Abstract
Masson pine (Pinus massoniana) is primarily present within subtropical and tropical areas in China, and a number of these regions have a severe deficiency in inorganic phosphate (Pi). As a macronutrient, phosphorus plays a crucial role in plant development. Although several studies have documented the responses of masson pine to Pi starvation at a global level using RNA-Seq and comparative proteomic analyses, the detailed features in the roots that primarily respond to low Pi stress have not yet been studied. Our study examined the response of masson pine roots to a deficiency in Pi. Approximately 1117 unigenes were shown to respond to Pi deficiency by differential expression when analyzed using RNA-Seq. A total of 819 and 298 of these transcripts were up- and down-regulated, respectively. Several transporters including phosphate transporters (PHT1, PHO88), ABC transporters and metal transporters were identified. The ethylene response factor (ERF) was found to be the most abundant transcription factor. Analyses of these genes, including gene ontology enrichment and the KEGG pathway analysis, indicated that the metabolic processes are the most enriched under abiotic stresses, including Pi deficiency. This study provided abundant transcriptomic information to functionally dissect the response of masson pine roots to Pi deficiency. This will additionally aid to elucidate the biological regulatory mechanisms that the pines use to respond to low Pi stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
The masson pine (Pinus massoniana) is a native gymnosperm from the southern portions of China. This tree has broad economic value for its pulp, resin, and timber production. The distribution area of this tree species is primarily in acid yellow soils that critically lack inorganic phosphate (Pi) (Zhang et al. 2010). Phosphorus, in the form of Pi, is an essential macronutrient in plants, which play a vital role in development and metabolic activities (Abel et al. 2002). Although the soil may contain large amounts of phosphorus, the quantity available for biological uptake is often limited as phosphorus has the tendency to complex with other soil constituents and then slowly be released (Shen et al. 2011). Thus, the deficiency of phosphorus in soil may constrain the growth and productivity of the plants that grow in these areas (Vance et al. 2003). Over the past several decades, Pi fertilizer has been widely used for crop and forestry plantations. This annually decreases the non-renewable phosphate rock reserves, which may lead to the inevitable loss of this resource (Deng et al. 2018). Furthermore, much of the Pi fertilizer applied results in environmental pollution, including the eutrophication of water systems (Cordell et al. 2009). Therefore, it is significant to understand the strategies that plants use to absorb and utilize phosphorus, and adapt the mechanisms of the plants that can efficiently utilize soil Pi.
Plants have evolved a number of different strategies that enable them to adapt and manage a deficiency of Pi, involving modifications in developmental programs and metabolic networks (Fan et al. 2014). These adaptive strategies are thought to be mediated synergistically by a series of genes. Extensive research on the response of model plants to Pi starvation, that have been conducted over the past several decades, have considerably enhanced the knowledge of the phosphorus signaling and response pathway (Hammond et al. 2003; Woo et al. 2012; Gho et al. 2018). A series of genes enable the plant to adapt to a deficiency of Pi, primarily through the regulation of signal transduction, internal remobilization, phosphorus acquisition, and metabolic changes (Fang et al. 2009). Previous studies have identified the key role of the PHOSPHATE STARVATION RESPONSE1 (PHR1) gene in transcriptional responses that are related to a deficiency of Pi (Zhao et al. 2018). This research also revealed that the proteins contained in the SYG1/PHO81/XPR1 (SPX) domain are vital for the plant to sense Pi and maintain homeostasis (Zhang et al. 2014; Wild et al. 2016; Zhao et al. 2018). The expression of the genes that encode the purple acid phosphatases (PAPs) and Pi transporters were found to be up-regulated under Pi deficient conditions (Duff et al. 1994; Poirier and Jung 2018). In addition, a number of genes involved in the metabolism of lipids, including UDP sulfoquinovose synthases and nonspecific phospholipases, were induced under low-Pi stress (Lan et al. 2012).
