Abstract
Strigolactones (SLs) are a small class of diverse metabolites derived from the carotenoid pathway. These active biomolecules are a recent inclusion to the list of non-traditional phytohormones or plant growth regulators. Previous reports and articles have discussed their pro-regulatory roles in plant growth, development, signaling and delay of senescence. However, the multi-level control of SL biosynthesis is less known. The anabolic genes are strictly regulated through synchronized co-operation between crucial phytohormones. Epigenetic and microRNA-mediated post-transcriptional regulation fine tunes the cellular accumulation of these putative phytohormones. The question now arises that why such multi-level intricate regulation at all is required for SLs, which were originally detected as under-rated germination and rhizosphere stimulants. This review answers the question in the backdrop of the positive roles of SLs in promoting abiotic stress resilience across diverse plant species. SLs reportedly accumulate in the plant tissues in response to environmental sub-optimal conditions like drought, salinity, temperature, nutrient deprivation and oxidative stresses. Fluctuations in the light quality and intensity also trigger variable accumulation of SLs, indicating their potential in regulating light stress as well. Though the exact roles of SLs have not yet been characterized, it is predicted that they possibly induce the expression of downstream osmolytes to maintain metabolic homeostasis in the stressed cells. Thus, exogenous treatments or transgenic approaches for higher SL bioaccumulation can be potential strategies for developing multiple abiotic stress tolerance in crops and plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are sessile organisms tolerating ecological constraints for sustainable survival. The mechanisms for tolerance have eventually developed as a result of the evolutionary tinkering of the adaptive signaling processes (Banerjee et al. 2017). Phytohormones are the critical signaling molecules in plants. They act locally or distantly and regulate stress responses even at very low concentrations of 10−6–10−5 mol L−1 (Wang and Irving 2011). These are traditional plant growth regulators (PGRs) consisting of auxins, gibberellins (GAs), cytokinins (CKs), abscisic acid (ABA), ethylene (ET) and brassinosteroids (BRs) (Banerjee et al. 2017). Additionally, other metabolites like salicylic acid (SA), jasmonic acid (JA), nitric oxide (NO) and polyamines (PAs) are regarded as non-traditional PGRs. Strigolactones (SLs) are the most recent introduction to this categorical list (Smith and Li 2014). These PGRs often exhibit interactions among each other to form elaborate signaling networks and to regulate developmental growth under hostile environmental conditions (Wang and Irving 2011). This review presents an exhaustive discussion on the diverse molecular networks regulating the biosynthesis of SLs. The non-traditional PGRs also play crucial roles in generating tolerance towards reactive oxygen species (ROS)-mediated abiotic stresses in plants.
SLs are derivatives of the carotenoid pathway. They are exuded from the roots of terrestrial plants involved in symbiotic associations with soil arbuscular mycorrhiza (AM). Mycorrhizal symbiosis between plants and Glomeromycota fungi first revealed that SLs act as an external stimulant and induce hyphal branching, leading to AM symbiosis. This phenomenon has been held responsible for the gradual evolution of terrestrial plants and their ability to tolerate different stress conditions (Liu et al. 2007). SLs also accelerate the root-mediated absorption of soil nutrients like phosphates, which have low edaphic mobility. Identification and characterization of shoot branching mutants across plant species has established SLs as potential phytohormones (Pandey et al. 2016). These mutants include more axillary growth 1-4 (max1-4) in Arabidopsis thaliana, dwarf and high tillering dwarf (d/htd) in Oryza sativa, decreased apical dominance 1 (dad1) in Petunia hybrida, ramosus 1 (rms1) to rms5 in Pisum sativum (Leyser 2009; Beveridge and Kyozuka 2010). Thus, by virtue of their roles as endogenous phytohormones as well as exogenous rhizosphere stimulants, SLs might be innovatively targeted to promote sustainable neo-domestication of plants under harsh environmental conditions.
Biosynthesis and general roles of SLs
Cook et al. (1972) reported the first natural SL, strigol as a germination stimulant of the parasitic weed, Striga lutea. SLs also co-ordinate the growth and developmental architecture in plants based upon the soil nutrient availability. The positive effects of SL, promoting root growth and root hair elongation have been observed (Koltai 2011). In spite of inhibiting secondary branching in shoots, SLs accelerate secondary growth in stems by increasing the internodal length via an interaction with the growth promoting PGR, auxin (Yamada et al. 2014).
SL biosynthesis has been evolutionarily conserved across several higher plants and even in some algae and bryophytes. Hence, it can be hypothesized that these diverse molecules play essential roles, considered indispensable by the forces of evolution. The structural and functional variations in SLs (containing four rings named: A–D) occur due to the different chemical groups attached to the A and B rings (Boyer et al. 2012). The C and D rings show maximum conservation. The A rings contain one or two methyl groups, whereas one or more hydroxyl or acetylonyl groups remain attached to both the A and B rings (Pandey et al. 2016). The A, B and C together form the central tricyclic lactone connecting the D ring which represents a butenolide group (Xie et al. 2010).
