Abstract
Plants being mostly sessile are exposed to several adverse environmental conditions. Many endogenous and exogenous factors play a vital role in acclimatizing plants in such varying environments. Plant growth regulators (PGRs) are one such endogenous factor that regulates the phenomenon of growth and development in plants. Strigolactone (SL) has been accepted as a new class of phytohormones or PGRs. It has contributed to different aspects of plant growth and development such as root growth and shoot branching as well as the response of plants to several biotic and abiotic stresses. Lately, a deep understanding of the SL biosynthetic pathway has been revealed. Transcriptomics and genetic analysis showed that SLs are derived from an intermediate carotenoid biosynthesis pathway, all-trans-β-carotene. Carlactone (CL) is formed from all-trans-β-carotene by the subsequent action of a set of core enzymes DWARF27 (D27), and carotenoid cleavage dioxygenases (CCD7 and CCD8). CL is the ultimate biosynthetic precursor of all naturally occurring SLs. The investigation has been also carried out on signal perception and downstream cascade involved in SL signaling by utilizing various mutants from different plant species. D14, AtD14, and DAD2 are identified as orthologous SL receptors of Oryza sativa, Arabidopsis thaliana, and Petunia, respectively. These are identified as αβ hydrolase, having the activity of both receptor and enzyme. The present review summarizes the current perception of the nature and biosynthesis of SL and the deciphering of the mechanism involved in its signal transduction cascade.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants naturally produce chemicals called plant growth regulators (PGRs). They control growth and other functions in different parts of the plant, even far from where they are made. They work in very small amounts and are important for influencing the growth, yield, and quality of crops (Desta and Amare 2021). Strigolactone (SL) is a butenolide PGR (Yang et al. 2019). It is an evolutionary old plant signaling molecule, having multiple roles in organisms such as AMF (arbuscular mycorrhizal fungi), bryophytes, and angiosperms (Lopez-Obando et al. 2015). The SLs were brought to light eight years after the explication of auxin structure, the first phytohormone (Bouwmeester et al. 2019). The SLs were discovered almost 50 years ago originally as a stimulant in root exudate that induces germination in several parasitic plant species (Brewer et al. 2013; Wani et al. 2020). The term ‘strigolactone’ is derived from its role as a stimulant of ‘Striga’ seed germination and its chemical structure containing a ‘lactone’ ring (Lopez-Obando et al. 2015; Wani et al. 2020). The name ‘strigolactone’ is proposed by Butler, a research protagonist in 1995 (Faizan et al. 2022).
Structurally, SLs are categorized into canonical and non-canonical types (Table 1). Canonical SLs comprise four ring structure in which the ABC-ring is fused with the D-ring by an enol-ether bond. Non-canonical SLs lack basic ABC-ring but D strigol-ring with enol bridge is strictly present (Guercio et al. 2023). The C- and D-ring must be a lactone and a methyl furanone, respectively, while AB-ring shows variation (Zorrilla et al. 2022). As a novel PGR, SL administers various aspects of growth and development in plants. It generally inhibits shoot outgrowth. Additionally, as a signaling molecule from root exudates, it also acts as a communication signal between plants and microorganisms in the rhizosphere (Yoneyama and Brewer 2021; Wang et al. 2022). Earlier, SL was considered baleful because of its role in stimulating germination in parasitic weeds; however, it was later regarded as useful because of its role in providing a defensive response against pathogens (Wani et al. 2021), AMF colonization, alleviation of abiotic stresses in the plant (Bouwmeester et al. 2019), as well as regulation of root and flower development and senescence of leaf (Zhou et al. 2013).
To discover more functional aspects of SLs, detailed knowledge of their biosynthesis, as well as signaling cascade in plants, is required. The biosynthetic study was initiated when root exudates from carotenoid biosynthesis mutant of maize and fluoridone treated wild-type seedling of maize reduced the Striga germination. From these findings, it has been concluded that SLs are derived from a carotenoid (Ruyter-Spira et al. 2013). Mutants of several plant species (Arabidopsis, pea, rice, and Petunia) have been investigated to elucidate the key steps and various enzymes involved in SL biosynthesis (Jia et al. 2018). The main biosynthetic steps include reversible isomerization, cleavage, and oxidation. The key enzymes involved are D27, CCD7, and CCD8, the activity of which forms a core pathway, leading to the formation of carlactone (CL). CL is the key intermediate precursor of all SL. Further modification of CL to form structurally diverse SLs is accomplished through other enzymes, MAX1, carlactone oxidase (CO), orobanchol synthase (OS), and Lateral Branching Oxidoreductase or LBO (Zhang et al. 2014; Brewer et al. 2016; Haider et al. 2023).
The signaling mechanism of SL is similar to auxin, gibberellin, and jasmonic acid in which proteolytic cleavage occurs due to activation by phytohormones. They require SCF (Skp1-Cullin-F-box) E3 ubiquitin ligase complex, an F-box protein component, which specifically targets proteins, ubiquitinylates them, and subsequently causes their degradation. However, signaling in each case differs in the type of protein it targets. In the case of SL signaling, it’s binding to its receptor subsequently causes the ubiquitinylation and degradation of D53/SMXL by the 26S proteasomal complex (Waters et al. 2017). The present review focuses on different steps involved in the biosynthesis of natural SLs, involving various enzymes and their genes identified in different plant species will be discussed. The following review will also discuss the current findings related to SL signal perception and the signaling cascade involved in regulating SL-responsive genes.
