Abstract
Aluminum (Al) toxicity is a major limiting factor that inhibits root elongation and decreases crop production in acidic soils. The symptoms of inhibited root growth include a reduced uptake of nutrients because the roots become stubby and brittle. The release of organic anions from roots can protect a plant from Al toxicity. The mechanism relies on the efflux of organic anions, such as malate or citrate, which protect roots by chelating the Al3+. In this study, homologs of TaALMT1, a Camelina gene that encodes an aluminum-activated malate transporter, were investigated. The expression of this gene was induced by Al in the root, but not in the shoots. Using green fluorescent protein (GFP) fusion constructs and Western-blot analysis, we observed that CsALMT1 was localized in the plasma membrane. Also, to determine the degree to which Al tolerance was affected by malate secretion in Camelina root, we generated CsALMT1 overexpressing plants. CsALMT1 overexpressing transgenic plants showed a higher root elongation rate than the wild-type plant. Damaged cell staining analysis by hematoxylin under 25 µM Al treatment for 2, 4, and 6 h showed a pattern of less damage in CsALMT1 transgenic plants than in wild-type plant, especially in the root elongation zone. Furthermore, the rate of increase of secretion of organic acid in overexpressed plants after Al treatment was higher than that in the wild-type plant. In addition, in the Al-specific dye morin staining on root protoplast under 50 µM Al treatment, less Al accumulation was observed in the CsALMT1 transgenic plants than in the wild-type plant. The Al contents in the roots of the transgenic plants were at a lower level than those in the wild-type plant. These results show that the overexpression of CsALMT1 improves Al tolerance by increasing the release of malate from the root to the soil and, thereby, detoxifies the Al3+.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
At soil pH values below 5.0, soluble aluminum (Al) in the soil exists as a toxic trivalent cation that affects plant growth and function (Kochian et al. 2004). One of the most significant symptoms of Al toxicity in the soil inhibited root growth and root elongation via impaired cell division and cell elongation. The root apex is a primary target and plays a central role in the mechanism of Al toxicity and tolerance (Ryan et al. 1993; Kochian 1995; Ahn et al. 2001). However, many plant species have developed mechanisms to detoxify Al, both internally and externally. Internal detoxification involves the sequestration of Al into the plant’s vacuoles and chelation with an organic acid. The most documented and generally known mechanism of external detoxification in both monocotyledons and dicotyledons is the secretion of organic acid anions, such as malate, citrate, and oxalate, from the cytoplasm of root cells (Sasaki et al. 2004; Kochian et al. 2005; Ma 2007). These organic anions can be released through an anion transporter and can chelate toxic Al3+ into a nonphytotoxic Al form in the rhizosphere. This external tolerance of Al toxicity is a general mechanism in many species (Ryan and Delhaize 2010). The first tolerance gene, designated as an aluminum-activated malate transporter (TaALMT1), was isolated from wheat (Triticum aestivum) by Sasaki et al. (2004). Other members of Al-activated secretion of malate (ALMT1) have been identified in Arabidopsis thaliana (AtALMT1), rapeseed (BnALMT1), barley (HvALMT1), and maze (ZmALMT1) (Hoekenga et al. 2006; Ligaba et al. 2006; Gruber et al. 2010; Pineros et al. 2008).
Patterns of organic acid anion secretion were summarized by Ma (2000), and can be grouped into two patterns depending on the timing of secretion. In pattern I, the secretion of organic acid anions immediately occurs when the plant is exposed to Al, as observed in wheat (Delhaize et al. 1993) and buckwheat (Zheng et al. 2005). Pattern II exhibits a significant lag phase until the onset of organic acid anion release, as in Cassia tora (Ma et al. 1997b), Sorghum bicolor (Magalhaes et al. 2007), and Zea mays (Maron et al. 2008). These results indicate that the genetically engineered protein that modulates the organic acid anion release in the plasma membrane is an effective way to enhance Al tolerance in plants. Although the function of ALMT1 from other plant species has been well studied, the regulatory role of CsALMT1 (Genbank accession number: KF021618) from Camelina in malate exudation remains unknown.
Camelina sativa L. is an emerging bioenergy crop that can be grown under harsh soil conditions (Knorzer 1978; Putnam et al. 1993; Vollmann et al. 1996; Zubr 1997; Park et al. 2014). Furthermore, Camelina ALMT1 (CsALMT1), which is a homolog of the Arabidopsis ALMT1, was isolated. This gene may mediate Al tolerance via malate exudation in Camelina roots under Al treatment. Therefore, overexpression of CsALMT1 in Camelina plants may improve the Al tolerance. In this study, we conducted tests to investigate the effect of CsALMT1 on the detoxification of aluminum toxicity by comparing with Camelina wild-type and CsALMT1 overexpressing transgenic plants.
Materials and methods
Plant material and growth conditions
Camelina (Camelina sativa L.) seeds were sown on nylon mesh at 25 °C under dark conditions for 2 days with half-strength Hoagland solution [contained 1.25 mM KNO3, 1.5 mM Ca(NO3)2, 0.75 mM MgSO4, 0.5 mM KH2PO4, 75 µM FeEDTA, 50 µM H3BO3, 10 µM MnCl, 2 µM ZnSO4, 1.5 µM CuSO4, and 0.75 µM (NH4)6Mo7O24]. After germination, seedlings were grown under cycles of 16 h light/8 h dark for 3 days. The Hoagland solution (HS) was changed every day. To create a hydroponic culture for several experiments, 4-day-old seedlings were transferred to a modified HS adjusted to pH 4.5 without phosphate at least 12 h prior to treatment. To analyze the seedling growth rate under Al stresses, plants were grown vertically on 125 × 125 × 20 mm polystyrene Petri dishes containing half-strength nutrient medium (without phosphate) containing 1.25 mM KNO3, 1.5 mM Ca(NO3)2, 0.75 mM MgSO4, 75 µM FeEDTA, 50 µM H3BO3, 10 µM MnCl, 2 µM ZnSO4, 1.5 µM CuSO4, and 0.075 µM (NH4)6Mo7O24 containing 0.8% (w/v) phytoagar (Duchefa Biochemie) after sterilization with 75% ethanol for 10 min. Al was supplemented to the corresponding media at the concentrations indicated in Fig. 4.