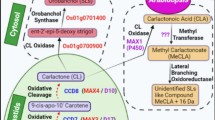
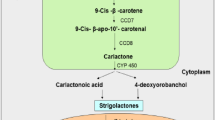
Treatment of Zea mays plants with fluridone (a carotenoid biosynthetic inhibitor), resulted in low accumulation of SLs, which directly showed that SLs must be carotenoid derivatives (Matusova et al. 2005). In another report, it was revealed that SLs are apocarotenoids formed by the carotenoid cleavage dioxygenase (CCD)-mediated cleavage of target carotenoid molecules (Booker et al. 2005). Apocarotenoid biosynthesis is initiated in the plastids. The three plastid localized enzymes, DWARF 27 (D27), CCD7 and CCD8 sequentially transform plastidial all-trans-β-carotene to carlactone (CL) (Alder et al. 2012). Production of 9-cis-β-carotene is catalysed by the carotenoid isomerase, D27. The product is then converted to 9-cis-β-apo-10′-carotenal by CCD7. The CCD8 finally catalyzes the conversion of the compound to CL (Alder et al. 2012) (Fig. 1). The cytochrome P450 monooxygenase in Arabidopsis, MAX1 and related homologs then oxidise CLs into diverse SLs via unidentified intermittent steps, catalyzed by novel enzymes (Mishra et al. 2017). The rice genome encodes for five MAX1 homologs consisting of Os900 (Os01g0700900), Os1400 (Os01g0701400), Os1500 (Os01g0701500), Os1900 (Os02g0221900), and Os5100 (Os06g0565100) (Cardoso et al. 2014; Pandey et al. 2016). Os900 and Os1400 expression have been detected in high tillering cultivars which produce low amounts of SLs. The encoded enzymes catalyze two distinct steps downstream of CL synthesis. Os900 oxidizes CL to form ent-2′-epi-5-deoxystrigol (DS) which is in turn hydroxylated by Os1400 to produce orobanchol (Cardoso et al. 2014) (Fig. 1). The MAX2/D3/RMS4 (F-box protein) and D14/AtD14/DAD2 (α/77β-hydrolase) proteins possibly play roles during plant perception of SLs (Mishra et al. 2017). D14 contains a strictly conserved catalytic triad composed of Ser-His-Asp, which is quintessential for SL hydrolysis and signaling. Structural evidence for an allosteric signaling model is lacking, as there are no substantial differences between the crystal structures of apo-D14 and D14 in complex with intact SL, 2,4,4,-trihydroxy-3-methyl-3-butenal, or 5-hydroxy-3-methylbutenolide (Waters et al. 2017).
The biosynthetic pathway of the SLs: strigol and orobanchol. The catalysis and production of carlactone from carotenoids occurs in plastids. The formed carlactone is processed via several unidentified intermittent enzymes before being shuttled into the cytoplasm. Further breakdown of the compound ultimately synthesizes SLs
SL accumulation has been recorded to be much higher in the roots compared to other tissues. In Arabidopsis, high expression of MAX1 was observed in the cells of the root vasculature; whereas AtCCD8 expression was reported in the root cap columella cells of primary and lateral roots (Cheng et al. 2013). The root elongation zone exhibited high expression of AtMAX2 and OsD14 in Arabidopsis and rice plants respectively (Cheng et al. 2013). In rice, transcript levels of OsCCD7 and OsCCD8 were high in the vascular parenchyma cells of the roots (Cheng et al. 2013). High accumulation of SLs in the roots stimulates germination under hostile edaphic conditions, whereas low production of SLs in the shoots is corroborated with the normal branching pattern. Additionally, SLs might play unidentified roles during fruit ripening and seed development in a species-specific manner, as high levels of SlCCD7 were recorded in the immature green fruits of Solanum lycopersicum (Vogel et al. 2010). SCFMAX2 targets members of the SUPPRESSOR OF MAX2 1 (SMAX1) or D53 protein family, evolutionarily associated with the ClpB/HEAT SHOCK PROTEIN 100 (HSP100) class of HSPs. D53 orthologs, SMAX1-LIKE6 (SMXL6), SMXL7 and SMXL8 directly regulate SL-related aspects of the max2 phenotype (Waters et al. 2017). Other targets of MAX2 include the BR target protein, BRl1-EMS SUPPRESSOR1 and the DELLA family of GRAS transcription factors (TFs). SMXL/D53 proteins are often regarded as potential TFs due to the consensus presence of a C-terminal Ethylene Response Factor-Associated Amphiphilic Repression (EAR) motif. Such motifs have been detected in the Aux/IAA and jasmonate ZIM-domain (JAZ) proteins which promote target gene silencing via interactions with TOPLESS-RELATED (TPR) proteins (Waters et al. 2017).
SL-mediated regulation has also been verified in the legume–rhizobia interactions. Knockdown of CCD7 in Lotus japonicus directly reduced the number of nodules without hampering their developmental morphologies (Liu et al. 2013). The Pisum sativum mutants for CCD8 similarly produced fewer nodules compared to the wild type plants (Foo et al. 2013). It was argued that SLs act independently of the autoregulation of nodulation (AON) pathway to promote nodule formation (Foo et al. 2014). GRAS-type TFs like NODULATION SIGNALING PATHWAY 1 (NSP1) and NSP2 are required for Nod-factor-dependent nodulation in legumes. These TFs are crucial for SL biosynthesis in Medicago truncatula as well as in non-nodulating plants like rice (Liu et al. 2011).
Physiological regulation of SL biosynthesis
Regulation of the metabolite sink is a quintessential process of plant survival. It is an important strategy by which the system maintains the equilibrium of the cellular energy equivalents. SLs being crucial PGRs are evidently synthesized in a condition-dependent context only after all the regulatory gateways have been check listed.
Phytohormone-mediated regulation
Phytohormones are the essential carriers of signaling cues. They trigger stress responses via the molecular regulation of an entire cascade. We have mentioned earlier that phytohormones often interact and co-operate among themselves to co-ordinate systemic responses. SLs being novel hormone also support this notion.
Molecular interactions between SLs and ABA
ABA is considered as the universal stress phytohormone due its participation and regulation in almost all types of abiotic stresses (Banerjee and Roychoudhury 2017). Interestingly, like ABA, SLs are also apocarotenoids. Thus, SLs themselves might collectively act as stress-induced phytohormones (Lopez-Raez et al. 2010). Reduced levels of SLs were observed in the tomato plants treated with abamineSG [inhibitor of 9-cis-epoxycarotenoid dioxygenase (NCED), the rate limiting enzyme in ABA biosynthetic pathway] (Lopez-Raez et al. 2010). Similar consequences were also noted in ABA mutants like notabilis (loss-of-function of NCED), sitiens and flacca (loss-of-function of aldehyde oxidase). The tomato ABA mutants exhibited down-regulated expression of SL biosynthetic genes like LeCCD7 and LeCCD8 (Lopez-Raez et al. 2010). This directly highlights the close synchronization between ABA and SL anabolic pathways. SL-deficient mutants of Arabidopsis plants showed ABA hyposensitivity due to down-regulation of ABA import genes, ABCG22 and ABCG40 (Ha et al. 2014). The max2 mutants exhibited an entirely different expression pattern of ABA-inducible genes during drought stress (Bu et al. 2014). The gene expression profile revealed that a number of stress-responsive ABA-inducible genes like Responsive to Dehydration 29A (Rd29A), Rd29B, Cold-Regulated Protein 47 (Cor47) and KIN1 (cold inducible) were all down-regulated in the mutants. Low transcript levels of even ABA biosynthesis-, transport- and signaling-associated genes like NCED3, ABCG22, ABA Insensitive 1 (ABI1), cytochrome P450 707A3 and Hypersensitive to ABA 1 could be detected (Pandey et al. 2016, Bu et al. 2014) (Fig. 2).
