJ, Sakr S, Lagrimini LM (2015) Differential role for trehalose metabolism in salt-stressed maize. Plant Physiol 169:1072–1089
Hong C, Chao Y, Yang M, Cheng S, Cho S, Kao C (2009) NaCl-induced expression of glutathione reductase in roots of rice (Oryza sativa L.) seedlings is mediated through hydrogen peroxide but not abscisic acid. Plant Soil 320:103–115
Hong Y, Zhang H, Huang L, Li D, Song F (2016) Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front Plant Sci 7:4
Igarashi Y, Yoshiba Y, Takeshita T, Nomura S, Otomo J, Yamaguchi-Shinozaki K, Shinozaki K (2000) Molecular cloning and characterization of a cDNA encoding proline transporter in rice. Plant Cell Physiol 41:750–756
Islam MO, Kato H, Shima S, Tezuka D, Matsui H, Imai R (2019) Functional identification of a rice trehalase gene involved in salt stress tolerance. Gene 685:42–49
Jang IC, Oh SJ, Seo JS, Choi WB, Song SI, Kim CH, Kim YS, Seo HS, Choi YD, Nahm BH, Kim JK (2003) Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth. Plant Physiol 131:516–524
Li H, Zang B, Deng X, Wang X (2011a) Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 234:1007–1018
Li J, Jiang DG, Zhou H, Li F, Yang J, Hong LF, Fu X, Li ZB, Liu ZL, Li JM, Zhuang CX (2011b) Expression of RNA-interference/antisense transgenes by the cognate promoters of target genes is a better gene-silencing strategy to study gene functions in rice. PLoS One 6:e17444
Liu SC, Jin JQ, Ma JQ, Yao MZ, Ma CL, Li CF, Ding ZT, Chen L (2016) Transcriptomic analysis of tea plant responding to drought stress and recovery. PLoS One 11:e0147306
Mu M, Lu X, Wang J, Wang D, Yin Z, Wang S, Fan L, Ye W (2016) Genome-wide Identification and analysis of the stress-resistance function of the TPS (trehalose-6-phosphate synthase) gene family in cotton. BMC Genet 17:54
Nuccio ML, Wu J, Mowers R, Zhou H, Meghji M, Primavesi LF, Paul MJ, Chen X, Gao Y, Haque E, Basu SS, Lagrimini LM (2015) Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat Biotechnol 33:862–869
Paul M (2007) Trehalose 6-phosphate. Curr Opin Plant Biol 10:303–309
Paul MJ, Primavesi LF, Jhurreea D, Zhang Y (2008) Trehalose metabolism and signaling. Annu Rev Plant Biol 59:417–441
Ramon M, Rolland F (2007) Plant development: introducing trehalose metabolism. Trends Plant Sci 12:1360–1385
Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D (2006) A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441:227–230
Shima S, Matsui H, Tahara S, Imai R (2007) Biochemical characterization of rice trehalose-6-phosphate phosphatases supports distinctive functions of these plant enzymes. FEBS J 274:1192–1201
Smeekens S, Ma JK, Hanson J, Rolland F (2010) Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol 13:274–279
Trijatmiko KR, Gabunada LFM, Alam R, Jimenez R, Mendioro MS, Slamet-Loedin IH, Sreenivasulu N, Bailey-Serres J, Ismaill AM, Mackill DJ, Septiningsih EM (2015) A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat Plants 1:15124
Vishal B, Krishnamurthy P, Ramamoorthy R, Kumar PP (2019) OsTPS8 controls yield-related traits and confers salt stress tolerance in rice by enhancing suberin deposition. New Phytol 221:1369–1386
Wang CL, Zhang SC, Qi SD, Zheng CC, Wu CA (2016) Delayed germination of Arabidopsis seeds under chilling stress by overexpressing an abiotic stress inducible GhTPS11. Gene 575:206–212
Ward SP, Leyser O (2004) Shoot branching. Curr Opin Plant Biol 7:73–78
Xie D, Wang X, Fu L, Sun J, Zheng W, Li Z (2015) Identification of the trehalose-6-phosphate synthase gene family in winter wheat and expression analysis under conditions of freezing. J Genet 94:55–65
Zang B, Li H, Li W, Deng X, Wang X (2011) Analysis of trehalose-6-phosphate synthase (TPS) gene family suggests the formation of TPS complexes in rice. Plant Mol Biol 76:507–522
Zhai H, Wang F, Si Z, Huo J, Xing L, An Y, He S, Liu Q (2016) A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol J 14:592–602
Zhang Z, Li J, Li F, Liu H, Yang W, Chong K, Xu Y (2017) OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev Cell 43:731–743
Zhang C, Peng X, Guo X, Tang G, Sun F, Liu S, Xi Y (2018a) Transcriptional and physiological data reveal the dehydration memory behavior in switchgrass (Panicum virgatum L.). Biotechnol Biofuels 11:91
Zhang Q, Song X, Bartels D (2018b) Sugar metabolism in the desiccation tolerant grass Oropetium thomaeum in response to environmental stresses. Plant Sci 270:30–36
Zhou H, Zhou M, Yang YZ, Li J, Zhu LY, Jiang DG, Dong JF, Liu QJ, Gu LF, Zhou LY, Feng MJ, Qin P, Hu XC, Song CL, Shi JF, Song XW, Ni ED, Wu XJ, Deng QJ, Liu ZL, Chen MS, Liu YG, Cao XF, Zhuang CX (2014) RNase ZS1 processes Ub L40 mRNAs and controls thermosensitive genic male sterility in rice. Nat Commun 5:4884
Zhu G, Ye N, Zhang J (2009) Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol 50:644–651
Acknowledgements
This study was supported by the National Natural Science Foundation of China (31100872) and Genetically Modified Breeding Major Projects (2016ZX08001-004).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, D., Chen, W., Gao, J. et al. Overexpression of the trehalose-6-phosphate phosphatase OsTPP3 increases drought tolerance in rice. Plant Biotechnol Rep 13, 285–292 (2019). https://doi.org/10.1007/s11816-019-00541-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-019-00541-4