Strigolactone: Nature and Biosynthesis
Nature of Strigolactones
The first identified SL (1966) was ‘strigol’ from Gossypium hirsutum L. (cotton) root exudate, which triggered the germination in the parasitic witchweed Striga lutea Lour., a root parasite, belonging to the plant family Orobanchaceae (Brun et al. 2018; Bouwmeester et al. 2019; Faizan et al. 2020; Wani et al. 2020). Since the discovery of the first SL, more than 30 SLs from various plant species have been reported (Yoneyama and Brewer 2021). SLs were discovered formerly as stimulants of seed germination in parasitic plant species but are now considered PGRs (Xie 2016). The absolute stereochemistry of the first naturally occurring SL ‘strigol’ was done by X-ray diffraction analysis (Zwanenburg and Pospíšil. 2013; Faizan et al. 2022). The basic structure of naturally occurring SLs consists of a 4-ring system, i.e., tricyclic lactone system (ABC-ring) and a butenolide part (D-ring or a methyl butenolide ring) which are joined via an enol-ether linkage (Seto et al. 2012; Nakamura et al. 2013; Rani et al. 2023). The AB-rings are variant because of the presence of double bonds and various functional groups. The A-ring also has a varied number of C-atoms (Zorilla et al. 2022). The D-ring is invariable α, β-unsaturated furanone moiety and is important for SLs activity (Lopez-Obando et al. 2015; Mashiguchi et al. 2021). Although SLs have a common structural pattern, however, structural diversity is striking and the main reason behind this may be symbiosis (Kramna et al. 2019). Based on structural chemistry, naturally occurring SLs are categorized as canonical (classical) and non-canonical SLs (Fig. 1) (Guercio et al. 2023; Mashiguchi et al. 2021).
Chemical structures of some naturally occurring Strigolactones (SLs) (canonical and non-canonical), and GR24, a synthetic SL. Canonical SLs are characterized by the presence of ABC-ring system. Non-canonical SLs include an enol-ether connected to a methylbutenolide D-ring, which is vital for their biological activity
Canonical Strigolactones
Canonical SLs comprise tricyclic lactone rings (ABC- rings) and a butenolide ring (D- ring), linked via 2’R configured enol-ether bridge. Depending upon the stereochemistry of the C-ring, canonical SLs are further subdivided into strigol-type and orobanchol-type. The strigol-type SLs have β-oriented C-ring and are derived from 5-deoxystrigol (5-DS) while orobanchol-type has α-oriented C-ring and are derived from 4-deoxyorobanchol (4-DO) (Mashiguchi et al. 2021; Guercio et al. 2023). 5-DS and 4-DO were detected in the root exudates of the burley tobacco cultivar – Michinoku No. 1 and the bright yellow tobacco cultivar- Tsukuba No. 1 (Xie 2016). Further derivation of SLs is possible by methylation, acetylation, hydroxylation, epoxidation, and ketolation of AB-ring (Mashiguchi et al. 2021; Guercio et al. 2023). The canonical SLs includes (Table 1) strigol, strigyl acetate, strigone, sorgolactone, alectrol, orobanchol, 5-DS, 2-epi-orobanchol, orobanchyl acetate, 7-oxo-orobanchol, 7-oxo-orobanchyl acetate, hydroxy-orobanchyl acetate, sorgomol, fabacyl acetate, solanacol, etc. (Zwanenburg and Pospíšil 2013).
Non-canonical Strigolactones
Non-canonical SLs are comparatively simple in structure as they lack a characteristic ABC-ring system but contain 2’R configured enol-ether bridge and D-ring moiety. Examples of non-canonical SLs are CL, methyl carlactonate, methyl heliolactonate, avenaol, and methyl zealactonate (zealactone) (Table 1) (Mashiguchi et al. 2021; Guercio et al. 2023). Despite the fact that both canonical and non-canonical SLs are chemically unstable, canonical SLs are comparatively more stable (Yoneyama et al. 2018). Although there is a structural variation among canonical and non-canonical SLs, both show similar bioactivities such as stimulation of seed germination in parasitic plant species and this is due to the presence of D-ring in both (Zorrilla et al. 2022).
Due to the diverse biological role of SL from growth and development to symbiosis, this hormone is at the frontline in plant science research (Seto et al. 2012). For a detailed investigation of SLs activity as well as their application in the agricultural field, their availability in enough amount is important (Kountche et al. 2018). Plants produce SL in very small quantities, i.e., pg-ng/g root fresh weight (Yoneyama et al. 2021). However, the synthesis of natural SLs is challenging because of their complicated structure and their secretion in very minute quantities. Thus, synthetic analogs of SLs are lately developed namely, GR24 (Fig. 1) and methyl phenlactonoates (MP), and are widely used in research (Kountche et al. 2018). The GR24 is a great tool for investigating SL’s role in plant physiology (Wani et al. 2020).
