Abstract
Automatic segmentation of 3D coronary arteries from computed tomography angiography (CTA) is an indispensable part of accurate and efficient coronary artery disease (CAD) diagnosis. However, it remains challenging due to the complex anatomy of coronary arteries. Inspired by the denoising diffusion probabilistic model (DDPM), we propose a diffusion-based multi-attention network for 3D coronary artery segmentation from CTA. The proposed method is called DiffCAS in short. DiffCAS utilizes the denoising diffusion of the diffusion model to yield segmentation results. During the denoising diffusion, the Swin Transformer is adopted to extract semantic information from CTA images, and an adaptive residual feature enhancement (ARFE) module is proposed as denoising encoder in the diffusion model, a feature fusion attention (FFA) module is coined to fuse the features from Swin Transformer and ARFE encoders, so as to improve the segmentation performance. Experimental results and comparisons on the ASOCA and ImageCAS datasets show that the proposed DiffCAS outperforms some SOTA networks in terms of Dice coefficient that are 84.41% and 84.59%, on ASOCA dataset and ImageCAS dataset, respectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
In recent years, coronary artery disease (CAD) has remained one of the major causes of death [1], with a high incidence and mortality rate. Therefore, accurate diagnosis and risk assessment of CAD are crucial. Computed tomography angiography (CTA) is a widely used non-invasive imaging technique for routine clinical diagnosis of CAD, and the image quality is comparable to that of invasive coronary angiography (ICA). However, in CTA images, the location and severity of stenosis require manual assessment by specialized physicians. This assessment process is not only time-consuming, but also prone to misdiagnosis and missed diagnosis. Hence, achieving high-quality and fully automated coronary artery segmentation is of paramount importance. Currently, automated segmentation of CTA images still faces various challenges. Firstly, the complex structure of coronary arteries consists of branches of varying sizes, and their shapes and positions vary among individuals. Secondly, the similarity in structure and appearance between coronary arteries and other vessels can lead to misidentification. Lastly, inherent noise and artifacts introduced during image acquisition can further complicate the segmentation task.
In the realm of medical image segmentation, significant strides have been witnessed in recent years through the application of deep learning methods [2,3,4]. For instance, Yu et al. [5]. proposed the DenseVoxNet, a densely connected convolutional neural network, for automatically segmenting vascular structures from 3D MRI. Mou et al. [6]. introduced the CS2-Net, a segmentation network leveraging a two-channel attention mechanism to grasp intricate representations of curvilinear structures. Song et al. [7]. used dense blocks and residual blocks to extract representative features for coronary artery segmentation. Xia et al. [8]. proposed the ER-Net, which preserved spatial edge information and improved segmentation performance. Duan et al. [9]. designed the ECA-UNet that enabled cross-channel interaction and effectively improved the segmentation performance of coronary artery. Dong et al. [10]. proposed a multi-level 3D deep learning network for automatic coronary artery segmentation and yielded promising segmentation accuracy. These methods do not take into account the complex anatomy of coronary arteries and the effect of noise, which severely limits segmentation performance.
Self-attention mechanism derived from Transformer [11] has been applied to computer vision tasks, modeling distant dependencies in sequence-to-sequence tasks. Within this framework, the Vision Transformer [12] was proposed. This method achieves remarkable performance by using Transformer. Many works, such as TransUNet [13], TransBTS [14], TransFuse [15], UNETR [16], MISSFormer [17], MedT [18], and MS-TransUnet++ [19], employed Transformer to model long-range semantic information present in medical images. Zhao et al. [20]. proposed the CA-Net for semi-supervised left atrium segmentation from 3D MRI. The CA-Net is composed of V-Net and Transformer, which can better learn contextual information. Xiang et al. [21]. proposed the DeTr-V consisting of V-Net and Transformer to segment left atrium. However, due to the computational complexity of Transformer structures, there is still room for improvement for these methods to extract multi-scale features. The Swin UNETR [22] leverages the Swin Transformer [23] as an encoder to extract multi-scale features and adopts a CNN-based decoder to generate outputs. The Swin UNETR effectively captures global dependencies and spatial details in extracting multi-scale features.