RNA extraction and Cloning of CsALMT1 from cDNA
The roots of 5-day-old seedlings treated with 50 µM Al were harvested from Camelina, homogenized in liquid nitrogen, and RNA extracted using Trizol (Invitrogen, Carlsbad, CA) and an RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. The quantity of RNA was measured using a spectrophotometer. RNA was treated with RNase-free DNase-I for 45 min (Ambion, Austin, TX) followed by DNase-I removal according to the manufacturer’s instructions. Total RNA was synthesized to cDNA using a PrimeScript 1st strand cDNA Synthesis Kit (Takara).
The degenerate primers (5′-CAARGTSGGMCTKGCKCTYG-3′ and 5′- CYTSYCCRGCCCARACVGG-3′) were designed according to the conserved domains of the Aluminum-activated Malate Transporter 1 (ALMT1s) from Arabidopsis (Arabidopsis thaliana), maize (Zea mays), and wheat (Triticum aestivum). A fragment of 475 bp was PCR amplified using the degenerate primers from the cDNA. cDNA was synthesized using a SMARTertm RACE cDNA Amplification Kit (Clonetech). CsALMT1 was amplified using Rapid Amplification of cDNA Ends (RACE) PCR followed by primers (5′-GTCCTTTCCCAAGTGTGGCCCCAACAG-3′, 5′-GGTCA -TGTTGGTCTTTGTGCAAGCTGC-3′). All the PCR products were cloned into the pGEM T-Easy vector (Promega, Madison, WI, USA) for sequence analysis.
Northern-blot hybridization
For Northern-blot hybridization, 10 µg of RNA was denatured in formamide and formaldehyde, and separated by electrophoresis on 1.2% (w/v) agarose gels. A full-length gene-containing Open Reading Frame (ORF) region of the CsALMT1 probe was used. RNA was transferred to a nylon membrane (Amersham Hybond™-N+, GE Healthcare) and a cDNA probe was radioactively labeled as [α-32P]dCTP using the Prime-α-Gene-Labeling System (Invitrogen). Northern blot was hybridized at 60 °C in 10 ml hybridization buffer for 18 h and washed at 65 °C four times with 0.1× SSC, 0.1% SDS for 15 min. Hybridization was detected using a phosphoimager (Typoon FLA7000, Fujifilm-Life Science).
Characterization of CsALMT1 expression in different tissues using the qRT-PCR
5-day-old and 6-week-old plants were used for the ALMT1 expression pattern analysis of different tissues such as the flower, silique, stem, leaf, total root, basal root, and root tip. For comparative quantification of the CsALMT1 gene, real-time quantitative PCR (qRT-PCR) was performed in a Bio-Rad CFX96 Real-Time PCR system (Bio-Rad Laboratories Inc., Hercules, CA, USA). The 20-µL PCR mixture contained 4 µL of diluted cDNA, 10 µL of SYBR Premix (Takara), 2 µL of a mix of the primer pair, and 4 µL of water. The real-time PCR condition was followed by a three-step protocol with the melting curve. The data were analyzed using the Bio-Rad CFX manager (version 1.0) after normalization to CsActin. Triplicate PCR and three biological replicates were analyzed. The primer pairs used for this experiment were as follows: CsALMT1 5′-CCGTCAGTGTGCCTACAGAA-3′ (forward) and 5′-TGCAGGCAGATTGTGAGTTC-3′ (reverse), CsActin 5′- ACAATTTCCCGCTCTGCTGTTGTG-3′ (forward) and 5′-AGGGTTTCTCTCTTCCACATGCCA-3′ (reverse).
Subcellular localization of CsALMT1::GFP fusion protein
To determine the subcellular localization of the CsALMT1::GFP fusion protein, a green fluorescent protein (GFP) gene was fused in-frame to the C-terminus of the CsALMT1 gene and was subsequently expressed under the control of the CaMV 35S promoter. The CsALMT1::GFP fusion protein was transiently introduced to tobacco leaves. After 48 h of fusion protein infiltration, the fluorescence was observed by Laser Confocal Scanning Microscope (TCS NT, Leica Instruments).