Strigolactones: Biosynthetic Pathway in Plants
SLs are basically sesquiterpene lactones and their chemical structure is somewhat related to isoprenoids/ terpenoids (Souri et al. 2020). However, the results from root exudate analysis of carotenoid biosynthetic mutants of maize in which the efficiency to induce germination in Striga is reduced, and the results from the use of fluoridone, an inhibitor in carotenoid biosynthetic pathway, suggest that SLs are derivatives of carotenoid-dependent pathway (Tsuchiya and McCourt 2009; Ruyter-Spira et al. 2013; Jia et al. 2018; Wani et al. 2020). Modification of A and B rings through oxidation, ketolation, epoxidation, and hydroxylation led to structural diversity among SLs. Many biosynthetic genes are involved leading to this structural diversity (Wani et al. 2021). Several mutants such as max (more axillary growth) in A. thaliana, rms (ramosus) in P. sativum L., htd (high tillering dwarf) in O. sativa L., and dad (decreased apical dominance) in P. hybrida were examined for the identification of key genes involved in SL biosynthesis. The biosynthetic genes identified are MAX1, MAX3, MAX4, and LBO in A. thaliana, RMS1 and RMS5 in P. sativum, D10, D17(HTD1), and D27 in O. sativa, and DAD1 and DAD3 in P. hybrida (Wang et al. 2017). The general biosynthetic pathway of SLs has been clarified by using molecular mechanisms: reverse genetics and transcriptomics (Yoneyama and Brewer 2021).
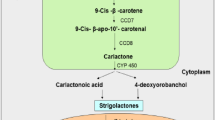
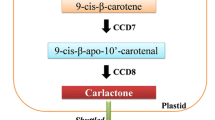
The biosynthesis of SL primarily occurs in chloroplast (plastid) until the formation of CL which later gets transported to cytoplasm for further processing (Fig. 2) (Wani et al. 2020). Carotenoids are isoprenoids that are produced by all photosynthetic organisms (plants, algae, and blue green algae), aphids, fungi, and bacteria which play crucial role in processes of lipid peroxidation, photosynthesis, and in the biosynthesis of plant growth regulators such as SL and abscisic acid (Sun et al. 2022). During carotenoid biosynthesis pathway, geranylgeranyl pyrophosphate (GGPP), phytoene, lycopene, alpha carotene, beta carotene, and gamma carotene are some of the intermediates that are formed (Nimish et al. 2016). The biosynthetic pathway of SL sequentially includes isomerization, cleavage, and oxidation. These steps are catalyzed by enzymes beta carotene isomerase (DWARF27/D27), carotenoid cleavage dioxygenase 7/8 (CCD7 and CCD8), and cytochrome P450 (CYP), respectively (Wang et al. 2017). A newly identified enzyme LBO in A. thaliana is responsible for the SL diversity and it catalyzes the last step of biosynthesis (Jia et al. 2018).
An overview of biosynthesis of different types of naturally occurring Strigolactones (SL). Carlactone (CL), a biosynthetic precursor of all SLs, is produced in plastid (chloroplast) and then moves to cytoplasm. In cytoplasm, several enzymatic actions convert CL into various canonical and non-canonical SLs through intermediates, involving different genes. D DWARF, MAX more axillary growth, HTD high tillering dwarf, DAD decreased apical dominance, RMS ramosus, CCD carotenoid cleavage dioxygenase, LBO lateral branching oxidoreductase, Os Oryza sativa, Vu Vigna unguiculata, Sl Solanum lycopersicum, Ga Gossypium arboretum, Lj Lotus japonicus, CYP cytochrome P450, SL Strigolactone, C carbon. (Figure created with www.biorender.com).
Isomerization by 9-cis/all-trans-β-Carotene Isomerase
The primary step in the biosynthesis of SL is the reversible interconversion of all-trans-β-carotene (an intermediate of the carotenoid biosynthetic pathway) into 9-cis-β-carotene (Ito et al. 2022). It has a C40 skeleton with an extended conjugated double-bond system (Wani et al. 2020). Firstly, the targeted double bond is broken, the molecule is rotated around the single bond, and, finally, a double bond is reintroduced (Jia et al. 2018). An iron-containing enzyme 9-cis/all-trans-β-carotene isomerase catalyzes this step of isomerization. A non-heme iron cofactor is needed for the redox activity of this enzyme (Waters et al. 2012a). It was first identified in rice as DWARF27 (D27/ OsD27) followed by A. thaliana as AtD27. In A. thaliana, three genes with similar phenotypic expression to D27 in rice are reported which suggests the existence of other D27-like enzymes (Wang et al. 2017). Phylogenetic analysis shows that At1g03055, At1g64680, and At4g01995 are three orthologs of OsD27 in A. thaliana. The AtD27 is subcellularly localized in the plastid which is confirmed by GFP fluorescence which is not detected outside the plastid (Waters et al. 2012a). In the SL biosynthesis pathway, AtD27 works upstream of MAX1 and is confirmed by grafting experiments. The transcription of the AtD27 gene is induced by auxin, ABA, and deficiency of phosphate (Kramna et al. 2019).