The denoising diffusion probabilistic model (DDPM) [23, 24] has recently demonstrated exceptional performance in various tasks, including image coloring [25], super-resolution [26], inpainting [27], and image generation [28]. Moreover, diffusion model has proven their utility in semantic segmentation. Baranchuk et al. [29]. showcased the applicability of diffusion model in semantic segmentation by examining the representations learned by DDPM, which captured valuable high-level semantic information for downstream visual tasks. Pinaya et al. [30]. proposed a method based on diffusion model to detect and segment anomalies in brain image. Amit et al. [31]. proposed the SegDiff, based on DDPM, for image segmentation. Wu et al. [32]. proposed the MedSegDiff by taking use of a DenoisingUNet for 2D medical image segmentation. Similarly, Wolleb et al. [33]. employed the diffusion model for 2D medical image segmentation by aggregating output results from various diffusion steps using summation during testing. Wu et al. [34]. later proposed the MedSegDiff-V2, which incorporates the Transformer into the DDPM to improve segmentation performance. These studies underscore the substantial potential of diffusion model in medical image segmentation and yielded satisfactory outcomes. However, diffusion model for coronary artery segmentation is seldomly studied and all these methods are for segmenting 2D images. Therefore, we introduce the DiffCAS, a network that combines diffusion model with attention mechanisms for 3D coronary artery segmentation in this work.
In the proposed DiffCAS, the denoising diffusion of the diffusion model is utilized to yield segmentation results. There are two encoders, a decoder and a feature fusion module in the DiffCAS. During denoising diffusion, the Swin Transformer that serves as a feature encoder extracts semantic information from input images. An adaptive residual feature enhancement (ARFE) module in the denoising encoder receives the features from the noisy image and the Swin Transformer and augments the perceptual sensitivity of the network to coronary artery details. Since not all features are effective in extracting the vessel structure, a feature fusion attention (FFA) module is used at the end of the two encoders to select effective features. In the denoising decoder, we adopt standard convolution operations while preserving skip connections to prevent information loss, and finally segmentation accuracy is refined further. In summary, the main contributions of this work are as follows:
-
1)
Based on the diffusion model, a novel network, DiffCAS in short, is proposed for segmentation of the 3D coronary artery from CT angiography (CTA), and the Swin Transformer is employed to act as a feature encoder in the DiffCAS.
-
2)
An adaptive residual feature enhancement (ARFE) module is coined and there are four ARFE modules in the denoising encoder of denoising diffusion, the denoising encoder aims to enhance the perceptual sensitivity of coronary artery details and capture both global and local image features.
-
3)
A feature fusion attention (FFA) module is proposed to fuse the features from the feature encoder and denoising encoder, so as to select important features from two encoders and enhance the weight of the detailed portion of coronary artery.
-
4)
The proposed DiffCAS is validated on two publicly available datasets, i.e., the ASOCA and ImageCAS datasets, and the results are promising.
The remainder of this paper is organized as follows: the details of the proposed DiffCAS are presented in Section II. Following that, experiment settings and experimental results are reported in Section III, and the conclusion is drawn in Section IV.
2 The proposed DiffCAS
2.1 Overview of the DiffCAS
We design our model based on the diffusion model. Generally, the diffusion model comprises a forward process and a reverse process. During the forward process, the segmentation label \({x}_{0}\) gradually adds Gaussian noise through a series of steps T. Conversely, in the reverse process, the neural network is trained to recover the original data by reversing the noise process, which can be mathematically expressed as:
where \(\theta\) represents reverse process parameters. Starting from a Gaussian noise \({x}_{T}\), \({p}_{\theta }\left({x}_{T}\right)=N({x}_{T};0,{I}_{n\times n})\), where \(I\) is the original image, the reverse process transforms the latent variable distribution \({p}_{\theta }\left({x}_{T}\right)\) into the data distribution \({p}_{\theta }\left({x}_{0}\right)\). In order to be symmetric with the forward process, the backward process gradually restores the noisy image to achieve the final clear segmentation.
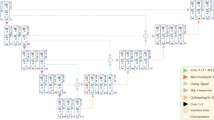
The architecture of the proposed DiffCAS is depicted in Fig. 1, which includes two encoders, a single decoder, and a feature fusion module. During the denoising diffusion process, the Swin Transformer that serves as a feature encoder sends the semantic information from input images to the denoising encoder. An adaptive residual feature enhancement (ARFE) in the denoising encoder receives the features from the noisy image and Swin Transformer and enhances the intricate coronary artery details. The feature fusion attention (FFA) module is added at the end of the two encoders to select important features from two encoders, thereby enhancing the weight of the detailed portion of coronary artery. In the denoising decoder, we employ standard convolution operations while preserving skip connections to mitigate information loss, and consequently segmentation accuracy is refined further.
As illustrated in Fig. 1, the denoising process runs in an iterative manner, and the prediction result after each iteration can be represented as:
where \(I\) and \({x}_{t}\) are inputs at t epoch; \({{x}_{t-1}(x}_{t},I,t)\) represents the prediction results at t epoch; \({E}_{t}^{I}\) and \({E}_{t}^{x}\) denote the feature encoder and the denoising encoder at t epoch, respectively; D means the denoising decoder. Finally, after T iterations, the ultimate prediction result \({x}_{0}\) is obtained.