Isolation of the plasma membrane vesicles and Western-blot analysis
Plasma membrane (PM) vesicles were prepared at exactly 4 °C following the method by Palmgren et al. (1990). PM was isolated from 2-month-old tobacco leaves with the addition of CsALMT1 fusion protein. The leaves were ground in an ice-cold homogenization buffer containing 50 mM MOPs-BTP (pH 7.5), 330 mM sucrose, 5 mM EDTA, 5 mM dithiothreitol (DTT), 0.5 mM PMSF, 0.2% (w/v) bovine serum albumin (BSA), 0.2% (w/v) casein, and 0.5% polyvinylpyrrolidine (PVP). The homogenate was filtered through four layers of cheesecloth and centrifuged at 10,000 g for 15 min at 4 °C. The supernatant was centrifuged at 21,000 rpm for 45 min, and the resulting precipitate was resuspended in suspension buffer consisting of 330 mM sucrose, 5 mM potassium phosphate (KH2PO4; pH 7.8), 5 mM KCl, 1 mM DDT, and 0.1 mM EDTA. The homogenate was loaded into a two-phase system containing 6.5% (w/w) Dextran T-500, 6.5% (w/w) polyethylene glycol (PEG)-3350, 250 mM sucrose, 4 mM KCl, and 5 mM KH2PO4. After the batch procedure, the resulting upper phase and low phase were collected and mixed with a dilution buffer containing 5 mM 3-(N-morpholino) propanesulphonic acid-Bis–tris Propane (MOPs-BTP) (pH 7.5), 330 mM sucrose, and 5 mM KCl, and then centrifuged at 100,000g for 60 min. The resulting pellet was resuspended in the dilution buffer. PM samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli 1970) consisting of a 5.6% (w/v) stacking gel and a 12% (w/v) separating gel. For Western-blot analysis, the SDS-PAGE resolved proteins were transferred to a polyvinylidene fluoride (PVDF) membrane and blocked in a phosphate buffered saline–Tween (PBST) buffer. The PVDF membrane was incubated at room temperature for 1 h with rabbit polyclonal GFP antibody (1:10,000, Invitrogen) and rabbit polyclonal PM H+-ATPase. After rinsing in PBST buffer, each membrane was incubated for 1 h with a 1:10,000 dilution of horseradish peroxidase (HRP)-conjugated anti-rabbit IgG as secondary antibody (Promega, Madison, WI, USA). The signal detection was performed with enhanced chemiluminescence (ECL) solution (ECL Western-blotting substrate, Promega) using G-Box iCHEMi XL (Syngene, Cambridge, UK).
Hematoxylin staining of Al-treated roots
5-day-old intact seedlings were grown in HS that included 25 µM Al. After 0, 2, 4, and 6 h exposure to Al, the seedlings were removed from the HS and stained for the visual detection of Al using the method proposed by Polle et al. (1978). The seedlings were washed with distilled water (DW) for about 5 min and stained with 0.2% hematoxylin solution (sigma) for 10 min. The seedlings were washed again for 15 min with DW to remove excess stain and were then photographed.
Organic acid assay
The 4-day-old seedlings were transferred to a modified HS adjusted to pH 4.5 without phosphate at least 12 h prior to treatment. The 5-day-old seedlings were subjected to the same solution condition containing 25 µM AlCl3 (pH 4.5) and the root exudates were collected every 2 h (0, 2, 4, 6, and 12 h after Al treatment) for organic acid analysis. The collected solutions containing the root exudates were passed through a cation exchange column (16 × 14 mm) filled with 5 g of Amberlite IR-120B (H+ form) resin, and then through an anion-exchange column (16 × 14 mm) filled with 2.5 g of Bio-Rad AG 1-X8 resin in a cold room. Organic acids retained in the anion-exchange resin were eluted with 2 M HCl, and the eluate was concentrated using a rotary evaporator at 40 °C. The residue was re-dissolved in 1 ml of water, and the concentrations of organic acids were analyzed by HPLC equipped with an ion-exclusion column (Shim-pack SCR-102 H, 8.0 mm × 30 cm, Shimadzu, Kyoto, Japan) as described in detail by Ma et al. (1997a) and Yang et al. (2001).
Vector construction and Camelina transformation
Two recombination sites, attB1 and attB2 at 5′-ends, were added to the forward and reverse primer, respectively. The PCR products were purified and cloned into the pGEM-T Easy vector (Promega, Madison, WI), followed by sequencing. Products were cloned in pDONR207 using the Gateway cloning system (Invitrogen). The binary vector pCB302-3 (Xiang et al. 1999) contains the promoter and terminator of the CaMV 35S promoter to overexpress CsALMT1 cDNA in Camelina. pCB302-3 was modified to be compatible with the Gateway system as described by the manufacturer. The construct was transformed into the Agrobacterium tumefaciens strain GV3101, which was used for Camelina transformation conducted via vacuum infiltration (Bechtold and Pelletier 1998). Antibiotic selection was performed in E. coli and Agrobacterium tumefaciens with kanamycin (50 mg/ml in MS medium containing 1% agar) and in transgenic plants with DL-phosphinotricin (PPT, 0.03% was sprayed on seedlings) (Duchefa, Harlem, The Netherlands).
Protoplast isolation
Protoplasts were isolated from antibiotic-selected transgenic plants and wild-type Camelina roots. The roots were excised using a blade and digested in 10 ml of enzyme solution. The enzyme solution was prepared before the roots were excised. The enzyme solution consisted of Mannitol 0.72 g (final 1.5%), 0.2 M MES 1 ml (final 20 mm), Cellulase 150 mg (final 1.5%), Macerozyme 40 mg (final 0.4%), 1 M CaCl2 100 µl (final 10 mM), 10% BSA 100 µl (final 0.4%), and DW to a final volume of 10 ml, all of which were filtered (0.45 µm) into a 50-ml falcon tube. Digested roots were subjected to vacuum for 30 min and incubated for 3 h (at room temperature (RT), in the dark, and covered with aluminum foil). During incubation, WI and W5 solutions were prepared. The WI solution (50 ml) consisted of 4.5 g Mannitol (final 2 mM), 0.2 M MES (pH 5.7) 1 ml (final 4 mM), 2 M KCl 0.5 ml (final 20 mM), and DW added to a final volume of 50 ml adjusted to pH 4.5 (for Al treatment). W5 consisted of 0.2 M MES (pH 5.7) 0.5 ml (final 2 mM), 1 M CaCl2 6.25 ml (final 125 mM), 1 N NaCl 7.7 ml (final 154 mM), 2 M KCl 125 µl (final 5 mM), and DW added to a final volume of 50 ml. After incubating for 3 h, 10 ml (1× vol.) of W5 buffer was added to the enzyme solution and mixed by swirling. The solution was filtered through a mesh into a 50-ml conical tube and centrifuged at 500 rpm for 4 min. The supernatant was then removed. 4 ml of W5 was added for re-suspension, which was incubated on ice for 30 min. After incubation, W5 was removed without touching the pellet, which was resuspended with 3–4 ml of WI buffer adjusted to pH 4.5.