Cleavage by Carotenoid Cleavage Dioxygenases (CCD7 & CCD8)
Following the isomerization reaction, the cleavage of 9-cis-β-carotene into CL is sequentially performed by carotenoid cleavage dioxygenases, i.e., CCD7 and CCD8 (Wang et al. 2017). This group of enzymes forms aldehyde and carbonyl products by the introduction of oxygen molecules into the double bonds of apocarotenoids (or carotenoids) (Al-Babili and Bouwmeester 2015). The 9-cis-β-carotene (C40) is cleaved primarily at C9’-C10’ double bond by CCD7 into C27 and C13 apocarotenoids, i.e., 9-cis-β-apo-10-carotenal and β-ionone, respectively. Subsequently, CCD8 rearranges 9-cis-β-apo-10-carotenal by adding three oxygens and forms C19 and C8 compounds, i.e., CL and ω-OH-(4-CH3) heptanal, respectively (Jia et al. 2019). In Arabidopsis, rice, pea, and Petunia, CCD7 enzyme is encoded by genes MAX3, D17/HTD1, RMS5, and DAD3, respectively, while MAX4, D10, RMS1, and DAD1 encode the CCD8 enzyme in Arabidopsis, rice, pea, and Petunia, respectively (Al-Babili and Bouwmeester 2015; Wang et al. 2017). CCD7 has been analyzed in several plant species and has been regarded as a critical enzyme of SL biosynthesis as loss in function of the CCD7 gene causes hyperbranched phenotype (Yang et al. 2022b). D17 and D10 have been reported to be the ortholog of MAX3 and MAX4, respectively. Biosynthesis of SL is altered in rice by targeted disruption of CCD7 using CRIPR/Cas9, ccd7 mutant thus produced showed reduced height with high tillering phenotype. The mutant phenotype was rescued using rac-GR24, a synthetic SL analog (Butt et al. 2018).
Oxidation by Cytochrome P450 (CYP)
The CL formed by the subsequent action of CCDs in plastid is transported to cytosol for further processing and subsequent biosynthesis of SLs (Sun et al. 2020). CL is the first non-canonical SL that was reported and has been suggested as a biosynthetic precursor of SL (Yoneyama et al. 2018). In case of SL biosynthetic mutants of rice (d10 and d27) with high tillering phenotypes and branching in max3, mutants of Arabidopsis are suppressed by CL (Wang et al. 2017). A group of scientists studied the conversion of CL into SLs in rice by using 13C-labeled CL, which gets converted into labeled SL. Exogenously applied CL converts into SL, suggesting its role as an endogenous biosynthetic precursor for SL (Seto et al. 2014; Wani et al. 2020). The CL is oxidized by the action of an enzyme CYP. It is encoded by the gene MAX1 (Yang et al. 2019).
In Arabidopsis, MAX1 is suggested to be localized in cytoplasm (Wang et al. 2017). In Arabidopsis, CYP enzyme converts the CL into carlactonoic acid (CLA), a non-canonical SL, while in rice, it forms 4-deoxyorobanchol (4-DO). The 4-DO is further converted into orobanchol by the action of orobanchol synthase, homolog of MAX1 (Baz et al. 2018). The cytochrome P450 oxygenases subfamily CYP711A is responsible for converting CL into canonical and non-canonical SLs (Alvi et al. 2022). Starting from all-trans-beta-carotene upto the formation of CLA, all steps are highly conserved (Yang et al. 2019). In Arabidopsis, through a parallel pathway, methylation of CLA occurs via unknown methyl transferase enzyme (SABATH methyltransferase) via 19-hydroxy-carlactone as an intermediate. The final product formed is methyl carlactonoate (MeCLA) (Wu et al. 2019; Alvi et al. 2022). This reaction is MAX1 independent as confirmed from Arabidopsis mutant analysis (Wani et al. 2020).
In Arabidopsis, CYP is encoded by one MAX1 gene. In rice (O. sativa L.), total five homologs of MAX 1 are reported – Os01g0700900 (Os900), Os01g0701400 (Os1400), Os01g0701500 (Os1500), Os02g0221900 (Os1900), and Os06g0565100 (Os5100). Reportedly, two of these homologs catalyze the conversion of CL to SL. The CL is first converted into 4-DO by Os900 (carlactone oxidase/CO), whose subsequent hydroxylation at C4 of 4-DO forms orobanchol by Os1400 (orobanchol synthase/OS or 4-DO hydroxylase) (Zhang et al. 2014; Jia et al. 2018; Wu et al. 2019). Carlactone oxidase stereo-specifically catalyzes the formation of BC-ring. It acts by adding a proton to hydroxyl group (-OH) present at C18 and remove a proton from the carboxyl group (COOH) present at C19. This results in the closure of BC-ring and a molecule of water is released. Following these data, it has been suggested that these enzymes D27, CCD7, CCD8, and CYP are enough to produce orobanchol-like molecule from all-trans-beta-carotene (Jia et al. 2018).