2.2 Swin Transformer: feature encoder
In order to improve the ability of the model to extract semantic features from the original image, the Swin Transformer module is employed as the feature encoder. This choice leverages robust feature extraction and representation capabilities of Swin Transformer, which allows the model to learn global dependencies and contextual information.
Swin Transformer modifies the original multi-head self-attention (MSA) structure of Transformer. Swin Transformer proposes a window-based multi-head attention (W-MSA) structure, which decomposes the image into multiple non-overlapping windows, and calculates its attention separately in each window. W-MSA significantly reduces the computational complexity compared with the attention calculation for the whole image in ViT. At the same time, in order to solve the problem that features cannot be transferred between different windows, Swin Transformer also proposes a shifted window-based MSA (SW-MSA) structure, which transfers feature information in different windows by moving the position of these windows. Compared with the traditional Transformer module, it replaces the MSA part with W-MSA and SW-MSA structures. The structure of the Swin Transformer module is depicted in Fig. 2, which includes two similar Swin Transformer sub-modules. In the first Swin Transformer sub-module, W-MSA and multi-layer perceptron (MLP) are utilized. In addition, a Linear Norm (LN) layer is applied before the W-MSA module and the MLP module, and a residual connection is applied after each module. The output \({z}^{l}\) of the \(l\)th layer can be expressed as:
where \({\widehat{z}}^{l}\) and \({z}^{l}\) denote the output features of the W-MSA module and the MLP module, respectively; W-MSA denotes window based multi-head self-attention using regular window partitioning configurations.
Similarly, in the second Swin Transformer sub-module, denoted as SW-MSA. The output \({z}^{l+1}\) of the \((l+1)\)th layer can be expressed as:
where \({\widehat{z}}^{l+1}\) and \({z}^{l+1}\) denote the output features of the SW-MSA module and the MLP module, respectively; SW-MSA denotes window based multi-head self-attention using shifted window partitioning configurations.
2.3 ARFE: adaptive residual feature enhancement module
In deep learning, as the depth of the network increases, the model can capture a broader range of high-level features that embody rich semantic information. These features play a pivotal role in recognizing semantically significant regions in lower-level features. Similarly, lower-level features encapsulate abundant spatial information that is helpful to reconstruct accurate details in high-level features. Given the intricate anatomical structure of coronary artery, both high-level and low-level features in the model are crucial for accurate vessel prediction. To this end, we introduce the ARFE module that serves as the denoising encoder. This ARFE module leverages both low-level and high-level features presented in the network, and adaptively enhances semantic features to improve accuracy. The ARFE module consists of a dual-layer residual (DLR) component and an adaptive feature selection (AFS) part, as illustrated in Fig. 3.
The DLR component incorporates two short skip connections on the top of convolutions. This improvement better exploits both low-level and high-level features to enhance the feature perception capability of the network. Following the DLR, the AFS takes the feature from the DLR as input and augments the input features. The AFS includes channel attention and spatial attention, and the detailed structure of the two attention modules is shown in Fig. 4. Specifically, there are two convolution blocks following an input feature \({F}_{i}\in {R}^{C\times H\times W\times D}(i=\text{1,2},\text{3,4})\). Each block comprises a 3 × 3 × 3 convolutional layer, a normalization layer and a ReLU activation function. Finally, two residual operations are used to yield feature \({X}_{3}\). The process can be summarized as:
where \({conv}_{3\times 3\times 3}\) represents a convolution operation with a \(3\times 3\times 3\) kernel, \(BN\) denotes the batch normalization, \({conv}_{1\times 1\times 1}\) changes the channel size.
The channel attention can reweight different channels with adaptive selection of task-relevant information. Spatial attention can reweight different spatial positions, so that the network can focus on different regions. Through these two modules, more details of the coronary artery can be captured, and more important features can be adaptively enhanced. Specifically, \({X}_{3}\) that obtained from the DLR component serves as the input to the AFS component, and then channel attention and spatial attention are applied to \({X}_{3}\) in turn. The overall process can be summarized as:
where Υ denotes element-wise multiplication, Channel represents the channel attention module, and Spatial represents the spatial attention module. The channel attention module takes the feature \({X}_{3}\) and separately feeds it into max-pooling and average-pooling operations to aggregate feature information. The features are then input into a shared MLP layer to generate channel attention features. The results are summed and passed through a sigmoid function to obtain the output. This process can be represented by the following formula:
The spatial attention module takes the feature \({X}_{4}\) and yields two feature maps using two pooling operations. These two feature maps are then concatenated, passed through a convolutional layer, and further processed through a sigmoid function to obtain the spatial attention features. This process can be summarized as:
where σ represents the sigmoid activation function, and \({f}^{7\times 7\times 7}\) denotes the convolution operation with a 7 × 7 × 7 kernel. Finally, the result is obtained through a residual operation. This process can be described as:
2.4 FFA: feature fusion attention module
To facilitate better fusion of features from the feature encoder and the denoising encoder, inspired by the Efficient Attention [35], we design an FFA module. This module performs attention fusion on the last layers of both the feature encoder and the denoising encoder, so that the network captures both global and detail information effectively, and the coronary artery segmentation accuracy is improved.