Masson pine seedlings under low-Pi stress were the source of transcriptomes, which were analyzed using a combination of 8 × 60 K DNA microarrays and RNA-Seq (Fan et al. 2014). The results indicated a strong representation of genes and transcription factors that were associated with stress. In addition, genes whose functions were unknown were subjected to up- and down-regulation. The results suggested that Pi-deficiency stress involves the cumulative implication of a number of genes and multiple metabolic pathways. In another study, Fan et al (2016) compared proteomes using seedlings of an elite line of masson pine and identified 98 proteins that were differentially expressed. These proteins that responded to Pi starvation were involved in defense, photosynthesis, biosynthesis, cellular organization, secondary metabolism, energy metabolism, and signal transduction. Although these studies identified both global transcriptomic and proteomic responses to Pi deficiency in masson pine, the roots that exhibit primary responses to low-Pi stress have been studied less extensively. Therefore, to address this dearth of information, functional analyses of root responsive genes are merited. In our study, the transcriptomic profile of masson pine root responses to a deficiency of Pi was demonstrated. It showed how the responsive genes for phosphate starvation bolster the efficiency of the acquisition, recycling, and remobilization of phosphorus in roots.
Methods
Materials and treatment
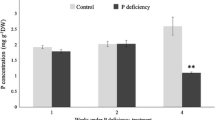
Masson pine full-sibling seeds were collected from an elite tree, which was shown to confer a tolerance to Pi deficiency in our previous study, in the Duyun Forest Farm in the Guizhou Province in China. The seeds were disinfected by treating them with 5% sodium hypochlorite for 5 min, followed by 70% ethanol for 10 min. Next, the seeds were soaked in water maintained at room temperature. The germinated seeds were planted in plastic pots and grown at a day/night temperature of 28 ºC/18 ºC (14 h day/10 h night) in a plant growth chamber with 70% relative humidity and a light intensity of 2000 lx. At 15 days, after emergence, the seedlings were treated with a modified Hoagland nutrient solution (Fan et al. 2014) that was deficient in Pi (0.01 mM KH2PO4) and sufficient in Pi (0.5 mM KH2PO4) as the treatment and control, respectively. The experiment utilized six seedlings per treatment that were replicated three times. The nutrient solution was added at 3-day intervals. The seedling roots were harvested 36 days after treatment, snap frozen with liquid nitrogen, followed by storage at – 80 ºC.
RNA isolation, cDNA library construction and sequencing
Total root RNA was isolated with the Invitrogen Plant RNA Isolation Kit based on provided directions. RNA was assessed for quality and was quantified through a combination of agarose gel electrophoresis together with an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). mRNA was specifically isolated from the extracted RNA through the use of Oligo (dT) magnetic beads, followed by fragmentation and cDNA generation. The short fragments were connected using adapters following their purification, and agarose gel electrophoresis was performed. Fragments of 200 bp in length were isolated for PCR amplification followed by sequencing on the Hi-Seq 2000 platform (Illumina, San Diego, CA, USA). The software for the platform was used to conduct the primary analysis of the data and the base calling.
Transcriptome assembly and assessment of the expression
To generate a set of transcripts that were non-redundant, adaptor sequences and empty reads were removed, and high-quality reads were selected using Q20 and Q30 as the basis. Trinity software was used for clean read assembly, after filtering, to construct the unigenes. The levels of expression for each unique transcript were calculated by quantifying the reads based on the RPKM approach (reads per kilobase per million reads) (Mortazavi et al. 2008). DEG-Seq was used to judge the differentially expressed genes (DEGs) with an FDR (false discovery rate) < 0.05 and the absolute value of log2 RPKM ratio ≥ 1 as the threshold (Anders and Huber 2010).
Gene enrichment analyses
The GO and KEGG analyses were conducted with the Blast2GO program. Biological process, molecular function, and cellular component were graphed with the second level of the GO hierarchy. The adjusted standard of significantly substantiated GO terms and enriched pathway was a p value set to less than 0.05.
Validation of transcriptome sequencing using quantitative real-time PCR (qRT-PCR)
Ten DEGs were randomly selected and quantitatively analyzed using qRT-PCR to confirm RNA sequencing expression profiles. Primers were designed using Primer Premier 5.0 (Table S1). SYBR Select Master Mix Kit (Applied Biosystems, CA, USA) was used for triplicate qRT-PCR reactions with a 7500 Fast Real-Time PCR System (Applied Biosystems) based on provided directions. The cycle threshold (Ct) was utilized to assess relative transcript levels for each gene, which were normalized using 18 s-rRNA and UBQ as internal controls (Fan et al. 2014).