Depending upon the similarity in the basic skeleton of canonical SLs, the tricyclic skeleton of 5-DS and 4-DO is formed from CLA that later get modified to produce strigol, orobanchol, and various other SLs (Wakabayashi et al. 2022). Recent research suggests that a subfamily CYP722C is responsible for strigol and orobanchol biosynthesis. GaCYP722C in Gossypium arboretum (cotton) and LjCYP722C in Lotus japonicus (lotus) reportedly convert CLA to 5-DS (Alvi et al. 2022). 5-DS is responsible for the production of strigol-type SLs while 4-DO is responsible for the production of orobanchol-type SLs (Wang and Bouwmeester 2018). The simplest form of SL is 5-DS and is present in both dicots and monocots. It either forms strigol or orobanchol by allylic hydroxylation or sorgomol by homoallylic hydroxylation. Sorgomol undergoes sequential oxidation and decarboxylation to form sorgolactone (Wani et al. 2020).
Based on the data obtained from the orobanchol-producing plants such as red bell pepper, red clover, and cowpea, it has been suggested that apart from indirect synthesis of orobanchol from CLA, direct conversion of CLA to orobanchol is also possible (Wakabayashi et al. 2022). For instance, a study reported that in Vigna unguiculata (cowpea) and Solanum lycopersicum (tomato), CLA converts to orobanchol in the presence of VuCYP722C and SlCYP722C, respectively, through an intermediate 18-hydroxy-carlactonoic acid (Alvi et al. 2022).
The diversified presence of SL in nature suggests the involvement of more enzymes in the biosynthetic pathway of SL (Wang et al. 2017). In view of this, another gene LBO has been discovered in A. thaliana that encodes 2-oxoglutarate-dependent dioxygenase enzyme (2OGD). It converts the MecLA to 1’-hydroxymethyl carlactonoate (1’-HO-MeCLA), identified as an unstable product that get converted into CLA (Yoneyama et al. 2020; Wani et al. 2020; Alvi et al. 2022). LBO is like oxido-reductase enzyme and belongs to 2-oxoglutarate and FE (II)-dependent dioxygenase superfamily (Jia et al. 2018). It is an important late-acting enzyme in biosynthetic pathway of SL. In Arabidopsis, lbo mutants with loss of function result in the accumulation of MeCLA, having a phenotype intermediate of wild and max4 mutant type. LBO mutant with overexpression was not able to inhibit or reduce branching (Yang et al. 2022a).
SL Biosynthetic Pathway Inhibitors
Biosynthetic inhibitors act as tools to investigate SL biosynthesis and develop better understanding of its biological roles in different plant species. Specific inhibitors of biosynthetic pathway are useful in identifying and characterizing new biosynthetic mutants and SL signal transduction (Ito et al. 2010). Although SL biosynthetic pathway is not fully identified, set of core enzymes (D27, CCDs, and CYP) involved are fully known. All these catalytic agents contain an iron atom in their molecule. Nitrogen (N2) contains a unshared pair of electrons which form an ionic or co-ordinate bond with the iron atom. Therefore, it has been suggested that any chemical with nitrogen in their structure with great affinity to substrate binding site of these enzymes can act as potent SL biosynthetic inhibitor (Nakamura and Asami 2014). Based on these data, SL biosynthesis inhibitors are basically classified into two classes – carotenoid cleavage dioxygenases (CCD) inhibitors and CYP superfamily inhibitors (Seto et al. 2012). CCDs are inhibited by hydroxamic acids such as D2, D4, D5, D6, D13, and D15 (Harrison et al. 2015).
Although the research data regarding SL inhibitors are limited, derivatives of hydroxamic acid, TIS 13, and tebuconazole are the three classes of chemicals that are identified to have inhibitory role in SL biosynthesis. Besides CCDs, another target of SL biosynthetic inhibitor is CYP. Heterocyclic compounds with nitrogen in them such as triazole and imidazole are reportedly shown to have inhibitory action on CYPs (Kawada et al. 2020). The triazole compound, TIS13, has great affinity toward CYP and is comparatively more potent inhibitor than hydroxamic acid (Ito et al. 2010). Chemical modification of TIS13 led to the recognition of TIS108 and KK5, more specific SL biosynthesis inhibitors. The fungicide tebuconazole also targets CYP (Nakamura and Asami 2014). Recently, a novel inhibitor of cytochrome P450 superfamily, triflumizole, was reported, which effectively reduces the 4-DO level in rice by inhibiting carlactone oxidase (Os900) (Kawada et al. 2020).
Additionally, carotenoid biosynthesis inhibitor fluridone was used to demonstrate that carotenoid biosynthesis is mandatory for SLs biosynthesis, and thus, it can be a SL biosynthetic inhibitor. However, it causes photo destruction of chlorophyll and thus cannot be used as an ideal inhibitor for study purposes (Ito et al. 2010).
Strigolactone: Current Understanding of Signal Perception and Downstream Cascade
The signaling pathway for SLs comprises the process of polyubiquitination and proteasomal degradation. This pathway involves three main components: (i) a hydrolase protein known as D14 which is found in rice, (ii) an F-box leucine-rich protein named MAX2/D3 (Stirnberg et al. 2002; Johnson et al. 2006), and (iii) D53 a repressor protein, associated with the SMAX1-like (SMXL) protein family (Jiang et al. 2013; Stanga et al. 2013). D14, which is the SL receptor protein, becomes activated after it binds to its ligand, resulting in the formation of a signaling complex with other molecules (Marzec et al. 2016). The SL hormone undergoes hydrolysis, deactivating the hormone. Several SL-insensitive mutants have been examined to recognize different SL signaling components.