Al accumulation in root protoplasts
The Al accumulation experiments were performed using morin. Morin reagent was dissolved in 0.1 M dimethyl sulfoxide (DMSO). Isolated root protoplasts were incubated with 50 µM morin for 1 h, and the 50 µM Al was then applied for 40 min. The green fluorescence signal occurred due to Al-morin complexes. The signal intensity was determined using Image J software (Schneider et al. 2012).
Determination of Al contents in roots
Three-week-old plants were treated with P 1/2 HS (pH 4.5) containing 25 µM Al. After 0, 2, 4, and 6 h treatments, the roots were washed with DW. Each of the individual excised roots was transferred to a test tube (16 × 124 mm) and dried at 80 °C for 48 h. The dried samples were weighed and digested with 12 N HCl at 200 °C overnight. The digested samples were diluted with 2 N HCl and then analyzed using inductively coupled plasma mass spectroscopy (IRIS-AP; Thermo Scientific, Pittsburgh, PA).
Statistical analysis
All biological data were analyzed using the Dunnett’s test after one-way ANOVA. Statistical analyses were carried out using SPSS version 20 software (IBM, Chicago, IL, USA). Statistical differences among data for different results were considered statistically significant at P ≤ 0.05.
Results
Isolation of CsALMT1 gene from Camelina sativa L
A CsALMT1 fragment (475 bp) was amplified from cDNA using degenerate primers designed according to the conserved domains of ALMT1 s from AtALMT1 (Hoekenga et al. 2006), TaALMT1-1 (Sasaki et al. 2004), and ZmALMT1 (Pineros et al. 2008). CsALMT1 (Gene accession number: KF021618) was amplified using RACE PCR, and the coding region of a full gene was 1482 bp in length and the proteins comprised 493 amino acid residues with five transmembrane domains according to SOUSI software (Fig S1). CLUSTAL 2.1 Multiple Sequence Alignments indicated that CsALMT1 was 86% similar to the At1g08430 (AtALMT1) in Arabidopsis. However, it was 36% similar to the TaALMT1-1 in wheat and 37% similar to the ZmALMT1 in the maze (Fig. 1). Fig. S1b presents a phylogenetic tree that shows the difference in homology between monocotyledons and dicotyledons.
Multiple sequence alignment of the ALMT1 gene from Wheat (TaALMT1-1), Camelina (CsALMT1), Arabidopsis (AtALMT1), and Maize (ZmALMT1). Alignments were carried out using CLUSTAL (http://clustalw.genome.jp). Identical amino acids and similar amino acids are indicated by dark shading and light shading, respectively, using BOXSHADE software (http://www.ch.embnet.org/software/BOX_form.html). The two plants indicate the position of degenerate primers
Expression of CsALMT1 gene affected by Al
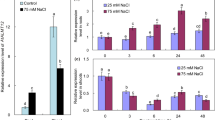
To determine the expression pattern of CsALMT1, it is investigated under various Al doses (0, 10, 25, 50, 75, and 150 µM Al) and time periods (0, 1, 3, 6, 12, and 24 h). Northern-blot analysis was used to analyze CsALMT1 expression patterns in Camelina seedling. CsALMT1 has Al-dependent expression (Fig. 2a). Very low levels of expression were observed within 10 µM Al concentration. However, the transcript levels of ALMT1 greatly increased at concentrations greater than 25 µM Al and then decreased to relatively low levels by 150 µM Al treatment. We also determined the expression pattern of CsALMT1 according to Al treatment time. CsALMT1 gene transcription level remained unchanged up to 1 h after Al treatment, but increased from 3 h after Al treatment, maintaining a steady state until 24 h of treatment. Furthermore, we tested the expression specificity of CsALMT1, which is expressed only in the roots, not in the shoots, at both vegetative and reproductive stages (Fig. 2b). These results suggested that CsALMT1 has Al-dependent and root-specific expression. Also, we determined the suitable experimental conditions of Al concentration and treatment time to identify CsALMT1 function.