The regulatory mechanisms of SL biosynthesis. The presence of cellular auxin promotes AXR1 to stabilize the SCFTIR1/AFB complex, which degrades the transcriptional repressor BDL. Interestingly, BDL and auxins both promote the accumulation of SLs. Thus, BDL acts as a transcriptional activator in case of SL biosynthesis and cellular auxin levels limit excessive accumulation of SLs. On the contrary, SLs induce AP2 to form auxin-containing clathrin coated vesicles and promote their mobilization through the PIN1 auxin transporters. This ensures root elongation and gravitropic responses. The auxin-induced accumulation of SLs also inhibits CK to release BRC1 from suppression. As a result, shoot branching is prevented. Condition-dependent SL–ABA interaction was observed. Stress stimuli induced simultaneous up-regulation of both the phytohormones in a mutually co-operative manner. However, during seed germination, SLs triggered demethylation of the CYP707A1 promoters. Rapid expression of this ABA catabolic gene reduced the cellular ABA level, thus turning the balance towards the accumulation of GAs
Mycorrhizal plants exposed to abiotic stresses accumulate high levels of ABA and SLs (Ruiz-Lozano et al. 2016). However, during mild stress, SL levels often decrease in the plant roots associated with AM fungi. This is probably because the mycorrhizal association itself confers some protection against hostile conditions. Hence, excessive SL synthesis can be prohibited to prevent excessive fungal colonization in the rhizosphere (Aroca et al. 2013; Lopez-Raez 2016). Negative correlation between ABA and SL accumulation was observed in the roots of L. japonicus where exogenous application of the SL analogue, GR24 down-regulated LjNCED2 expression and hence prevented ABA accumulation during osmotic stress (Liu et al. 2015). Similar observations showing increases in ABA levels and decreases in the levels of SLs have been reported in tomato and lettuce plants exposed to drought in the absence of mycorrhizal associations (Ruiz-Lozano et al. 2016). These results cumulatively indicate that site-specific lowering of SL levels promotes local accumulation of ABA, which is actually beneficial to tackle abiotic stress in the absence of mycorrhiza.
Molecular interactions between SLs and auxins
Auxins are crucial growth promoters which reportedly induce the expression of SL-associated biosynthetic genes like CCD7 and CCD8 in Arabidopsis, rice, pea and Chrysanthemum (Lopez-Raez 2016). Arabidopsis mutants of Auxin Resistant 1 (AXR1: encoding ubiquitin activating enzyme, E1) and Bodenlos (BDL: encoding the transcriptional repressor, INDOLEACETIC ACID RESPONSE 12) exhibited reduced expression of AtCCD7 and AtCCD8 (Hayward et al. 2009). Auxin-responsive elements were detected in the promoter sequences of AtCCD7 and AtCCD8 (Hayward et al. 2009). Thus, these cis acting elements attract auxin-induced silencers to down-regulate the expression of the respective genes. Usually, AXR1 stabilizes the S-PHASE KINASE-ASSOCIATED PROTEIN 1/CULLIN 1 (SKP1-CUL1)-E3 ubiquitin ligase complex. This complex formed of the F-box protein, TRANSPORT INHIBITOR RESPONSE 1 (TIR1) and the auxin receptor, AUXIN-RELATED F-BOX (AFB) is also termed as SCFTIR1/AFB, which directly degrades BDL (Fig. 2). Thus, in the axr1 mutants, the SCFTIR1/AFB complex is not formed and the transcriptional repressor, BDL inhibits auxin-mediated responses (Hayward et al. 2009). This establishes a direct connection between auxin and SL biosynthetic pathways. Interestingly, exogenously applied SLs suppressed the branching phenotype in the axr1, bdl and quadruple tir1/afb1/afb2/afb3 mutants of Arabidopsis (Hayward et al. 2009). From these observations, it can be inferred that SLs act downstream to the auxin signaling pathway (Fig. 2).
Auxin-induced factors transcriptionally regulate the expression of SL anabolic genes like MAX3 (CCD7), MAX4 (CCD8) and D27 in Arabidopsis, since their expression levels are substantially reduced in the decapitated and naphthylphthalamic acid-treated axr1 mutants with depleted stores of auxin (Pandey et al. 2016). It is presumed that the auxin-induced positive feedback regulation of SL biosynthesis triggers MAX3 and MAX4 expression in the Arabidopsis d27 mutants (Rasmussen et al. 2012). The transcript levels of AtCCD8 typically increased in the pro-vascular tissues of the primary roots and the cortical tissues in the elongation zone of the root apex after exogenous treatment with the synthetic auxin, 1-naphthalene acetic acid (NAA) (Rasmussen et al. 2012). A hormonal interaction has been visualized in maize roots exposed to acute N-starvation (Trevisan et al. 2015). Under control conditions, SLs promoted ADAPTOR PROTEIN 2 (AP2) to form clathrin-coated endocytic vesicles and facilitated auxin transport via PINFORMED 1 (PIN1). However, under N-deficient conditions, the genes determining SL biosynthesis and auxin transport were down-regulated in the root elongation zone, thus highlighting the synchronized co-operation between the phytohormones (Trevisan et al. 2015) (Fig. 2).
Molecular interactions among SLs, CK and auxin
The tripartite co-ordination among phytohormones has been less reported. The auxin-dependent AXR1 pathway triggers the SL biosynthesis with simultaneous suppression of the CK biosynthetic genes (Brewer et al. 2009; Dun et al. 2012). SL and CK control bud outgrowth by regulating the expression of a TF, BRANCHED 1 (BRC1). Thus, auxin promotes SL-mediated regulation of secondary growth in the aerial biomass by reducing the cellular level of CK. The TF belonging to the TEOSINTE BRANCHED-CYCLOIDEA-PCP (TCP) family inhibits shoot branching and acts downstream of the SL anabolic pathway (Braun et al. 2012). This is because the high branching phenotype was not restored in the Arabidopsis and pea brc1 mutants exogenously treated with SLs (Braun et al. 2012; Dun et al. 2012). The Arabidopsis max2 mutants exhibited down-regulated expression of the CK catabolic genes, CKX1, CKX2, CKX3 and CKX5 (Ha et al. 2014). This is another instance where negative correlation between SLs and CK can be observed (Fig. 2).