SLs act as signaling molecules by binding to a receptor complex composed of D14 and D3 proteins. D14 is a highly conserved α/β hydrolase fold protein that binds SLs with high affinity, and D3 is a transcriptional repressor that interacts with D14 to form a heterodimeric receptor complex (Takeuchi et al. 2021). Upon SL binding, the D14-D3 complex undergoes a conformational change that leads to the degradation of D3, resulting in the release of D14 from the receptor complex. This allows D14 to interact with other proteins and initiate downstream signaling (Hu et al. 2017).
In A. thaliana, O. sativa, and Petunia, AtD14/D14/DAD2 is SL orthologous receptors, respectively (Fig. 3). Similar to soluble gibberellic acid receptor GID1, these receptors contain a conserved catalytic triad of Ser, His, and Asp (Zhau et al. 2013; Wani et al. 2020; Takeuchi et al. 2021). Substitution mutation of Ser-to-Ala in the triad leads to loss DAD2 (petunia receptor) catalytic activity. As a result, receptor loses its contact with the F-box proteins, which inhibits shoot branching (Marzec et al. 2016). The hydrolysis of GR24 is probably caused by catalytic triad activity; GR24 and DADs2 go through slow hydrolysis; nonetheless, the dad2 mutant phenotype is not reversed (Zhau et al. 2013). This confirms DAD2 participation in SL signaling along with hydrolytic process is more significant than the final products (Seto and Yamaguchi 2014).
Diagrammatic representation of predicted model for Strigolactone (SL) signaling in plants. In the absence of SL, SMXL/D53, repressor of SL signaling, inhibits the expression of SL-responsive genes. In the presence of SL, SCF (Skp-Cullin-F-box) containing E3 ubiquitin ligase causes ubiquitination of SMXL/D53 that subsequently degrades the repressor and causes the expression of SL-responsive genes. D Dwarf, SL Strigolactone, At Arabidopsis thaliana, DAD decreased apical dominance, SMXL SUPPRESSOR OF MAX2 1-LIKE, MAX MORE AXILLARY GROWTH, SCF SKP1-Cullin-F-box complex, Ub E3 Ubiquitin ligase (Figure created with www.biorender.com).
SL signaling cascade is mediated by ligand binding to its receptor D14, an a/b-hydrolase. After binding, it causes the hydrolysis of SL ligand, causing its separation into two – the ABC-ring portion called ABC-formyl tricyclic lactone (ABC-FTL) and the D-ring called hydroxy methyl butenolide (HMB) (Wani et al. 2020). In O. sativa, the SL hormone interacts with the D14 receptor to cause SL cleavage and the formation of a CLIM (covalently linked intermediate molecule) which is attached to D14 (Bythell-Douglas et al. 2017). HMB continues to be covalently linked to the D14 receptor when ABC-FTL is released (Marzec and Brewer 2019). HMB-D14 intermediate is known as CLIM. The D14's conformation is altered by this process, enabling it to communicate with signaling elements present downstream. SL signaling is initiated when the receptor D14 binds with the F-box leucine-rich protein MAX2/D3/RMS4. The Skp-Cullin-F-box containing (SCF) E3 ubiquitin ligase complex contains MAX2. These orthologs' critical function in SL signaling is confirmed by mutations that cause SL insensitivity (Marzec et al. 2016; Wani et al.2020). It has been noticed that the D53 and D53-like SMXL repressor proteins are targeted for proteasomal degradation by the SCF complex (Jiang et al. 2013; Zhou et al. 2013; Bennett et al. 2016). SMXL6-8 in Arabidopsis is thought to be D53 orthologs as they affect shoot branching and other SL-regulated activities (Soundappan et al. 2015; Bennett et al. 2016; Ligerot et al. 2017). D53, with its EAR motifs, is predicted to interact with TPR (TOPLESS-related transcriptional corepressor) proteins to form complex called D53-TPR which then represses SL target gene expression (Smith and Li 2014). Upon GR24 treatment, the D53 repressor binds with the D14 receptor and is subsequently degraded through the SCF complex (Smith and Li 2014; Wani et al. 2020). The ligand binding causes conformational change in D14 that enables the receptor to recruit SMXL7 into the SCF complex (Liang et al. 2016). SMXL7 performs both transcriptional and non-transcriptional roles, but the molecular processes that happen after its decomposition are still not well understood (Waters et al. 2017; Bythell-Douglas et al. 2017). According to Song et al. (2017), IPA1, the main regulator of plant architecture in O. sativa, functions downstream of the D53 repressor and controls SL-induced gene expression. D53 interact with IPA1 both in vitro and in vivo and represses the transcriptional activation function of IPA1. Various fascinating hypotheses have been given to elucidate how D14 and D14-like receptor proteins evolved ligand and signaling specificity. An instance of convergent evolution has been seen in parasitic plants, where D14-like proteins, which are closely similar to D14 proteins, serve as receptors of SLs secreted by the host (Tsuchiya et al. 2015; Conn and Nelson 2015). Additionally, these D14-like protein subfamilies comprise subfunctionalized proteins that react to other ligands, notably d-lactone-containing substances like karrikins (Waters et al. 2012b; Saeed et al. 2017). The MAX2 F-box protein is required for the perception of both SLs and karrikins (Zhao et al. 2015). Since F-box proteins normally lack selectivity when recruiting target proteins, how MAX2 differentiates among the two routes to create different responses is still unknown (Nelson et al. 2011; Nakamura et al. 2013). According to Wang et al. (2020b), SMXL2 serves as the usual target for polyubiquitylation and degradation in Arabidopsis, and it does this in a manner that is D14 or KAI2-dependent. This suggests that the SL signaling pathways interact with Karrikin pathway at this particular protein.