Results
Transcriptome dataset of masson pine roots
Six libraries based on three Pi-sufficient biological replicates (CK36-R-1, CK36-R-2, CK36-R-3) and three Pi-deficient (P36-R-1, P36-R-2, P36-R-3) treatments were generated to reveal the gene expression profiles of the masson pine roots subjected to the stress of Pi deficiency. These libraries were used for further transcriptome sequencing analysis. After filtering the adaptor sequences and those that had a low base quality, high-quality paired end reads were generated, respectively (Table 1). The averages of Q20 and Q30 were more than 98.5% and 96.0%, respectively. A total of 95,658 unigenes were obtained, with an average length of 748 bp, and an N50 length of 1104 bp. The unigenes ranged from 201 to 12,246 bp in length. More specifically, the number of genes within the size ranges of 201–500, 501–1000, 1001–2000, 2001–3000 bp and > 3000 bp were 50,653, 22,918, 15,080, 4848 and 2159 unigenes, respectively. These results indicated that the experiment was successful in producing high-quality transcriptome data.
The resultant unigenes were run in the NR, KOG, SwissProt, and KEGG databases according to the sequence similarity analysis. Over 45,000 unigenes were identified in the plants, and the NR database provided the largest number of annotations, with a value of 43,168 (45.72%).
DEGs in response to a low Pi treatment
DE-Seq with the FDR set to less than 0.05 enabled differential expression analysis to be conducted using the raw data obtained from the Illumina sequencing. A total of 1117 unigenes responded to the treatment of Pi deficiency and were significantly differentially expressed. A total of 819 and 298 of these transcripts were up- and down-regulated (Fig. 1, Table S2), respectively. Previous research suggested that the DEGs related to phosphorus metabolism can be divided into six categories, including the genes that are involved in transcription, lipid metabolism, transport, phosphorylation/dephosphorylation, metabolism, and miscellaneous (Lan et al. 2015). In this study, several kinds of putative transcription factors (TFs) have been shown to be associated with low-Pi tolerance (Table 2). Of these, the ethylene response factor (ERF) comprised the most abundant TFs that responded to low-Pi stress. Beyond that, the MYB family TF (Unigene0011607), WRKY TF (Unigene0042859), and bZIP TFs (Unigene0042142 and Unigene0054957) were significantly expressed under low-Pi conditions. These identified TFs were found to be up-regulated with the exception of one, bZIP TF (Unigene0042142).
Differentially expressed genes under low-Pi stress. a Volcano plot represents fold change versus false discovery rate. The horizontal dashed line indicates a cutoff of statistical significance at 0.05. b Scatter plot x axis displays the average signal intensity for the control sample, while the y axis displays the average signal intensity for the treatment sample
From the transport overview, phosphate transporter genes, including PHT1 (Unigene0072509) and PHO (Unigene0093941), were dramatically up-regulated in Pi-limited roots. ATP-binding cassette (ABC) transporters (Unigene0067284 and Unigene0073352) were strongly induced. Other transporter subfamily genes, including organic cation, potassium, metal, and sugar transporters, were also identified to be highly responsive to the low-Pi stress in the masson pine roots. In terms of lipid metabolism, genes that could degrade phospholipids were induced under low-Pi stress, such as short-chain dehydrogenase/reductase (Unigene0002082), glycerol kinase (Unigene0008107 and Unigene0020667), and lipoxygenase (Unigene0065294). One iron-superoxide dismutase gene (Unigene0040690) was found to be dramatically down-regulated. Many of the genes that were up-regulated in response to phosphorus starvation were found to be involved in metabolic processes. These included pyruvate phosphate dikinase (Unigene0009113 and Unigene0065735), thioredoxin (Unigene0006520), cytochrome P450 (Unigene0008678), phosphoenolpyruvate carboxykinase (Unigene0036016), and phosphoglucomutase (Unigene0030703). With regards tophosphorylation/dephosphorylation, the DEGS included a subset of 26 kinases and one acid phosphatase (Table S2).
Previous studies indicated that abiotic stress conditions, including Pi deficiency, can generate reactive oxygen species (ROS) (Fan et al. 2014). We identified that some of the genes that were up-regulated, were involved in scavenging ROS, including glutathione peroxidase (Unigene0017511).
To validate the results of the transcriptome sequencing, ten differentially expressed unigenes were chosen for qRT-PCR analysis (Fig. 2). The results showed that there was consistency between the transcriptomic and the qRT-PCR analyses for all unigenes selected. This confirmed the reliability of the transcriptome data.