Role of SLs in epigenetics
Understanding the epigenomic landscape is crucial for visualizing the chromatin architecture and detecting its compatibility with the target gene expression (Banerjee and Roychoudhury 2018a). Epigenetic regulation of SL synthesis has been reported on the basis of DNA methylation during germination of Phelipanche ramosa seeds (Lechat et al. 2015). These require a dedicated four-day period of conditioning before sprouting. It has been observed that application of the SL analogue, GR24, markedly decreases the promoter DNA methylations of the ABA catabolic gene, PrCYP707A1 (Lechat et al. 2012, 2015). This triggers ABA breakdown and seed is released from its dormant stage. Such beneficial roles in seed germination indicate that SLs might have an unidentified interaction with the pro-germinating phytohormone, GA (Fig. 2). Treatment of the dormant P. ramosa seeds with methylating agent, hydroxyurea, hypermethylated PrCYP707A1 promoter and exhibited delayed germination. On the contrary, imbibitions of the seeds in the hypomethylating agent 5-azacytidine drastically reduced the conditioning period (Lechat et al. 2015). Further decryption of SL epigenomics is essential for better characterization of the molecular interplays of PGRs in the global context of plant systems biology (Makhzoum et al. 2017).
Post-transcriptional regulation of SL biosynthetic genes
Post-transcriptional regulation of gene expression in plants is centrally regulated by the non-coding small RNAs like small interfering RNAs (siRNAs) or microRNAs (miRNAs) (Banerjee et al. 2016). Recent reports have identified the involvement of miRNAs in the expression of SL anabolic genes (Pandey et al. 2016). It was found that a set of miRNAs (miRNA156a-g) specifically recognized MAX1, MAX2 and MAX3 in Arabidopsis; and D27, D3 and D10 in rice (Chen et al. 2015). These 21–24 nucleotide-long RNA molecules bind to the complementary target sequences to promote their degradation, resulting in post-transcriptional gene silencing (Banerjee et al. 2016). The positive feedback loop of SL biosynthesis in Arabidopsis d27 mutants, discussed above (Rasmussen et al. 2012) was also observed in the rice transgenics overexpressing osa-miRNA156e (Chen et al. 2015). Accordingly, the plants exhibited low transcript abundance of D27, though D3 and D14 transcription was maintained due to the feedback effect. The transgenic rice plants as well as maize and Arabidopsis seedlings overexpressing miRNA156 exhibited increased branching among shoots due to down-regulated SL biosynthesis (Chen et al. 2015). Thus, it appears that miRNA156 and its variants might control the tillering outgrowth phenotype, independent of the SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) regulation, especially in cereals like rice and maize (Pandey et al. 2016; Luo et al. 2012).
Roles of SLs in plant abiotic stresses
Abiotic stresses like salinity, drought, extremes of temperatures, nutrient unavailability, etc. cumulatively pose as a serious threat to the global food security. Such sub-optimal conditions are directly responsible for worldwide crop losses. Thus, to feed the ever-growing population, it is necessary to design novel strategies to improve crop production under hostile environments (Banerjee and Roychoudhury 2017). Due to their strong correlation with ABA (Roychoudhury and Banerjee 2017), it has been observed that under many situations, SLs can confer abiotic stress tolerance across plant species.
Roles of SLs during salinity and drought
Terrestrial plants often overcome the hazardous effects of salt stress by establishing symbiotic associations with soil AM fungi. The symbiosis facilitates the accumulation of compatible solutes, equilibration of ion uptake via roots and water import through aquaporins and compartmentalization of excess ions. This effectively maintains the cellular osmoticum and the physiological processes essential for sustaining plant growth (Ruiz-Lozano et al. 2012). In symbiotic lettuce plants, it was observed that the rhizospheric AM fungi became more abundant in response to the salinity-induced SL exudation from the roots (Aroca et al. 2013). The drought-tolerant phenotype in Trifolium alexandrinum (berseem clover) was also due to AM symbiosis in the roots (Saia et al. 2014). SLs play roles in salinity and drought responses in Arabidopsis as the max3 and max4 mutants exhibited stress sensitivity with high stomatal density and delayed ABA-induced stomatal closure (Saeed et al. 2017).
In silico analysis of the promoter sequences of SL biosynthetic genes in Arabidopsis identified the cis acting motifs, ACGTATERD1 and MYBIAT, which specifically bind to drought-responsive TFs (Marzec and Muszynska 2015). An interesting study using complementary approach established the evolutionary conservation of MAX2 activity between parasitic and non-parasitic plants (Li et al. 2016). Heterologous expression of OaMAX2 from the parasitic plant, Orobanche aegyptiaca in Arabidopsis loss-of-function mutants of AtMAX2, recovered the transgenics exposed to drought (Li et al. 2016). L. japonicus plants with depleted SL stores exhibited delayed ABA-dependent stomatal closure, associated with drought (Liu et al. 2015). In another study, the implications of AtMAX2 in drought and salt stress were characterized during seed germination (Bu et al. 2014). Expression of this gene was induced by the ABA-inducible TFs, ABI3 and ABI5 during germination and seedling development. However, its expression is slightly suppressed by ABA in the adult stages (Bu et al. 2014). Hence, the ABA signaling cascade possibly acts upstream of AtMAX2. The Arabidopsis max2 mutants exhibited degenerated cuticle and wider stomatal apertures when exposed to drought stress (Bu et al. 2014).
Islam et al. (2013) demonstrated the positive effects of SLs in enhancing plant survival under multiple stresses. The Arabidopsis plants overexpressing the group 1 glycosyltransferase, SDG8i from the resurrection grass, Sporobolus stapfianus exhibited elevated levels of auxin. The enzyme acted downstream of ABA to minimize drought-induced senescence and even glycosylated exogenously applied GR24. This creates a direct link between the SDG8i and SL signaling pathways. The treated transgenic plants survived drought, salt and cold stresses much efficiently and also showed significantly increased yields under control conditions (Islam et al. 2013).