The model of SL signal transduction is supported by various lines of evidences as it suggests the conformational changes in the D14 receptor brought on by SL binding and hydrolysis, which are the basis for SL signal cascade. For instance, GR24 induces the D14 receptor to become thermally unstable, which relies on the presence of an intact D14 catalytic triad (Wani et al. 2020). Additionally, between MAX2/D3 and D14, physical contact is promoted by GR24 which further destabilizes the receptor. Notably, 2R stereoisomers of SL analogues have a greater effect on D14-D3 interaction in O. sativa than 2S stereoisomers do. Moreover, when 5-hydroxy-3-methylbutenolide or 2,4,4-trihydroxy-3-methyl-3-butenal is associated with SL, no significant structural variation is reported among D14 and apo-D14 (Banerjee and Roychoudhury. 2018; Wani et al. 2020; Rani et al. 2023).
Recently, SL-D14 multiple mode of interaction has been studied; however, it is still unknown how D14 works with D3 to ubiquitinate the repressor protein D53. The C-terminal α-helix of D3 can be either engaged or disengaged forms. The disengaged forms recognize unaltered D14 and blocks its enzymatic activity, whereas the engaged form permits the binding of D14 and D3 with a hydrolyzed SL intermediate. The D3 α-helix aids in D14's SL-dependent recruitment of D53, which activates the hydrolase. Through MAX2, SL causes self-inflicted D14 deterioration, which, via negative feedback subsequently, inhibits its own signaling.
Experimental evidence has challenged the validity of the CLIM model, which proposes that the active site of the D14 receptor binds hydrolyzed SLs (Wani et al. 2020). The small electron density of CLIM makes it unlikely to be fit in the active site of D14. Instead, it is suspected that iodine in the crystallization reagents binds the active site (Chen and Shukla 2022). Moreover, after SL treatment, SL hydrolysis by D14 is too slow in comparison with the quick degradation of target proteins, i.e., D53/SMXLs. Thus, the quick response of SLs cannot be fully defined by the CLIM model. Recent research suggests that D14 initiates the active receptor signaling upon binding to a complete SL molecule. D14 then catalyzes hydrolysis after completing the pathway. The two products of SL hydrolysis, ABC-FTL and HMB, with Kcat, Km, and Vmax values of 0.12 min−1, 4.9 µM, and 4.0 nmol/min/mg protein, respectively, were found from kinetic study of the D14-catalyzed hydrolysis of 5-DS (Wani et al. 2020). Additionally, in rice and Arabidopsis, the SL receptors do not hydrolyze 3,6′-dihydroGR24, having single bond rather than a double bond in the enol-ether bridge (Seto et al. 2019). Debranones, which are SL analogues lacking the enol-ether bridge, have relatively low D14 catalytic activity but produce the same outcomes as GR24. These findings raise concerns regarding function of hydrolysis in SL signaling. Thus, D14 performs binary function and a new method of perceiving SL signals may be suggested (Yao et al. 2016). According to a molecular analysis, the D14 conformational shift expands the catalytic pocket, permitting SL to migrate into it before the helical lid domain is closed (Shabek et al. 2018). The D14 receptor protein primarily adopts an unstable conformation because the catalytic triad assembly is interrupted when an SL molecule attaches to the D14 (Yao et al. 2016). The D14 receptor interacts with other elements in this altered conformation to complete the SL signaling cascade. Following activation, D14 interacts with the F-box protein MAX2/D3 via the surface of its rearranged lid domain, and subsequently, the binding of D53/SMXL repressor takes place around the region of the Asp loop (Seto et al. 2019). After the degradation of D53/SMXL, the D14 catalytic triad is again assembled, which carries out the crucial hydrolysis step. The study related to the evolution of SL signaling mechanism has provided valuable information. It is believed that the main function of SLs was to promote adequate nutrient uptake by the recruitment of AMF (Simon et al. 1993; Waldie et al. 2014). Interestingly, SLs have also been found to be present in algae, and their application causes elongation of rhizoids. Similar response has also been observed in bryophytes such as liverworts and mosses; however, this response is most likely independent of MAX2 (Delaux et al. 2012; Waldie et al. 2014). In higher plants also, MAX2-independent SL signaling has been reported. The max2 mutant can experience root development inhibition even at very low GR24 doses. A D14 member of charophytes has more resemblance to the KARRIKIN INSENSITIVE2 (KAI2) receptor in comparison with canonical D14 proteins (Waldie et al. 2014). Therefore, it is likely possible that to initiate responses, SLs use this receptor rather than MAX2. With the advent of land plants, it's noteworthy that the D14 and MAX2 clades appeared rapidly. D14 most likely originated due to duplication in the clade, and a subsequent duplication within D14 led to the formation of the D14-LIKE2 group (Waldie et al. 2014). The diversification of terrestrial plants and these duplication occurrences were connected with various purposes. A similar trend was followed in the development of the D53 protein. These clades underwent further duplications because the D53-like genes in mosses are more related to SMAX1 than to the D53/SMAXL7 clade (Zhou et al. 2013).