GO enrichment analysis of the DEGs
To obtain a more precise understanding of the DEG function in Pi-limited roots, GO term enrichment analysis was conducted. The up- and down-regulated transcripts were both assessed based upon cellular components, molecular functions, as well as biological processes (Fig. 3). For the biological process ontology, the dominant terms in this study included “metabolic process”, “cellular process”, and “single-organism process”, followed by “response to stimulus”. The dominant terms for the cellular component were “cell”, “cell part”, “macromolecular complex”, “organelle”, and “organelle part”. The processes represented by the “binding”, “catalytic activity”, and “structural molecule activity” GO terms accounted for the majority of the molecular function, followed by “transporter activity”.
KEGG enrichment analysis
To illustrate the function of the DEGs identified using RNA-Seq, a KEGG enrichment analysis was conducted. The results indicated that 267 of the 1117 DEGs could be assigned to 72 KEGG pathways (Table S3). Among them, four pathways (metabolic pathways, secondary metabolite biosynthesis, ribosome, and microbial metabolism in diverse environments) were the most abundant in the Pi-deficient roots. Furthermore, the KEGG terms that were over-represented (Q < 0.05) were classified as the “ribosome” (Fig. 4). However, as shown in Fig. 4, the degradation of aromatic compounds, carbon fixation in photosynthetic organisms, and glycolysis/gluconeogenesis pathways were over-represented among the DEGs, suggesting that these pathways could be involved in the response to low-Pi. Other pathways that were closely linked to phosphorus were also detected, such as oxidative phosphorylation, pyruvate metabolism, pentose phosphate pathway, phosphatidylinositol signaling system, citrate cycle, and plant hormone signal transduction.
Discussion
As a non-renewable nature resource, phosphate rock is being rapidly depleted because of the world’s reliance on Pi fertilizers. Due to this, extensive research based on microarrays and RNA-Seq methods have documented certain crop species and various model plant responses under Pi-deficient conditions (Thibaud et al. 2010; O’Rourke et al. 2013; Gho et al. 2018). Previous research has reported global transcriptome responses to Pi deficiency in masson pine seedlings. However, there have been very few functional studies on candidate root genes that exhibit primary responses to Pi deficiency. Our study used RNA-Seq to examine whole-transcriptomic responses to low-Pi stress in masson pine roots. We identified 1117 DEGs that responded to Pi-deficiency conditions, of which 10 were chosen at random for qRT-PCR validation (Fig. 2). qRT-PCR-based expression was found to correlate with the RNA-Seq data confirming reliability of the DEGs.
When the enriched GO biological process term was examined, it was found that “metabolic process” (terpenoid biosynthetic processes in particular), was most significantly induced in Pi deficient masson pine roots. A previous study showed that diterpene phytoalexins and allelochemicals increased in plant roots as a response to various stress conditions (Zhao et al. 2005; Kong et al. 2006). In Arabidopsis and rice roots, strigolactone resulting from terpenoid biosynthetic processes accumulated under Pi starvation. This is known to suppress the growth of shoots (Gomez-Roldan et al. 2008) and to enhance the development of roots (Arite et al. 2012). These results suggested that secondary metabolites in masson pine may play an essential role in enhancing the survival rate in low Pi conditions. Furthermore, the KEGG pathway analysis showed that the ribosome, the degradation of aromatic compounds, carbon fixation in photosynthetic organisms, and the glycolysis/gluconeogenesis pathways were over-represented among the DEGs, suggesting that these pathways may be involved in the response to low Pi. These annotations provide a valuable source to investigate the specific processes by examining the functions of the DEGs.