Roles of SLs during temperature stress
In our previous section, we had mentioned that possibly due to similar biochemical origins, ABA stimulates the accumulation of SLs. However, SL in turn has been found to lower ABA to counterbalance the cellular GA level for promoting germination in seeds (Mishra et al. 2017). It is possibly this mechanistic regulation by which SL acts as a germination stimulant in the seeds of parasitic weeds and other plants. The increased GA content triggers high accumulation of CK which activates the metabolome and accelerates germination in the later stages. Such SL-mediated regulation of germination has also been shown during temperature stress in the SL-defective Arabidopsis mutants (Tsuchiya et al. 2010). Exogenous application of GR24 enabled the seeds of these mutant plants to germinate even at high temperatures by down-regulating the ABA biosynthetic gene, NCED9 (Tsuchiya et al. 2010). The temperature-induced dormancy in P. ramosa seeds during warm stratification is reversed by the application of SLs. As a result, the seeds can germinate even after heat stress (Lechat et al. 2015).
The regulatory interplay between SL synthesis and light
Light is the ubiquitous environmental factor which unlike other stresses cannot be evaded. The fluctuations and periodicity of light quality and intensity dictates the variable growth pattern among plants (Banerjee and Roychoudhury 2016). These parameters also crucially regulate the level of SLs in plants, indirectly justifying the necessity of SLs in light signaling cascades. The Arabidopsis SL-deficient mutants exhibited down-regulated expression of the light-inducible genes. These were up-regulated when the plants were exposed to exogenous GR24 (Mashiguchi et al. 2009). Similarly, treatment of SL-deficient tomato mutants with GR24 restored the expression of genes associated with the light harvesting complex (LHC). These genes were down-regulated in the mutants (Mayzlish-Gati et al. 2012). ELONGATED HYPOCOTYL 5 (HY5) regulating light adapted seedling development in Arabidopsis is controlled by the ubiquitin ligase, CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1). It was observed that the nuclear localization of COP1 is controlled by SLs (Tsuchiya et al. 2010). COP1 also promotes the nuclear accumulation of UV RESISTANCE LOCUS 8 (UVR8), essential for UV-B acclimation and tolerance in Arabidopsis (Yin et al. 2016). This hypothesizes a potential role of SLs in mediating UV-B tolerance in plants. This aspect, however, requires more thorough investigations.
Light intensity directly affects SL-mediated phenotypic regulations. Arabidopsis plants exposed to low light intensity under crowded conditions had elongated leaves with long and slender stems. However, the max1 and max2 mutants grown under similar conditions were stunted and contained round leaves (Stirnberg et al. 2002). The max2 mutants exhibited differential germination pattern and photomorphogenesis (Tsuchiya et al. 2010). In another experiment, increasing light intensity up-regulated CCD7 expression in tomato plants (Koltai et al. 2011). Compared to wild-type plants, adventitious root production in the pea ccd7 and ccd8 mutants drastically reduced under dark conditions, but was equivalent when grown under lighted conditions (Tsuchiya et al. 2010).
SLs have been depicted to play roles in the phytochrome (PHY) signaling pathway. The Arabidopsis double mutants, phy B/max2 or phy B/max4 grown under high intensity, red light did not exhibit the phy B phenotype of reduced branching. Instead, the mutants were highly branched similar to the max2 and max4 single mutants (Finlayson et al. 2010). Thus, SLs must be acting downstream of the PHY B signaling pathway, and so simultaneous phy B and SL deficiency results in expression of only the SL-deficient phenotype. The lowering of the red light intensity, however, relieved the suppression of the phy B phenotype (Finlayson et al. 2010). This evidently shows the light intensity-dependent action of SLs downstream of PHY B signaling.
Roles of SLs during nutrient starvation
Inorganic nutrient deficiency in plants is mainly due to the dearth of soil phosphates and nitrates, the respective sources of inorganic phosphate (Pi) and nitrogen (N) (Mishra et al. 2017). SLs possibly act as messenger molecules during nutrient stress as Pi- and N-deficiencies induced their accumulation in roots and root exudates in legumes and also other plants (Sun et al. 2014). SLs promoted the surface motility of rhizobia like Sinorhizobium meliloti in the rhizosphere of M. truncatula (Pelaez-Vico et al. 2016). After aiding in the symbiotic establishment, SL levels declined to prevent excessive nodulation. In the same symbiotic model, SL biosynthesis was triggered by Pi-starvation, whereas N-deficiency abolished the positive correlation (Pelaez-Vico et al. 2016).
Accumulation of SLs was reported in Arabidopsis plants exposed to Pi-starvation. Suppression of signature genes like 5-acid phosphatase, phosphate transporter 1;5 (PHT1;5) and PHT1;4 expressed during Pi limitation, was observed in the max2 and max4 mutants (Mayzlish-Gati et al. 2012). Thus, SLs are positive regulators of Pi-starvation which accelerate Pi mobility and uptake during acute deficiency. The stressed wild-type plants exhibited inhibition of lateral bud outgrowth (Mayzlish-Gati et al. 2012). Altered anatomical architecture was also observed in rice plants exposed to Pi-starvation, where increased activity of DS led to the inhibition of tiller bud outgrowths (Umehara et al. 2010). Analysis of d3 and d10 rice mutants under similar conditions revealed no such inhibition and justified that such anatomical manipulations are governed by SLs in absence of Pi (Umehara et al. 2010). Up-regulated expression of D3, D10 and D27 was reported in wild type rice plants growing in the absence of both N and Pi (Sun et al. 2014). The root responsiveness to N- and Pi-stress was lost in the d3, d10 and d27 rice mutants where unlike wild type plants, no increase in the root length could be observed (Sun et al. 2014). Thus, it can be inferred that SLs effectively regulate the gravitropic responses in root tissues in response to nutrient stress. The SL-mediated localization of PIN proteins dictates auxin transport (Trevisan et al. 2015), and this regulation possibly controls the root responses during nutrient deficient conditions (Shinohara et al. 2013). Similar inhibition in root length and primary root branching was observed in Arabidopsis SL biosynthetic and signaling mutants grown in the absence of Pi. Exogenous application of GR24 rescued the Pi deficiency phenotypes in all genotypes except in the SL signaling mutants (Pandey et al. 2016). Therefore, it can be summarized that the SL biosynthesis dictates cellular auxin polarity to promote root elongation for the search of limiting nutrients. This illustrates an amazing aspect of plant systemic intelligence.