Analysis of CsALMT1 expression in Camelina. a Northern blot analysis of CsALMT1 expression in Camelina seedlings by dose and time course with AlCl3. Plants were exposed to 1/2 HS (Hoagland solution) without phosphate (pH 4.5). b Analysis of CsALMT1 expression in vegetative and reproductive stage of Camelina using quantitative real-time PCR. Total RNA was isolated from the leaves, the basal root (1–4 cm), and the root tip (0–1 cm) at vegetative stage and from the flower, silique, stem, leaf, and root at the reproductive stage. Expression (fold) was calculated by relative comparison with the expression of the CsALMT1 gene in the basal root (vegetative stage) or root (reproductive stage). CsActin was used as an internal control for normalization. Values are the averages of three replicates ± SE. Different lowercase letters in the figure indicate significant difference at P < 0.05 by Tukey’s test
Subcellular localization of CsALMT1 protein in tobacco
To examine the subcellular localization of CsALMT1, a fluorescent protein (GFP) was fused to the C-terminus of the full-length CsALMT1. CsALMT1::GFP was transiently introduced into tobacco leaf. The CsALMT1::GFP fusion green fluorescence was observed in the plasma membrane (Fig. 3a). Similar results have been observed in many ALMT1 s (Pineros et al. 2008; Gruber et al. 2010). We also performed Western-blot analysis with the GFP antibody to further substantiate the subcellular localization of CsALMT1. The transiently introduced plasma membrane of tobacco leaf was isolated for the Western blot. CsALMT1 fusion protein was detected in the membrane fraction (upper phase), but not in the control (Fig. 3b). The H+-ATPase antibody was used as a positive control. These results indicate that CsALMT1 is localized in the plasma membrane.
Subcellular localization and western blot of CsALMT1::GFP fusion proteins in Tobacco. a Laser confocal laser scanning microscope images of tobacco cells expressing CsALMT1::GFP fusions and GFP proteins [a and b GFP-fluorescence (green), c bright-field image, d merged images show bright-field image, chlorophyll fluorescence (red), and GFP-fluorescence (green)]. Scale bars represent 30 µm. b Western blot of CsALMT1::GFP in tobacco leaf. Plasma membranes were isolated from leaves using two-phase partitioning method. Upper phase and lower phase were separated by SDS-PAGE followed by analysis with anti-GFP and anti-H+-ATPase (positive control for plasma membrane control)
Overexpression of CsALMT1 in Camelina improved Al tolerance
To test whether CsALMT1 can be used to develop plants with enhanced Al tolerance, we generated transgenic Camelina expressing a 35S promoter, CsALMT1, using a gateway cloning system. CsALMT1 transcript levels of two CsALMT1 independent transgenic T2 plants (OX1 and OX2) and the wild-type plant were determined by RT-PCR using a CsALMT1-specific primer (Fig. 4a). To evaluate the Al tolerance of CsALMT1 transgenic plants, seeds of wild-type and two transgenic plants were germinated and grown on agar plates containing 10 and 25 µM Al for 4 days. No differences were observed in the germination between the wild-type and transgenic plants in treatments compared to the control. The root elongation rate was measured for 4 days after treatment. The growth of the transgenic plants was superior to the wild-type plants under Al stress. In particular, the transgenic plants showed enhanced Al tolerance in 25 µM Al concentration. However, in the control medium, the growth of each plant was similar (Fig. 4b). When the roots exhibited the first signs of Al toxicity, root growth was inhibited as the concentration of Al ions increases and root elongation was clearly inhibited when 25 µM Al was applied (Fig. 4c).
Effect of Al treatments on root growth in wild-type and CsALMT1-overexpressing plants. a CsALMT1-overexpressing plants identified by RT-PCR using CsALMT1-specific primer. One-week-old seedlings were transferred in agarose medium containing Al adjusted to pH 4.5 without phosphate. b Photographs of Camelina wild-type and two CsALMT1-overexpressing lines grown on agar plates with or without 25 µM Al concentration for 4 days. c Root elongation of the plants measured daily during 4 days after transferring to Al-mediums. Data represent mean ± SD (n = 30). Asterisks in the figure indicate significant difference from wild type subjected to the same treatment time at *P < 0.05 by Dunnett’s test
Pattern of Camelina sativa L. root cell disruption under Al stress
Camelina seedlings were exposed to 25 µM Al concentration with different durations (0, 2, 4, and 6 h). Al treatments were performed in hydroponic culture without phosphate. To compare the Al-induced root cell disruption, hematoxylin was used to stain the Al-treated roots of Camelina. No differences were observed in the germination between the wild-type and transgenic plants in normal condition. However, the difference in the degree of staining between the wild-type and the CsALMT1 transgenic plants gradually increased in the roots after 2 h of Al treatment. The swollen section in the root of the wild-type plant was observed, which is one of the symptoms of damage by Al stress. The difference of staining color formation in the root was distinct from the Al treatment for 4 h between the wild-type and the CsALMT1 transgenic plants. The blue color was more intense in the root of the wild-type plant than in the CsALMT1 transgenic plants. These results indicate that Al stress causes more damage to the cells in the vicinity of the growth point of wild-type roots than in CsALMT1 transgenic plants (Fig. 5).
Time period of malate secretion
Because several plants with aluminum tolerance are well known for their increased secretion of organic acids under Al stress, we analyzed the malate secretion in root exudates of the wild-type and CsALMT1 transgenic plants collected at 2, 4, and 6 h after initiation of Al treatment. A small amount of secretion of malate in the root exudate of the wild-type plant was maintained steadily up to 4 h after Al treatment, and then increased after 6 h Al treatment by about 1.5-fold compared to the initial secretion amount (Fig. 6). However, CsALMT1 transgenic plants secreted larger amounts of malate than the wild-type plant from the beginning of Al treatment (2 h), and continued to increase significantly up to 6 h after Al treatment (Fig. 6).