Previous studies suggested that the plant transcription factors were essential for the regulation of the downstream genes under low-Pi conditions. Fan et al. (2014) demonstrated that the MYB TF family represented the largest class of TFs that were induced by Pi deficiency in masson pine seedlings. PHR1 (a part of the MYB TF) was previously identified for its involvement in low-Pi stress. It activates a series of genes, including those that contained SPX domains that can be normally be activated by binding to promoter regions (Nilsson et al. 2007; Secco et al. 2012). In this study, only one putative MYB TF was dramatically induced under Pi deficiency in roots. Twelve years ago, Hernández et al. (2007) identified four TFs up-regulated in Pi-deficient common bean roots, and three of them were members of the MYB family. In later studies, numerous plant MYBs were identified as regulators involved in Pi starvation. Examples include AtMYB2 that regulates the plant response to Pi deficiency by controlling miR399 expression in Arabidopsis (Baek et al. 2013). It is well known that several morphological processes can be affected under Pi-starvation conditions, including a change in the root system architecture (RSA). Ethylene has previously been shown to be involved in remodeling the RSA, and it also has a role in other types of plant responses to Pi deficiency (Song and Liu 2015). As a group of AP2 (APETALA2) domain-containing transcription factors, ERFs serve as either repressors or activators of ethylene-mediated transcription. Chen et al. (2018) showed that the down-regulation of the JcERF035 gene contributed to RSA regulation as well as both the biosynthesis and accumulation of anthocyanins in aerial tissues of plants under Pi-starvation conditions. Some transcriptomic analyses have shown that the expression of several genes dramatically changed in plant roots (Thibaud et al. 2010; Kang et al. 2014; Zhao et al. 2018).Our study identified ERFs as the most abundant up-regulated TFs responding to low-Pi stress. This is different from the results of Fan et al (2014) who demonstrated that the MYBs represented the largest class. This may be due to the differences in time of sampling as well as the tissues sampled.
Previous studies showed that the uptake, transport and translocation of phosphorus in plants are generally carried out by phosphorus transporters (PHT1 and PHO1) (Nussaume et al. 2011; Hamburger et al. 2002). Most transporters that are in the PHT1 group are known to be differentially expressed in the roots of Zea mays, rice and Brachypodium distachyon (Nagy et al. 2006; Gho et al. 2018; Zhao et al. 2018). In our study, only one high-affinity phosphate transporter gene (PHT1) was up-regulated (Table S2), and one putative PHO88, rather than PHO1, was differentially expressed in masson pine roots. It has been suggested that PHO88 plays a role in the phosphate transport pathway via the regulation or maturation of secretory proteins in yeast (Hurto et al. 2007). As a putative membrane protein, it was rarely reported in plant roots that were Pi deficient, and additional genetic analyses will be necessary to determine its role in the phosphorus homeostasis in plant roots. In addition to phosphate transporters, some other transporter families were significantly changed under Pi deficiency. ABC transporters, which are known to be induced under various abiotic stress conditions (Huang et al. 2010; Xi et al. 2012), were dramatically induced in this study. An organic cation/carnitine transporter was also up-regulated, suggesting that it might be involved in the response to a Pi deficiency in plants, as was previously documented in soybean roots (Zeng et al. 2016). Under Pi deficiency, plants over-accumulated some metallic cations, including Fe3+, K+ or Al3+ (Hirsch et al. 2006; Abel 2017). In this study, one gene that encoded a metal transporter (Unigene0067271) was also up-regulated in the roots.
The lipid composition of the plant membranes may be dramatically altered under Pi-starvation conditions, with a decrease in phospholipids and an increase in lipids that do not contain phosphorus (Russo et al. 2007). Generally, the phospholipids of the plasma membrane and tonoplast are replaced by galactolipid digalactosyldiacylglycerol or glycolipids (Mehra et al. 2019; Tawaraya et al. 2018). In this study, some genes that may degrade phospholipids were induced under low-Pi stress, including a short-chain dehydrogenase/reductase, glycerol kinase and lipoxygenase. The induction of these genes suggested that the membrane lipid composition might change when subjected to the stress of low levels of Pi.
Conclusions
In our study, the whole transcriptomic profile of masson pine roots subjected to Pi deficiency was generated. Phosphate transporter genes, ERF TFs, and genes linked with lipid remodeling among 1,117 DEGs were detected. The results of this study revealed some different DEGs and enriched pathways from previous research in seedlings, and provided new perceptions on the responses of masson pine to a deficiency of Pi.
Author contributions statement
Xiaocheng Pan: experiment performance, data analysis and paper writing. Haibo Hu: experiment design, data analysis and paper revision.