Roles of SLs during oxidative stress
Abiotic stresses like salinity, drought, high or low temperature, light or nutrient deprivation trigger systemic damages via the accumulation of the toxic reactive oxygen species (ROS) like hydrogen peroxide (H2O2), hydroxyl (OH−), superoxide (O ·−2 ) radicals, etc. Such oxidative stress is the central disruptor of all essential physiological processes like photosynthesis, signaling, transport, reproduction and plant survival. ROS triggers the degradation of protein and nucleic acids and accelerates lipid peroxidation in the membranes (Foyer and Noctor 2005). ROS also act as secondary messengers which co-ordinate with phytohormone-mediated signaling to induce stress-responsive gene expression, thereby promoting stress acclimation (Banerjee and Roychoudhury 2018b).
The transposase-related TF, FAR-RED ELONGATED HYPOCOTYL 3 (FHY3) is a crucial component in the PHYA signaling of the circadian rhythm (Lin et al. 2007). FHY3 regulates far-red (FR) light response and also suppresses ROS production by inhibiting the NADPH oxidase, RESPIRATORY BURST OXIDASE HOMOLOG (RBOH) (Lin et al. 2007). An interesting SL-mediated regulation has been identified at this juncture as FHY3 was reported to inhibit the expression of MAX2 in Arabidopsis (Ouyang et al. 2011). The fhy3/max2 double mutants exhibited high expression of RBOH genes, which might be responsible for the inhibited branching phenotype (Ouyang et al. 2011). The RBOH-RNAi lines in tomato plants showed increased shoot branching, which proves that RBOHs regulate the branching phenotype in plants (Koltai et al. 2011). Recently stress acclimatization by SL-mediated stomatal closure was physiologically evaluated (Lv et al. 2017). The SL-associated genetic mutants exhibited large stomatal apertures which could be recovered by exogenous treatment of SLs. Interestingly, the SL-mediated stomatal closure during oxidative stress acted in an ABA-independent fashion. However, mutation in genes like D14, MAX2 and the anion channel, Slow Anion Channel-Associated 1 (SLAC1) completely disrupted SL-mediated stomatal closure. The SL accumulation in the guard cells stimulated the H2O2 and nitric oxide (NO) contents, which in turn promoted stomatal closure (Lv et al. 2017). Further molecular evaluation of such ABA-independent SL-mediated stomatal regulation could reveal novel strategies for tackling oxidative stress.
Nutrient deprivation also promotes the accumulation of toxic ROS (Banerjee and Roychoudhury 2018b). P- and N-limiting conditions activate NADPH oxidases in Medicago truncatula roots and these induce high expression of the SL biosynthetic genes (Bonneau et al. 2013). Such condition-specific accumulation of SLs indicates that these molecules might themselves have the capacity to scavenge ROS or SLs might promote the accumulation of downstream osmolytes and antioxidants to maintain the cellular osmotica.
Conclusion and future perspectives
SLs are a recent addition to the novel list of non-traditional PGRs. These endogenous metabolites are lactone-containing carotenoid derivatives sharing the same origin as the universal stress hormone, ABA. Thus, by virtue of its biosynthetic origin, SLs play diverse roles in plant growth, development and establishment of symbiotic associations. SL biosynthesis is regulated by a number of mutually interactive check-points. Thus, these metabolites have evolved as critical inter-communicative messengers of a physiologically vast signaling network. SLs act downstream of ABA, auxin as well as PHY B signaling pathways. It is possible that by accomplishing such a downstream location, SLs control the quality of several systemic response pathways. SLs might also act as molecular valves to regulate the extent of signaling and determine the level of their effects in plants. The epigenetic and post-transcriptional regulations highlight the condition-dependent inducible status of the SL biosynthetic genes, which respond to various stress stimuli. The escalating importance of SLs is due to their immense potential in mediating multiple abiotic stress tolerance in plants. In view of their interactions with regulatory growth phytohormones, SLs modify the physiological and anatomical architectures to promote plant survival even under sub-optimal conditions like salinity, drought, light stress, nutrient deficiency and oxidative stress.
Researches depicting the roles of SLs in plant abiotic stress biology require exhaustive dimensions. The dynamics of SL signaling in abiotic stress is yet to be elucidated. It would be interesting to detect how SLs interact with the ABA signaling pathway and sense the cellular ABA content, which lead to stress-dependent simultaneous up-regulation of the genes associated with both the phytohormones. The previous sections showing the communications between SLs and multiple PGRs have illustrated the essence of plant intelligence in physiological signaling. Hence, it is absolutely necessary to experimentally develop a signaling blueprint characterizing the interactome of the phytohormones. Understanding the global status of the epigenomic landscape in SL-biosynthetic genes is a crucial aspect of developing stress tolerance in plants. Little information on the small RNA regulation during SL biosynthesis is available. Therefore, investigations encompassing small RNAome interactions with the target genes should be performed. Genome-wide association studies followed by stringent mapping of putative SL-responsive gene loci can lead to the identification of novel molecular targets. Transgenic approaches can then be envisaged to overexpress these SL-anabolic genes in plants to generate multiple stress tolerance.
Author contribution statement
AB drafted the entire manuscript following extensive review on the topic. AR critically analyzed the manuscript, made suitable modifications and supervised the overall work.