Time course of malate secretion from roots of camelina wild-type and CsALMT1 overexpressing transgenic plants. Root exudates were collected at 0, 2, 4, 6, and 12 h after 25 µM Al treatment at pH 4.5. Organic acids were analyzed by HPLC. Asterisks in the figure indicate significant difference from wild type subjected to the same treatment time at *P < 0.05 by Dunnett’s test
Comparison of Al absorption in roots of wild-type and CsALMT1 transgenic plants
Protoplasts of the roots of wild-type and CsALMT1 transgenic plants were isolated to compare the Al absorption of the root protoplasts. 50 µM Al was treated for 40 min after morin staining for 1 h. Then, measurements of the green fluorescence signal made by the Al-morin complexes indicated that the signal intensity was brighter in wild-type protoplasts than in the CsALMT1 overexpressing plants after 40 min (Fig. 7). This indicates that wild-type protoplasts absorbed more Al than the CsALMT1 overexpressing protoplasts. Subsequently, a comparison between the wild-type and CsALMT1 transgenic plants in Al content was performed. Plants were treated with 25 µM Al, measured at the roots, and sampled at 0, 2, 4, and 6 h. The result confirmed that the roots of the wild-type plant absorbed more Al than the roots of the CsALMT1 transgenic plants at all times (Fig. 8).
Aluminum accumulation in Camelina root protoplast. Root protoplasts from the wild-type and CsALMT1-overexpressing plants were incubated with 50 µM morin for 1 h before 50 µM Al treatment. Then, 50 µM Al was applied for 40 min. a Green fluorescence signal occurring due to Al-morin complexes. b Signal intensities were quantitated by ImageJ
Al contents in roots of the wild-type and CsALMT1 transgenic lines. 3-week-old plants were grown hydroponically and treated with 25 µM Al for 2, 4, and 6 h without phosphate (pH4.5). Data represent mean ± SD (n = 3). Asterisks in the figure indicate significant difference from wild type at *P < 0.05 by Dunnett’s test
Discussion
Al-induced organic acid secretion is a major Al tolerance mechanism in both monocotyledons and dicotyledons (Sasaki et al. 2004; Kochian et al. 2004, 2005; Ligaba et al. 2006; Hoekenga et al. 2006; Ma 2007; Gruber et al. 2010). CsALMT1 was cloned and characterized in this study. CsALMT1 amino acid sequence was 86% similar to AtALMT1, but it was 36–37% similar to TaALMT1-1 or ZmALMT1 amino acid sequences (Fig. 1). These differences in homology may arise from the differences between monocotyledons and dicotyledons (Fig. S1B). In this study, CsALMT1 expression was increased with the Al concentration and treatment time (Fig. 2). This is similar to BnALMT1 (Ligaba et al. 2006), AtALMT1 (Hoekenga et al. 2006), and ZmALMT1 (Pineros et al. 2008) but contrasts with TaALMT1 (Sasaki et al. 2004) and HvALMT1 (Gruber et al. 2010) that were unaffected by Al treatment. However, these differences in ALMT1 expression patterns observed from the various plants with or without Al treatment could not be correlated with the differences in homology between monocotyledons and dicotyledons.
Seven members of the ALMT family were characterized for two groups, where one group was characterized by little or no response to Al3+ and the other was strongly activated by Al3+, which includes CsALMT1. The first group includes HvALMT1 and ZmALMT1, and the second group includes AtALMT1, BnALMT1, BnALMT2, TaALMT1 (Gruber et al. 2010), and CsALMT1. Furthermore, CsALMT1 has a root-specific expression at both the vegetative and reproductive stages (Fig. 2b). The ALMT1 family of membrane proteins consists of 5-8 predicted transmembrane domains (Sasaki et al. 2004; Delhaize et al. 2007). The CsALMT1 has five putative transmembrane regions, which indicates that it is likely to be membrane-bound (Fig. S1). This result shows that CsALMT1 is probably localized to the plasma membrane. As expected, the CsAMT1::GFP fusion protein was localized to the plasma membrane, which was transiently introduced into tobacco leaf (Fig. 3a). This was also confirmed using Western blot, in which protein is localized in the plasma membrane (Fig. 3b).
The first sign of Al toxicity is the inhibition of root growth and cell division (Clarkson 1966; Klimashevski and Dedov 1975). In the present study, analysis of the root elongation and hematoxylin staining was used as an index for the Al tolerance in Camelina wild-type and CsALMT1 overexpressing plants (Figs. 4, 5, respectively). The results show that Camelina root elongation was clearly inhibited when 25 µM Al was applied because the Camelina seedling roots were very soft (Fig. 4). Interestingly, hematoxylin staining showed that the roots of the wild-type plant were more damaged than the roots of the CsALMT1 overexpressing plants. The root apex has been recognized as a primary site of Al toxicity in plants (Ryan et al. 1993; Kochian 1995; Ahn et al. 2001). However, in the case of Camelina, Al-induced injury occurred in the entire root, not only in the root apex, with 25 µM Al treatment (Fig. 5). This result indicates that the all the root cell membranes of Camelina were disrupted simultaneously.