Abbreviations
- Pi:
-
Inorganic phosphate
- ERF:
-
Ethylene response factor
- PHR1:
-
Phosphate starvation response1
- SPX:
-
SYG1/PHO81/XPR1
- PAPs:
-
Purple acid phosphatases
- DEGs:
-
Differentially expressed genes
- FDR:
-
False discovery rate
- ABC:
-
ATP-binding cassette
- ROS:
-
Reactive oxygen species
- AP2 :
-
Apetala2
References
Abel A (2017) Phosphate scouting by root tips. Curr Opin Plant Biol 39:168–177
Abel S, Ticconi CA, Delatorre CA (2002) Phosphate sensing in higher plants. Physiol Plant 115(1):1–8
Anders S, Huber W (2010) Differential expression analysis for sequence count data. J Genome Biol 11(10):R106
Arite T, Kameoka H, Kyozuka J (2012) Strigolactone positively controls crown root elongation in rice. J Plant Growth Regul 31:165
Baek D, Kim MC, Chun HJ, Kang S, Park HC, Shin G, Park J, Shen M, Hong H, Kim WY, Kim DH, Lee SY, Bressan RA, Bohnert HJ, Yun DJ (2013) Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol 161(1):362–373
Chen Y, Wu P, Zhao Q, Tang Y, Chen Y, Li M, Jiang H, Wu G (2018) Overexpression of a phosphate starvation response AP2/ERF gene from physic nut in Arabidopsis alters root morphological traits and phosphate starvation-induced anthocyanin accumulation. Front Plant Sci 9:1186
Cordell D, Drangert J, White S (2009) The story of phosphorus: global food security and food for thought. Glob Environ Change 19(2):292–305
Deng QW, Luo XD, Chen YL, Zhou Y, Zhang FT, Hu BL, Xie JK (2018) Transcriptome analysis of phosphorus stress responsiveness in the seedlings of Dongxiang wide rice (Oryza rufipogon Griff.). Biol Res 51:7
Duff SMG, Sarath G, Plaxton WC (1994) The role of acid-phosphatases in plant phosphorus-metabolism. Physiol Plant 4:791–800
Fan F, Cui B, Zhang T, Qiao G, Ding G, Wen X (2014) The temporal transcriptomic response of Pinus massoniana seedlings to phosphorus deificiency. PLoS ONE 9(8):e105068
Fan F, Ding G, Wen X (2016) Proteomic analyses provide new insights into the responses of Pinus massoniana seedlings to phosphorus deficiency. Proteomics 16:504–515
Fang ZY, Shao C, Meng YJ, Wu P, Chen M (2009) Phosphate signaling in Arabidopsis and Oryza sativa. Plant Sci 176(2):170–180
Gho YS, An G, Park HM, Jung KH (2018) A systemic view of phosphate starvation-responsive genes in rice roots to enhance phosphate use efficiency in rice. Plant Biot Rep 12:249–264
Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pages V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, Bouwmeester H, Becard G, Beveridge CA, Rameau C, Rochange SF (2008) Strigolactone inhibition of shoot branching. Nature 455:189–194
Hamburger D, Rezzonico E, Petetot JMC, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14(4):889–902
Hammond JP, Bennett MJ, Bowen HC, Broadley MR (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132:578–596
Hernández G, Ramírez M, Valdés-López O, Tesfaye M, Graham MA, Czechowski T, Schlereth A, Wandrey M, Erban A, Cheung F, Wu HC, Lara M, Town CD, Kopka J, Udvardi MK, Vance CP (2007) Phosphorus stress in common bean: Root transcript and metabolic responses. Plant Physiol 144(2):752–767
Hirsch J, Marin E, Floriani M, Chiarenza S, Richaud P, Nussaume L, Thibaud MC (2006) Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88(11):1767–1771
Huang CF, Yamaji N, Ma JF (2010) Knockout of a bacterial-type ATP-binding cassette transporter gene, AtSTAR1, results in increased aluminum sensitivity in Arabidopsis. Plant Physiol 153(4):1669–1677
Hurto RL, Tong AHY, Boone C, Hopper AK (2007) Inorganic phosphate deprivation causes tRNA nuclear accumulation via retrograde transport in Saccharomyces Cerevisiae. Genetics 176(2):841–852
Kang J, Yu H, Tian C, Zhou W, Li C, Jiao Y, Liu D (2014) Suppression of photosynthetic gene expression in roots is required for sustained root growth under phosphate deficiency. Plant Physiol 165:1156–1170
Kong CH, Li HB, Hu F, Xu XH, Wang P (2006) Allelochemicals released by rice roots and residues in soil. Plant Soil 288:47–56
Lan P, Li W, Schmidt W (2012) Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol Cell Proteom 11(11):1156–1166
Lan P, Li W, Schmidt W (2015) Omics approaches towards understanding plant phosphorus acquisition and use. Ann Plant Rev 48:65–97
Mehra P, Pandey BK, Verma L, Giri J (2019) A novel glycerophosphodiester phosphodiesterase improves phosphate deficiency tolerance in rice. Plant Cell Environ 42:1167–1179
Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628
Nagy R, Vasconcelos MJ, Zhao S, McElver J, Bruce W, Amrhein N, Raghothama KG, Bucher M (2006) Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol 8(2):186–197
Nilsson L, Müller R, Nielsen TH (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 301(12):1499–1512
Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC (2011) Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci 2:83
O’Rourke JA, Yang SS, Miller SS, Bucciarelli B, Liu J, Rydeen A, Bozsoki Z, Uhde-Stone C, Tu ZJ, Allan D, Gronwald JW, Vance CP (2013) An RNASeq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol 161(2):705–724
Poirier Y, Jung JY (2018) Phosphate transporters. Annu Plant Rev online, pp 125–158.