Figure 5 illustrates the architecture of the FFA module. In this module, \(X\in {R}^{n\times d}\) is derived from the feature encoder, while \(Y\in {R}^{n\times d}\) comes from the denoising encoder. \(X\) and \(Y\) are inputs of the FFA module. \(X\) passes through two linear layers to form the queries \(Q\in {R}^{n\times {d}_{k}}\) and keys \(K\in {R}^{{d}_{k}\times n}\), and \(Y\) passes through one linear layer to form the values \(V\in {R}^{n\times {d}_{v}}\). Unlike self-attention mechanisms, a multiplication operation is applied to the softmax-transformed \(K\) and \(V\) to obtain \(G\in {R}^{{d}_{k}\times {d}_{v}}\) in FFA. Subsequently, the softmax-transformed \(Q\) is multiplied with \(G\) to yield the final output \(E\in {R}^{n\times {d}_{v}}\). Through these operations, the module better fuses feature from multiple encoders, so that the accuracy of coronary artery segmentation is improved.
The following equation characterizes the FFA module:
where \(E\left(Q,K,V\right)\) represents output of the FFA; \({\rho }_{q}\) and \({\rho }_{k}\) are normalization functions for the \(Q\) and \(K\) features, respectively.
2.5 Loss function
To assess the similarity between labels and segmentation results, binary cross entropy (BCE) is employed as the loss function, which is formulated as follows:
where N denotes the number of voxels, \({y}_{i}\) represents the label and \({{y}_{i}}^{{\prime }}\) represents the segmentation result.
In coronary artery segmentation, due to the relatively small proportion of coronary artery compared to the large background region, dice loss is introduced to mitigate the influence of the background on the loss function. Dice loss does not allocate different weights to different classes, but directly optimizes the loss function value of the target region. By suppressing the impact of the background on the loss function, the training effectiveness of the model is improved. Dice loss is calculated as follows:
where TP represents true positives, FP represents false positives, and FN represents false negatives.
While dice loss can handle class imbalance better and focus on foreground information mining, it exhibits high sensitivity, causing substantial fluctuations in loss values and gradients upon prediction errors. Therefore, the final loss function is a combine of dice loss and BCE, as follows:
3 Experiments
3.1 Dataset
In this study, two publicly available datasets are employed for experiment, one is the ASOCA dataset and the other is the ImageCAS dataset.
The automated segmentation of coronary artery (ASOCA) [36] challenge, during MICCAI in 2020, focused on the automatic segmentation of coronary artery. This competition provided a dataset comprising 40 cases. The images were acquired using a GE LightSpeed 64-slice CT scanner. The plane resolution of each image ranged from 0.3 to 0.4 mm, with an out-of-plane resolution of 0.625 mm. All labels were independently annotated by three experts. Among the 40 cases, 25 are used for training, 5 for validation, and 10 for testing.
ImageCAS is another public large-scale dataset proposed by Zeng et al. [37]. , there are a total of 1000 CTA images, that were acquired by Siemens 128-slice dual-source scanner. Each 3D CTA image have 512 × 512 × (206–275) voxels in size, with a planar resolution of 0.29–0.43 \({mm}^{2}\) and a spacing of 0.25–0.45 mm. The left coronary artery and the right coronary artery were independently labelled by two radiologists in each image, and their results were cross-validated. Out of 1000 cases, 700 are used for training, 100 for validation and 200 for testing.
3.2 Evaluation metrics
Four metrics are employed to evaluate the segmentation performance. Firstly, three classical metrics are quantitatively used to evaluate the results: dice similarity coefficient (DSC), recall, and precision. All three values range between 0 and 1, with higher values indicating better segmentation quality. Furthermore, acknowledging the critical importance of vessel boundaries in coronary artery segmentation results, the quantification process additionally incorporates the use of hausdorff distance (HD) [38] metric. HD is the maximum distance between the boundary of the reference object and the boundary of the automatically segmented object, defined as follows:
where X and Y represent the boundaries of the segmented object and the reference object, respectively.