The patterns of organic acid anion secretion can be grouped into two patterns (Ma 2000). In pattern I, the secretion of organic acid anions immediately occurs when exposed to Al. Pattern II exhibits a significant lag phase until the onset of organic acid anion release. We propose a hypothetical model of Al-activated malate secretion from Camelina root, which is typical of pattern II. Al3+ interacts with a receptor on the plasma membrane to initiate a signal transduction pathway. In Arabidopsis, Al3+ and/or protons interact with a receptor on the plasma membrane to initiate a signal transduction pathway. A protein kinase phosphorylate sensitive to proton rhizotoxicity1 (STOP1) is used to convert the STOP1 from an inactive to active form (Iuchi et al. 2007; Sawaki et al. 2009). This activated transcription factor in Camelina may interact with sequences in the promoter of CsALMT1 s. Then, CsALMT1 s mRNA translates into the protein CsALMT1 s that are located on the plasma membrane of root cells, where they may transport malate to the external medium. The secreted malate binds and detoxifies Al3+ to protect the root. This result is similar to the regulation of the citrate secretion step in the rice bean (Liu et al. 2013). We demonstrated the simple regulation of malate secretion in response to Al exposure in the Camelina root. A lag phase of 0–4 h occurs for the secretion of malate from the Camelina root in response to Al treatment (Fig. 6), which is typical of pattern II organic acid release, as proposed by Ma (2000). A high level of malate secretion required almost 6 h of Al treatment. Interestingly, CsALMT1 overexpressing plants can secrete a substantial amount of malate secretion as pattern I, unlike wild-type plant (Fig. 6). This result suggested that high malate secretion enhanced the Al tolerance of the CsALMT1 overexpressing line. In addition, oxalic acid is an important organic acid in Al detoxifying mechanisms, both internally and externally (Zheng et al. 1998; Ma et al. 1998). Therefore, further study is required on the regulation of oxalic acid secretion in response to Al exposure in the CsALMT1 overexpressing line.
The roots undergo Al toxicity both externally and internally. When the root absorbs Al3+, internal toxicity occurs. To identify the amount of Al absorbed by the Camelina root, 25 µM Al was applied for 1, 3, 6, 12, and 24 h. The Al contents in 1 cm root segments from the root tip were compared with the basal root (2–3 cm). The accumulation of Al did not differ significantly in each root segment but increased slightly in the root tips after 12 h of treatment (data not shown). Al is rarely translocated from the root to shoots (data not shown). Furthermore, the root protoplasts from the wild-type Camelina absorbed more Al than in the CsALMT1 overexpressing plants (Fig. 7), which eventually led to lower aluminum content in the roots of the CsALMT1 overexpressing plants (Fig. 8). These results suggest that when CsALMT1 transgenic plants are stressed by Al, CsALMT1 as a malate transporter can contribute to induce the malate secretion and enhance the Al detoxification function by limiting Al absorption.
In conclusion, we isolated CsALMT1 from Camelina, which has Al tolerance enabled by malate secretion. The function of CsALMT1 was investigated using CsALMT1 overexpressing plants. CsALMT1 transgenic plants have Al tolerance compared with wild-type plants. This genetically induced plant changes in CsALMT1 expression, which may lead to increased malate exudation in the Camelina roots. Therefore, we suggest that CsALMT1 might be a malate transporter localized to the root plasma membrane that plays a critical role in Camelina Al tolerance.
Abbreviations
- HS:
-
Hoagland solution
References
Ahn SJ, Sivaguru M, Osawa H, Chung GC, Matsumoto H (2001) Aluminum inhibits the H+-ATPase activity by permanently altering the plasma membrane surface potentials in squash roots. Plant Physiol 126:1381–1390. doi:10.1104/pp.126.4.1381
Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82:259–266. doi:10.1385/0-89603-391-0:259
Clarkson DT (1966) Effect of aluminum on the uptake and metabolism of phosphorus by barley seedlings. Plant Physiol 41:165–172. doi:10.1104/pp.41.1.165
Delhaize E, Ryan PR, Randall PJ (1993) Aluminum tolerance in wheat (Triticum aestivum L.) (II. Aluminum-stimulated excretion of malic acid from root apices). Plant Physiol 103:695–702
Delhaize E, Gruber BD, Ryan PR (2007) The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Lett 581:2255–2262. doi:10.1016/j.febslet.2007.03.057
Gruber BD, Ryan PR, Richardson AE, Tyerman SD, Ramesh S, Hebb DM, Howitt SM, Delhaize E (2010) HvALMT1 from barley is involved in the transport of organic anions. J Exp Bot 61:1455–1467. doi:10.1093/jxb/erq023
Hoekenga OA, Maron LG, Pineros MA, Cancado GM, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, Matsumoto H, Yamamoto Y, Koyama H, Kochian LV (2006) AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc Natl Acad Sci USA 103:9738–9743. doi:10.1073/pnas.0602868103
Iuchi S, Koyama H, Iuchi A, Kobayashi Y, Kitabayashi S, Kobayashi Y, Ikka T, Hirayama T, Shinozaki K, Kobayashi M (2007) Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc Natl Acad Sci USA 104(23):9900–9905. doi:10.1073/pnas.0700117104
Klimashevski EL, Dedov VM (1975) Localization of the mechanism of growth inhibiting action of Al3+ in elongating cell walls. Fiziol Rast (Soviet Plant Physiol) 22:1040–1046
Knorzer KH (1978) Evolution and spreading of gold of pleasure (Camelina sativa S.L.). Ber Deutsch Bot Ges. 91:187–195
Kochian LV (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46:237–260. doi:10.1146/annurev.pp.46.060195.001321