Russo MA, Quartacci MF, Izzo R, Belligno A, Navari-Izzo F (2007) Long- and short-term phosphate deprivation in bean roots: plasma membrane lipid alterations and transient stimulation of phospholipase. Phytochemistry 68(11):1564–1571
Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J (2012) The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol 193(4):842–851
Shen J, Yan L, Zhang J, Li H, Bai Z, Chen X, Zhang W, Zhang F (2011) Phosphorus dynamics: from soil to plant. Plant Physiol 156(3):997–1005
Song L, Liu D (2015) Ethylene and plant responses to phosphate deficiency. Front Plant Sci 6:796
Tawaraya K, Honda S, Cheng W, Chuba M, Okazaki Y, Saito K, Oikawa A, Maruyama H, Wasaki J, Wagatsuma T (2018) Ancient rice cultivar extensively replaces phospholipids with non-phosphorus glycolipid under phosphorus deficiency. Physiol Plantarum 163(3):297–305
Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64(5):775–789
Vance CP, Uhde-Stone C, Allan DLP (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157(3):423–447
Wild R, Gerasimaite R, Jung JY, Truffault V, Pavlovic I, Schmidt A, Saiardi A, Jessen HJ, Poirier Y, Hothorn M, Mayer A (2016) Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352:986–990
Woo J, MacPherson CR, Liu J, Wang H, Kiba T, Hannah MA, Wang XJ, Bajic VB, Chua NH (2012) The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Bio 12(1):62
Xi J, Xu P, Xiang CB (2012) Loss of AtPDR11, a plasma membrane-localized ABC transporter, confers paraquat tolerance in Arabidopsis thaliana. Plant J 69(5):782–791
Zeng H, Wang G, Zhang Y, Hu X, Pi E, Zhu Y, Wang H, Du L (2016) Genome-wide identification of phosphate-deficiency-responsive genes in soybean roots by high-throughput sequencing. Plant Soil 398:207–227
Zhang Y, Zhou Z, Ma X, Jin G (2010) Foraging ability and growth performance of four subtropical tree species in response to heterogeneous nutrient environments. J For Res 15:91–98
Zhang Z, Liao H, Lucas WJ (2014) Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J Integr Plant Biol 56:192–220
Zhao H, Li H, Kong C, Xu X, Liang W (2005) Chemical response of allelopathic rice seedling under varying environmental conditions. Allelopathy J 15:105–110
Zhao P, Wang L, Yin H (2018) Transcriptional responses to phosphate starvation in Brachypodium distachyon roots. Plant Physiol Bioch 122:113–120
Acknowledgements
This research was supported by the Zhejiang Provincial Natural Science Foundation of China (LY15C160005). The funder in this study played a major role in designing experiments, analyzing data, and writing the manuscript. Our thanks to the English native speaker professional with a science background at MJ editor for revising our manuscript. We hereby express our thanks to ‘Research Square’ for sharing our work as a preprint (https://doi.org/10.21203/rs.2.14498/v1).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Enrico Schleiff.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pan, X., Hu, H. Transcriptome analysis of low phosphate stress response in the roots of masson pine (Pinus massoniana) seedlings. Acta Physiol Plant 42, 176 (2020). https://doi.org/10.1007/s11738-020-03167-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-020-03167-0