Among these metrics, DSC primarily reflects the overlap of pixels between the estimated result and the ground truth, and it can be seen as the most crucial evaluation measure for segmentation tasks. In this study, performance is primarily ranked based on DSC results, followed by 95%HD, recall, and precision.
3.3 Experimental setting and results
The proposed DiffCAS is implemented using the PyTorch framework on an RTX 3090Ti GPU. During each training step, a random cropping strategy is employed to crop CTA images into patches of size 96 × 96 × 96. Standard data augmentation techniques, such as random flips, rotations, and noise augmentation, are employed to prevent overfitting. The Adam optimizer is utilized to update network parameters, with an initial learning rate set to 0.0001 and no pre-trained weights are employed.
Six methods are employed for comparison, including 3D U-Net [3], V-Net [4], DenseVoxNet [5], CS2-Net [6], UNETR [16], and Swin UNETR [22]. To ensure fair comparisons, all experiments undergo the same preprocessing procedures, and all models are retrained using identical learning strategies to achieve optimal performance.
3.3.1 Segmentation results on ASOCA dataset
Table 1 presents quantitative results of DiffCAS and other methods on the ASOCA dataset. The experimental results demonstrate that the DiffCAS outperforms other methods. Specifically, the DiffCAS achieves an average DSC of 84.41%, 95%HD of 11.10 mm, Recall of 92.04%, and Precision of 78.49%. Among all methods, except for recall which is slightly suboptimal, the DiffCAS outperforms others in the remaining three metrics. When compared with the classical 3D U-Net, the DiffCAS shows DSC increase of 5%, 19.1 mm reduction in 95%HD, and 8.31% increase in Precision, that the effectiveness of DiffCAS is confirmed. Compared to the other methods, DiffCAS achieves improvements of 4.87%, 3.97%, 3.45%, 2.79%, and 1.65% in DSC, respectively. Overall, the DiffCAS exhibits superior performance compared to other methods.
We visualize the experimental results of the various methods, as shown in Fig. 6. For the results, 3D Slicer software is employed to perform three-dimensional visualization and qualitative analysis. As shown in Fig. 6, the 3D morphology of coronary artery varies widely. However, for certain intricate branches of coronary artery, over-segmentation and under-segmentation often arise, as indicated by the blue boxes in Fig. 6. In comparison with the other methods, the DiffCAS achieves more accurate segmentation of coronary artery details. Moreover, the DiffCAS substantially alleviates the impact of similar structures present in the input images and irrelevant background noise on the final segmentation results. In order to better observe the segmentation results of DiffCAS, we visualize the segmentation results from three perspectives: coronal plane, transverse plane, and sagittal plane, as shown in Fig. 7. Through quantitative comparisons and qualitative analyses, it is evident that the DiffCAS holds a significant performance advantage. These results also highlight the ability of DiffCAS to effectively perform complex coronary artery segmentation tasks.
3.3.2 Segmentation results on ImageCAS dataset
The DiffCAS is also verified on ImageCAS dataset. The quantitative results are shown in Table 2. For the ImageCAS, the DiffCAS achieves the best segmentation accuracy. The DiffCAS achieves 84.59%, 11.92 mm, 84.04% and 86.68% in terms of DSC, 95%HD, Recall and Precision, respectively. In detail, the DiffCAS outperforms 3D U-Net, V-Net, DenseVoxNet, CS2-Net, UNETR and Swin UNETR by 2.91%, 3.33%, 2.82%, 2.48%, 1.94% and 1.56% in terms of DSC, respectively. This indicates that the DiffCAS acquires more vessel detail and is able to discriminate coronary artery from complex background noise in CTA images.
Similarly, we visualize the results of the experiments on the ImageCAS dataset to facilitate an intuitive comparison. The results of the visualization experiment are shown in Fig. 8 that shows the segmentation results of three randomly selected CTA images from the ImageCAS dataset. It can be seen from Fig. 8 that some methods suffer from over-segmentation. The reason behind this observation is that these methods cannot deal well with the problem of noise present in the image and similar vascular structures. In contrast, the proposed DiffCAS can overcome over-segmentation considerately. For the ImageCAS dataset, we also select two cases from the visualization experiment to perform the multi-view visualization experiments, as shown in Fig. 9. The experimental results indicate that the DiffCAS possesses significant advantages in segmenting the 3D structure of coronary artery.
3.4 Ablation studies
3.4.1 Effectiveness of each component in DiffCAS
To validate the effectiveness and feasibility of DiffCAS, a comprehensive analysis is conducted on each module of DiffCAS. Building upon U-Net (baseline), further investigations are carried out to assess the performance of various modules, including DDPM, Swin Transformer (SwinT), ARFE and FFA. A series of ablation experiments are conducted on the ASOCA dataset, with the average values of DSC, 95% HD, Recall, and Precision metrics being presented. The experimental parameters and hardware environment settings remain identical to that in the comparative experiments.