References
Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P et al (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335:1348–1351
Aroca R, Ruiz-Lozano JM, Zamarreno AM, Paz JA, Garcia-Mina JM et al (2013) Arbuscular mycorrhizal symbiosis influences strigolactone production under salinity and alleviates salt stress in lettuce plants. J Plant Physiol 170:47–55
Banerjee A, Roychoudhury A (2016) Plant responses to light stress: oxidative damages, photoprotection and role of phytohormones. In: Ahammed GJ, Yu J-Q (eds) Plant hormones under challenging environmental factors. Springer, Dordrecht, pp 181–213
Banerjee A, Roychoudhury A (2017) Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254:3–16
Banerjee A, Roychoudhury A (2018a) The gymnastics of epigenomics in rice. Plant Cell Rep 37:25–49
Banerjee A, Roychoudhury A (2018b) Abiotic stress, generation of reactive oxygen species, and their consequences: an overview. In: Singh VP, Singh S, Tripathi D, Mohan Prasad S, Chauhan DK (eds) Revisiting the role of reactive oxygen species (ROS) in plants: ROS Boon or bane for plants?. Wiley, pp 23–50
Banerjee A, Roychoudhury A, Krishnamoorthi S (2016) Emerging techniques to decipher microRNAs (miRNAs) and their regulatory role in conferring abiotic stress tolerance in plants. Plant Biotechnol Rep 10:185–205
Banerjee A, Wani SH, Roychoudhury A (2017) Epigenetic control of plant cold responses. Front Plant Sci 8:1643
Beveridge CA, Kyozuka J (2010) New genes in the strigolactone-related shoot branching pathway. Curr Opin Plant Biol 13:34–39
Bonneau L, Huguet S, Wipf D, Pauly N, Truong HN (2013) Combined phosphate and nitrogen limitation generates a nutrient stress transcriptome favourable for arbuscular mycorrhizal symbiosis in Medicago truncatula. New Phytol 199:188–202
Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P et al (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell 8:443–449
Boyer FD, de Saint Germain A, Pillot JP, Pouvreau JB, Chen VX, Ramos S et al (2012) Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: molecule design for shoot branching. Plant Physiol 159:1524–1544
Braun N, de Saint Germain A, Pillot JP, Boutet-Mercey S, Dalmais M, Antoniadi I et al (2012) The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol 158:225–238
Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA (2009) Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150:482–493
Bu Q, Lv T, Shen H, Luong P, Wang J, Wang Z et al (2014) Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol 164:424–439
Cardoso C, Zhang Y, Jamil M, Hepworth J, Charnikhova T, Dimkpa SO et al (2014) Natural variation of rice strigolactone biosynthesis is associated with the deletion of two MAX1orthologs. Proc Natl Acad Sci USA 111:2379–2384
Chen Z, Gao X, Zhang J (2015) Alteration of osa-miR156e expression affects rice plant architecture and strigolactones (SLs) pathway. Plant Cell Rep 34:767–871
Cheng X, Ruyter-Spira C, Bouwmeester H (2013) The interaction between strigolactones and other plant hormones in the regulation of plant development. Front Plant Sci 4:199
Cook CE, Whichard LP, Wall ME, Egley GH, Coggon P et al (1972) Germination stimulants. II. Structure of strigol, a potent seed germination stimulant for witch weed (Striga lutea). J Am Chem Soc 94:6198–6199
Dun EA, de Saint Germain A, Rameau C, Beveridge CA (2012) Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158:487–498
Finlayson SA, Krishnareddy SR, Kebrom TH, Casal JJ (2010) Phytochrome regulation of branching in Arabidopsis. Plant Physiol 152:1914–1927
Foo E, Yoneyama K, Hugill CJ, Quittenden LJ, Reid JB (2013) Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol Plant 6:76–87
Foo E, Ferguson BJ, Reid JB (2014) The potential roles of strigolactones and brassinosteroids in the autoregulation of nodulation pathway. Ann Bot 113:1037–1045
Foyer CH, Noctor G (2005) Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ 28:1056–1071
Ha CV, Leyva-Gonzalez MA, Osakabe Y, Tran UT, Nishiyama R et al (2014) Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc Natl Acad Sci USA 111:851–856
Hayward A, Stirnberg P, Beveridge C, Leyser O (2009) Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151:400–412
Islam S, Griffiths CA, Blomstedt CK, Le T-N, Gaff DF et al (2013) Increased biomass, seed yield and stress tolerance is conferred in Arabidopsis by a novel enzyme from the resurrection grass Sporobolus stapfianus that glycosylates the strigolactone analogue GR24. PLoS One 8:e80035
Koltai H (2011) Strigolactones are regulators of root development. New Phytol 190:545–549
Koltai H, Cohen M, Chesin O, Mayzlish-Gati E, Bécard G, Puech V et al (2011) Light is a positive regulator of strigolactone levels in tomato roots. J Plant Physiol 168:1993–1996
Lechat MM, Pouvreau JB, Péron T, Gauthier M, Montiel G et al (2012) PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. J Exp Bot 63:5311–5322
Lechat MM, Brun G, Montiel G, Véronési C, Simier P, Thoiron S et al (2015) Seed response to strigolactone is controlled by abscisic acid-independent DNA methylation in obligate root parasitic plant, Phelipanche ramosa L. Pomel. J Exp Bot 66:3129–3140
Leyser O (2009) The control of shoot branching: an example of plant information processing. Plant Cell Environ 32:694–703
Li W, Nguyen KH, Watanabe Y, Yamaguchi S, Tran LS (2016) OaMAX2 of Orobanche aegyptiaca and Arabidopsis AtMAX2 share conserved functions in both development and drought responses. Biochem Biophys Res Commun 478:521–526
Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318:1302–1305
Liu J, Maldonado-Mendoza I, Lepez-Meyer M, Cheung F, Town CD, Harrison MJ (2007) Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J 50:529–544
Liu W, Kohlen W, Lillo A, den Camp RO, Ivanov S et al (2011) Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23:3853–3865
Liu J, Novero M, Charnikhova T, Ferrandino A, Schubert A et al (2013) CAROTENOID CLEAVAGE DIOXYGENASE 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model Lotus japonicus. J Exp Bot 64:1967–1981