Kochian LV, Hoekenga OA, Pineros MA (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Physiol Plant Mol Biol 55:459–493. doi:10.1146/annurev.arplant.55.031903.141655
Kochian LV, Pineros MA, Hoekenga OA (2005) The Physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 274:175–195. doi:10.1007/1-4020-4099-7_9
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi:10.1038/227680a0
Ligaba A, Katsuhara M, Ryan PR, Shibasaka M, Matsumoto H (2006) The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol 142:1294–1303. doi:10.1104/pp.106.085233
Liu MY, Chen WW, Xu JM, Fan W, Yang JL, Zheng SJ (2013) The role of VuMATE1 expression in aluminium-inducible citrate secretion in rice bean (Vigna umbellata) roots. J Exp Bot 64:1795–1804. doi:10.1093/jxb/ert039
Ma JF (2000) Role of organic acids in detoxification of aluminum in higher plants. Plant Cell Physiol 41:383–390. doi:10.1093/pcp/41.4.383
Ma JF (2007) Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int Rev Cytol 264:225–252. doi:10.1016/S0074-7696(07)64005-4
Ma JF, Hiradate S, Nomoto K, Iwashita T, Matsumoto H (1997a) Internal detoxification mechanism of Al in hydrangea. Identification of Al form in the leaves. Plant Physiol 113:1033–1039. doi:10.1104/pp.113.4.1033
Ma JF, Zheng SJ, Matsumoto H (1997b) Specific secretion of citric acid induced by Al stress in Cassia tora L. Plant Cell Physiol 38:1019–1025
Ma JF, Hiradate S, Matsumoto H (1998) High aluminum resistance in buckwheat—II. Oxalic acid detoxifies aluminum internally. Plant Physiol 117:753–759. doi:10.1104/pp.117.3.753
Magalhaes JV, Liu J, Guimaraes CT, Lana UGP, Alves VMC, Wang YH, Schaffert RE, Hoekenga OA, Pineros MA, Shaff JE, Klein PE, Carneiro NP, Coelho CM, Trick HN, Kochian LV (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet 39:1156–1161. doi:10.1038/ng2074
Maron LG, Kirst M, Mao C, Milner MJ, Menossi M, Kochian LV (2008) Transcriptional profiling of aluminum toxicity and tolerance responses in maize roots. New Phytol 179:116–128. doi:10.1111/j.1469-8137.2008.02440.x
Palmgren MG, Larsson C, Sommarin M (1990) Proteolytic activation of the plant plasma membrarne H+-ATPase by removal of a terminal segment. J Biol Chem 266:133–140
Park W, Feng Y, Ahn SJ (2014) Alteration of leaf shape, improved metal tolerance, and productivity of seed by overexpression of CsHMA3 in Camelina sativa. Biotechnol Biofuels 7:96. doi:10.1186/1754-6834-7-96
Pineros MA, Cancado GM, Maron LG, Lyi SM, Menossi M, Kochian LV (2008) Not all ALMT1-type transporters mediate aluminum-activated organic acid responses: the case of ZmALMT1—an anion-selective transporter. Plant J 53:352–367. doi:10.1111/j.1365-313X.2007.03344.x
Polle E, Konzak CF, Kittrick JA (1978) Visual detection of aluminum tolerance levels in wheat by hematoxylin staining of seedling roots. Crop Sci 18:823–827. doi:10.2135/cropsci1978.0011183X001800050035x
Putnam D, Budin J, Field L, Breene W (1993) Camelina: a promising low-input oilseed. In: Janick J, Simon J (eds) New Crops. Wiley, New York, pp 314–322
Ryan PR, Delhaize E (2010) The convergent evolution of aluminium resistance in plants exploits a convenient currency. Funct Plant Biol 37:275–284. doi:10.1071/FP09261
Ryan RP, Ditomaso JM, Kochian LV (1993) Aluminium toxicity in roots: an investigation of spatial sensitivity and the role of the root cap. J Exp Bot 44:437–446. doi:10.1093/jxb/44.2.437
Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37:645–653. doi:10.1111/j.1365-313X.2003.01991.x
Sawaki Y, Iuchi S, Kobayashi Y, Kobayashi Y, Ikka T, Sakurai N, Fujita M, Shinozaki K, Shibata D, Kobayashi M (2009) STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol 150:281–294
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Meth 9:671–675
Vollmann J, Damboeck A, Eckl A, Schrems H, Ruckenbauer P (1996) Improvement of Camelina sativa, an underexploited oilseed (ed. J. Janick). Progress in new crops. ASHS Press, Alexandria, VA 1:357–362
Xiang C, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40:711–717. doi:10.1023/A:1006201910593
Yang ZM, Nian H, Sivaguru M, Tanakamaru S, Matsumoto H (2001) Characterization of aluminium-induced citrate secretion in aluminium-tolerant soybean (Glycine max) plants. Physiol Plant 113:64–71. doi:10.1034/j.1399-3054.2001.1130109.x
Zheng SJ, Ma JF, Matsumoto H (1998) High aluminum resistance in buckwheat—I. Al-induced specific secretion of oxalic acid from root tips. Plant Physiol 117:745–751. doi:10.1104/pp.117.3.745
Zheng SJ, Yang JL, He YF, Yu XH, Zhang L, You JF, Shen RF, Matsumoto H (2005) Immobilization of aluminum with phosphorus in roots is associated with high aluminum resistance in buckwheat. Plant Physiol 138:297–303. doi:10.1104/pp.105.059667
Zubr J (1997) Oil-seed crop: Camelina sativa. Ind Crops Prod 6:113–119. doi:10.1016/S0926-6690(96)00203-8
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea, funded by the Ministry of Education (20141A1A4A01009621). This work was also supported by a grant from the Next-Generation BioGreen 21 Program (SSAC, grant#: PJ01108102), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
In this study, WP and H-S Kim contributed to the experimental conception, design, and writing the article. Dr. S-J Ahn participated in designing the study and revised the manuscript. All authors contributed to the complete article.
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Park, W., Kim, HS., Park, TW. et al. Functional characterization of plasma membrane-localized organic acid transporter (CsALMT1) involved in aluminum tolerance in Camelina sativa L.. Plant Biotechnol Rep 11, 181–192 (2017). https://doi.org/10.1007/s11816-017-0441-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-017-0441-z