Table 3 presents the quantified results with different modules, from which the improvements of each module can be observed. Based on Table 3, Model 1 combines DDPM with the Baseline for the experiment. Comparison with the Baseline, it is evident that the inclusion of the DDPM module led to a significant enhancement in the DSC metric by 3.18%, which confirms the effectiveness of DDPM. Furthermore, by comparing the experimental results of Model 1, Model 2, Model 3, and Model 4 in Table 3, it can be observed that the Swin Transformer module, ARFE module, and FFA module improve the DSC metric by 0.48%, 0.82%, and 0.52%, respectively. Simultaneously, by observing Table 3, it becomes apparent that the DDPM module has the most substantial impact on model performance, followed by the ARFE module, the FFA module, and finally the Swin Transformer module.
Figure 10 illustrates the visual results of the ablation experiments conducted on the ASOCA dataset. From Fig. 10, it can be observed that the network utilizing the DDPM module can suppress background noise unrelated to the segmentation target while segmenting the three-dimensional structure of the coronary artery. Moreover, it is noticeable that the inclusion of the Swin Transformer, ARFE, and FFA modules allow the network to better segment small vessels, with a reduction in the occurrence of over-segmentation. In Fig. 8, the green boxes represent regions where the DiffCAS has reduced the occurrence of over-segmentation.
3.4.2 Effects of loss parameter settings
To investigate the influence of loss functions, ablation experiments are conducted on the ASOCA dataset. The experiments display the average values of DSC, 95%HD, Recall, and Precision metrics. The experimental parameters and hardware environment settings remain identical to that in the comparative experiments, and the results are presented in Table 4.
From Table 4, it is evident that the combination of binary cross-entropy (BCE) loss and Dice loss yields the best results in terms of DSC, 95%HD, Recall, and Precision on the ASOCA dataset. The incorporation of these two loss functions aids in better convergence of the network and compensates for the limitations of both BCE and Dice loss functions. This combination enhances the segmentation capability of the model.
4 Conclusion
In this paper, a diffusion model-based multi-attention deep network, called DiffCAS, is proposed for automatic 3D coronary artery segmentation. The proposed DiffCAS can effectively select useful information from complex CTA images and make full use of the fused features from different encoders to obtain more semantic representations. DDPM and Swin Transformer are fused as the feature encoder to extract semantic information, suppressing background noise. An ARFE (adaptive residual feature enhancement) module is proposed to enhance the perception of coronary artery details of DiffCAS. The FFA (feature fusion attention) module is designed to fuse features from two encoders and enhance feature representation. The experimental results on the ASOCA and the ImageCAS datasets show that the proposed DiffCAS achieves a good performance for segmentation of the coronary artery and outperforms some state-of the-art networks.
While the proposed method has achieved promising results through both quantitative and qualitative analyses, there are still certain limitations to consider. Due to the fusion of DDPM and Transformer, the amount of parameter of the DiffCAS is increased, which makes the DiffCAS more complex. On the other hand, due to class imbalance in the data, there is still room to improve the DiffCAS. This could involve optimizing the loss function, such as incorporating weighted loss functions, or utilizing methods like Focal Loss [39] to mitigate class imbalance.
Data availability
Data will be made available on request.