Liu J, He H, Vitali M, Visentin I, Charnikhova T et al (2015) Osmotic stress represses strigolactone biosynthesis in Lotus japonicus roots: exploring the interaction between strigolactones and ABA under abiotic stress. Planta 241:1435–1451
Lopez-Raez JA (2016) How drought and salinity affect arbuscular mycorrhizal symbiosis and strigolactone biosynthesis? Planta 243:1375–1385
Lopez-Raez JA, Kohlen W, Charnikhova T, Mulder P, Undas AK et al (2010) Does abscisic acid affect strigolactone biosynthesis? New Phytol 187:343–354
Luo L, Li W, Miura K, Ashikari M, Kyozuka J (2012) Control of tiller growth of rice by OsSPL14 and strigolactones, which work in two independent pathways. Plant Cell Physiol 53:1793–1801
Lv S, Zhang Y, Li C, Liu Z, Yang N et al (2017) Strigolactone-triggered stomatal closure requires hydrogen peroxide synthesis and nitric oxide production in an abscisic acid-independent manner. New Phytol. https://doi.org/10.1111/nph.14813
Makhzoum A, Yousefzadi M, Malik S, Gantet P, Tremouillaux-Guiller J (2017) Strigolactone biology: genes, functional genomics, epigenetics and applications. Crit Rev Biotechnol 37:151–162
Marzec M, Muszynska A (2015) In silico analysis of the genes encoding proteins that are involved in the biosynthesis of the RMS/MAX/D pathway revealed new roles of strigolactones in plants. Int J Mol Sci 16:6757–6782
Mashiguchi K, Sasaki E, Shimada Y, Nagae M, Ueno K, Nakano M et al (2009) Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci Biotechnol Biochem 73:2460–2465
Matusova R, Rani K, Verstappen FW, Franssen MC, Beale MH, Bouwmeester HJ (2005) The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol 139:920–934
Mayzlish-Gati E, De-Cuyper C, Goormachtig S, Beeckman T, Vuylsteke M, Brewer PB et al (2012) Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiol 160:1329–1341
Mishra S, Upadhyay S, Shukla RK (2017) The role of strigolactones and their potential cross-talk under hostile ecological conditions in plants. Front Physiol 7:691
Ouyang X, Li J, Li G, Li B, Chen B, Shen H et al (2011) Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL 3 reveals its novel function in Arabidopsis development. Plant Cell 23:2514–2535
Pandey A, Sharma M, Pandey GK (2016) Emerging roles of strigolactones in plant responses to stress and development. Front Plant Sci 7:434
Pelaez-Vico MA, Bernabeu-Roda L, Kohlen W, Soto MJ, Lopez-Raez JA (2016) Strigolactones in the Rhizobium-legume symbiosis: stimulatory effect on bacterial surface motility and down-regulation of their levels in nodulated plants. Plant Sci 245:119–127
Rasmussen A, Mason MG, De Cuyper C, Brewer PB, Herold S, Agusti J et al (2012) Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol 158:1976–1987
Roychoudhury A, Banerjee A (2017) Abscisic acid signaling and involvement of mitogen activated protein kinases and calcium-dependent protein kinases during plant abiotic stress. In: Pandey G (ed) Mechanism of plant hormone signaling under stress, vol 1. Wiley, pp 197–241
Ruiz-Lozano JM, Porcel R, Azcon C, Aroca R (2012) Regulation of arbuscular mycorrhizae of the integrated physiological response to salinity in plants: new challenges in physiological and molecular studies. J Exp Bot 11:4033–4044
Ruiz-Lozano JM, Aroca R, Zamarreno AM, Molina S, Andreo-Jimenez B et al (2016) Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Environ 39:441–452
Saeed W, Naseem S, Ali Z (2017) Strigolactones biosynthesis and their role in abiotic stress resilience in plants: a critical review. Front Plant Sci 8:1487
Saia S, Amato G, Frenda AS, Giambalvo D, Ruisi P (2014) Influence of arbuscular mycorrhizae on biomass production and nitrogen fixation of berseem clover plants subjected to water stress. PLoS One 9:e90738
Shinohara N, Taylor C, Leyser O (2013) Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 1:e1001474
Smith SM, Li J (2014) Signalling and responses to strigolactones and karrikins. Curr Opin Plant Biol 21:23–29
Stirnberg P, van de Sande K, Leyser HMO (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129:1131–1141
Sun H, Tao J, Liu S, Huang S, Chen S, Xie X et al (2014) Strigolactones are involved in phosphate-and nitrate-deficiency-induced root development and auxin transport in rice. J Exp Bot 65:6735–6746
Trevisan S, Manoli A, Ravazzolo L, Botton A, Pivato M, Quaggiotti S (2015) Nitrate sensing by the maize root apex transition zone: a merged transcriptomic and proteomic survey. J Exp Bot 66:3699–3715
Tsuchiya Y, Vidaurre D, Toh S, Hanada A, Nambara E, Kamiya Y et al (2010) A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat Chem Biol 6:741–749
Umehara M, Hanada A, Magome H, Takeda-Kamiya N, Yamaguchi S (2010) Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol 51:1118–1126
Vogel JT, Walter MH, Giavalisco P, Lytovchenko A, Kohlen W, Charnikhova T et al (2010) SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J 61:300–311
Wang YH, Irving HR (2011) Developing a model of plant hormone interactions. Plant Signal Behav 6:494–520
Waters MT, Gutjahr C, Bennett T, Nelson DC (2017) Strigolactone signaling and evolution. Annu Rev Plant Biol 68:291–322
Xie X, Yoneyama K, Yoneyama K (2010) The strigolactone story. Ann Rev Phytopathol 48:93–117
Yamada Y, Furusawa S, Nagasaka S, Shimomura K, Yamaguchi S, Umehara M (2014) Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240:399–408
Yin R, Skvortsova MY, Loubery S, Ulm R (2016) COP1 is required for UV-B-induced nuclear accumulation of the UVR8 photoreceptor. Proc Natl Acad Sci USA 113:E4415–E4422
Acknowledgements
Financial assistance from Council of Scientific and Industrial Research (CSIR), Government of India, through the research Grant [38(1387)/14/EMR-II] to Dr. Aryadeep Roychoudhury is gratefully acknowledged. The authors are also thankful to University Grants Commission, Government of India, for providing fellowship to Mr. Aditya Banerjee.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Huang.
Rights and permissions
About this article
Cite this article
Banerjee, A., Roychoudhury, A. Strigolactones: multi-level regulation of biosynthesis and diverse responses in plant abiotic stresses. Acta Physiol Plant 40, 86 (2018). https://doi.org/10.1007/s11738-018-2660-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-018-2660-5