References
Sleeman, K.E., de Brito, M., Etkind, S., Nkhoma, K., Guo, P., Higginison, I.J., et al.: The escalating global burden of serious health-related suffering: Projections to 2060 by world regions, age groups, and health conditions. Lancet Global Health. 7(7), 883–892 (2019). https://doi.org/10.1016/S2214-109X(19)30172-X
Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2015, pp. 234–241. Springer (2015). https://doi.org/10.1007/978-3-319-24574-4
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3d u-net: Learning ¨ dense volumetric segmentation from sparse annotation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016, pp. 424–432. Springer (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Milletari, F., Navab, N., Ahmadi, S.-A.: V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571 (2016). https://doi.org/10.1109/3DV.2016.79. IEEE
Yu, L., Cheng, J.-Z., Dou, Q., Yang, X., Chen, H., Qin, J., et al.: Automatic 3d cardiovascular mr segmentation with densely-connected volumetric convnets. In: Medical Image Computing and Computer-Assisted Intervention- MICCAI 2017, pp. 287–295. Springer (2017). https://doi.org/10.1007/978-3-319-66185-8_33
Mou, L., Zhao, Y., Fu, H., Liu, Y., Cheng, J., Zheng, Y., et al.: Cs2-net: Deep learning segmentation of curvilinear structures in medical imaging. Med. Image Anal. 67, 101874 (2021). https://doi.org/10.1016/j.media.2020.101874
Song, A., Xu, L., Wang, L., Wang, B., Yang, X., Xu, B., et al.: Automatic coronary artery segmentation of ccta images with an efficient feature-fusion-and-rectification 3d unet. IEEE J. Biomedical Health Inf. 26(8), 4044–4055 (2022). https://doi.org/10.1109/JBHI.2022.3169425
Xia, L., Zhang, H., Wu, Y., Song, R., Ma, Y., Mou, L., et al.: 3d vessel-like structure segmentation in medical images by an edge-reinforced network. Med. Image Anal. 82, 102581 (2022). https://doi.org/10.1016/j.media.2022.102581
Duan, X., Sun, Y., Wang, J.: ECA-UNet for coronary artery segmentation and three-dimensional reconstruction. SIViP. 17, 783–789 (2023). https://doi.org/10.1007/s11760-022-02288-y
Dong, C., Xu, S., Li, Z.: A novel multistage deep learning solution for automatic coronary arteries segmentation from ccta. J. Am. Coll. Cardiol. 77(18 Supplement 1), 3224–3224 (2021)
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A.N.: Attention is all you need. Adv. Neural Inf. Process. Syst. 30 (2017)
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., et al.: An image is worth 16x16 words: Transformers for image recognition at scale. (2020). arXiv preprint arXiv:2010.11929
Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., et al.: Transunet: Transformers make strong encoders for medical image segmentation. arXiv Preprint arXiv:210204306 (2021)
Wang, W., Chen, C., Ding, M., Yu, H., Zha, S., Li, J.: Transbts: Multimodal brain tumor segmentation using transformer. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, pp. 109–119. Springer (2021). https://doi.org/10.1007/978-3-030-87193-2_11
Zhang, Y., Liu, H., Hu, Q.: Transfuse: Fusing transformers and cnns for medical image segmentation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, pp. 14–24. Springer (2021). https://doi.org/10.1007/978-3-030-87193-2_2
Hatamizadeh, A., Tang, Y., Nath, V., Yang, D., Myronenko, A., Landman, B., et al.: Unetr: Transformers for 3d medical image segmentation. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 574–584 (2022). https://doi.org/10.48550/arXiv.2103.10504
Huang, X., Deng, Z., Li, D., Yuan, X., Fu, Y.: Missformer: An effective transformer for 2d medical image segmentation. IEEE Trans. Med. Imaging. 42(5), 1484–1494 (2023). https://doi.org/10.1109/TMI.2022.3230943
Valanarasu, J.M.J., Oza, P., Hacihaliloglu, I., Patel, V.M.: Medical transformer: Gated axial-attention for medical image segmentation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, pp. 36–46. Springer (2021). https://doi.org/10.1007/978-3-030-87193-2_4
Wang, B., Wang, F., Dong, P., Li, C.: Multiscale transunet + +: Dense hybrid U-Net with transformer for medical image segmentation. SIViP. 16, 1607–1614 (2022). https://doi.org/10.1007/s11760-021-02115-w
Zhao, C., Xiang, S., Wang, Y., Cai, Z., Shen, J., Zhou, S., et al.: Context-aware network fusing transformer and v-net for semi-supervised segmentation of 3d left atrium. Expert Syst. Appl. 214, 119105 (2023). https://doi.org/10.1016/j.eswa.2022.119105
Xiang, S., Li, N., Wang, Y., Zhou, S., Wei, J., Li, S.: Automatic Delineation of the 3D Left Atrium from LGE-MRI: Actor-Critic based Detection and Semi-Supervised Segmentation. IEEE Journal of Biomedical and Health Informatics. : 1–12 (2024). (2024)
Hatamizadeh, A., Nath, V., Tang, Y., Yang, D., Roth, H.R., Xu, D.: In: International, M.I.C.C.A.I.B., Workshop (eds.) Swin Unetr: Swin Transformers for Semantic Segmentation of Brain Tumors in mri Images, pp. 272–284. Springer (2022). https://doi.org/10.1007/978-3-031-08999-2_22
Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., et al.: Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 10012–10022 (2021). https://doi.org/10.48550/arXiv.2103.14030
Ho, J., Jain, A., Abbeel, P.: Denoising diffusion probabilistic models. Adv. Neural Inf. Process. Syst. 33, 6840–6851 (2020)
Song, Y., Sohl-Dickstein, J., Kingma, D.P., Kumar, A., Ermon, S., Poole, B.: Score-based generative modeling through stochastic differential equations. arXiv Preprint (2020). arXiv:2011.13456
Saharia, C., Ho, J., Chan, W., Salimans, T., Fleet, D.J., Norouzi, M.: Image superresolution via iterative refinement. IEEE Trans. Pattern Anal. Mach. Intell. 45(4), 4713–4726 (2023). https://doi.org/10.1109/TPAMI.2022.3204461
Lugmayr, A., Danelljan, M., Romero, A., Yu, F., Timofte, R., Van Gool, L.: Repaint: Inpainting using denoising diffusion probabilistic models. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 11461– 11471 (2022). https://doi.org/10.48550/arXiv.2201.09865
Dhariwal, P., Nichol, A.: Diffusion models beat gans on image synthesis. Adv. Neural Inf. Process. Syst. 34, 8780–8794 (2021)
Baranchuk, D., Rubachev, I., Voynov, A., Khrulkov, V., Babenko, A.: Label-efficient semantic segmentation with diffusion models. arXiv Preprint arXiv:211203126 (2022)
Pinaya, W.H., Graham, M.S., Gray, R., Da Costa, P.F., Tudosiu, P.-D., Wright, P., et al.: Fast unsupervised brain anomaly detection and segmentation with diffusion models. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 705–714 (2022). https://doi.org/10.1007/978-3-031-16452-1_67. Springer
Amit, T., Shaharbany, T., Nachmani, E., Wolf, L.: Segdiff: Image segmentation with diffusion probabilistic models. arXiv Preprint arXiv:211200390 (2022)
Wu, J., Fu, R., Fang, H., Zhang, Y., Yang, Y., Xiong, H., et al.: Medsegdiff: Medical image segmentation with diffusion probabilistic model. arXiv Preprint arXiv:221100611 (2022)
Wolleb, J., Sandk¨uhler, R., Bieder, F., Valmaggia, P., Cattin, P.C.: Diffusion models for implicit image segmentation ensembles. In: International Conference on Medical Imaging with Deep Learning, pp. 1336–1348 PMLR (2022)
Wu, J., Fu, R., Fang, H., Zhang, Y., Xu, Y.: Medsegdiff-v2: Diffusion based medical image segmentation with transformer. arXiv Preprint arXiv:230111798 (2023)
Shen, Z., Zhang, M., Zhao, H., Yi, S., Li, H.: Efficient attention: Attention with linear complexities. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 3531–3539 (2021). https://doi.org/10.48550/arXiv.1812.01243
Gharleghi, R., Adikari, D., Ellenberger, K., Ooi, S.-Y., Ellis, C., Chen, C.-M., et al.: Automated segmentation of normal and diseased coronary arteries–the asoca challenge. Comput. Med. Imaging Graph. 97, 102049 (2022). https://doi.org/10.1016/j.compmedimag.2022.102049
Zeng, A., Wu, C., Lin, G., Xie, W., Hong, J., Huang, M., et al.: Imagecas: A large-scale dataset and benchmark for coronary artery segmentation based on computed tomography angiography images. Comput. Med. Imaging Graph. 109, 102287 (2023). https://doi.org/10.1016/j.compmedimag.2023.102287
Huttenlocher, D.P., Klanderman, G.A., Rucklidge, W.J.: Comparing images using the hausdorff distance. Trans. Pattern Anal. Mach. Intell. 15(9), 850–863 (1993). https://doi.org/10.1109/34.232073
Lin, T.-Y., Goyal, P., Girshick, R., He, K., Doll´ar, P.: Focal loss for dense object detection. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 2980–2988 (2017). https://doi.org/10.48550/arXiv.1708.02002
Acknowledgements
This work was supported in part by the National Science Foundation of China (NSFC) under Grants 61976241, 81827805, in part by the National key R&D program of China under Grants 2018YFA0704102, the Basic Research Project of Shenzhen Science and Technology Innovation Commission under Grants JCYJ20200109114610201 and in part by the International Science and technology cooperation plan project of Zhenjiang under grant GJ2021008.
Funding
This work was supported in part by the National Science Foundation of China (NSFC) under Grants 61976241, 81827805, in part by the National key R&D program of China under Grants 2018YFA0704102, the Basic Research Project of Shenzhen Science and Technology Innovation Commission under Grants JCYJ20200109114610201 and in part by the International Science and technology cooperation plan project of Zhenjiang under grant GJ2021008.
Author information
Authors and Affiliations
Contributions
J.L. and Q.W. wrote the main manuscript text, prepared the figures, performed the experiments. Y.W. supervised the development of the script and wrote the main manuscript text. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical approval
Not applicable. This submission does not include human or animal research.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, J., Wu, Q., Wang, Y. et al. DiffCAS: diffusion based multi-attention network for segmentation of 3D coronary artery from CT angiography. SIViP 18, 7487–7498 (2024). https://doi.org/10.1007/s11760-024-03409-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-024-03409-5