Interestingly, the exact mechanism through which MAX2 enters the SL route is yet unknown. It is hypothesized that MAX2 was primarily exclusively involved in AM colonization and that its function in signaling of SLs was subsequently developed. The d3 rice mutant, which is incapable of being colonized by AMF in SL conditions, supports this (Seto et al. 2019). This process of hormonal breakdown is crucial for maintaining hormone homeostasis and may also be seen in other hormonal pathways such as GA (Yamaguchi 2008).
Strigolactone Receptors: Conserved Among Plant Species
The α/β hydrolase functional domain is conserved among SL receptors, as described by Bennett and Leyser (2014). At first, this domain was discovered in the O. sativa d14 mutant, which is SL-insensitive (Mashiguchi et al. 2021) and since then, it has been identified in other plants, including Petunia, pea, and Arabidopsis (Hamiaux et al. 2012; de Saint et al. 2016; Waters et al. 2012b). D14 homologs have been identified in various plant clades, like bryophytes (Marchantia polymorpha), pteridophytes (Selaginella moellendorfii) and gymnosperms, belonging to the D14-like subfamily, while the genes of angiosperm are categorized into the D14 subfamily of the α/β-hydrolase superfamily (Wani et al. 2020). According to Arite et al. (2009) and Nardini and Dijkstra (1999), these subfamilies have different sequences but have a conserved catalytic triad, nucleophilic residue, and acidic residue. Notably, the α/β hydrolase superfamily involves enzymes responsible for acetylcholine metabolism, such as acetylcholinesterase (AChE), and the inactive gibberellic acid receptor (Holmquist et al. 2000).
According to Hamiaux et al. (2012), the D14 protein of O. sativa, functions both as a receptor and as an enzyme which distinguishes it from other plant hormone receptors. This protein contains a functional domain called α/β hydrolase, which includes a Ser-His-Asp catalytic triad domain that forms the ligand binding pocket, while the cap of the protein is composed of four α helices. The protein comprises of 318 amino acids, and a similar protein called D14-like is also found in the genome of rice (Arite et al. 2009).
Although D14 has enzymatic activity, the rate of SL hydrolysis in vitro is low (~ 0.3 molecules per minute). Therefore, it is not considered the primary producer of bioactive SL-derived signals (Snowden and Janssen 2016). Additionally, neither the final byproducts of SL hydrolysis (tricyclic lactone and HMB) nor the intermediate molecule 2,4,4-trihydroxy-3-methyl-3-butenal serves as signals for the regulation of shoot branching (Waters et al. 2017).
The receptor of SL in A. thaliana (AtD14) is evolutionarily conserved, like the rice D14 receptor (Waters et al. 2012b; Arite et al. 2009). It has the activity of both receptor and enzyme, and its structure comprises a catalytic triad (Hamiaux et al. 2012). The hormone binds to the receptor through two amino acids in the triad, resulting in a change in receptor conformation (Yao et al. 2016). This change causes an increase in length of one α helix domain and unfolding of another, which forms a loop (Yao et al. 2016). The receptor lid comprises of four α helix domains, and it destabilizes when hormone binding occurs (Zhao et al. 2015; Snowden and Janssen 2016). The closure of the enzymatic active site reduces its volume, which makes D-ring separation hard without dissociating the complex. This phenomenon could describe the slow enzyme activity. AtD14L/KAI2, which is involved in karrikin signaling, is like AtD14, with 51% identity and 75.9% similarity, but belongs to a separate phylogenetic clade (Waters et al. 2012b).
In Petunia, the D14 receptor ortholog is called DAD2 (Rameau et al. 2015; Chesterfield et al. 2020). X-ray crystallography shows that its structure comprises a lid with four α helices and a hydrophobic cavity to accommodate SLs in it (Burger and Chory 2020). In the presence of hormone GR24, DAD2 interacts with PhMAX2A (F-box protein), leading to GR24 hydrolysis. Mutations in the catalytic triad of DAD2 result in the loss of enzymatic activity and failure to interact with PhMAX2A (Hamiaux et al. 2012). The dad2 mutants display a profuse branching phenotype like that of dad1 (CCD8) and dad3 (CCD7) biosynthetic mutants (Wani et al. 2020). DAD2 is responsible for locally controlling shoot branching, as shown by grafting and genetic studies (Hamiaux et al. 2012). DAD2 is not involved in SL biosynthesis, as confirmed by the study of dad2 mutants. When the biosynthetic mutant was grafted onto wild-type rootstocks, it reverts back to branching phenotype. The dad2 mutant, however, is not graft revertible. Hence, it indicates that DAD2 is not involved in the synthesis of graft-transmissible hormone (Hamiaux et al. 2012).
The HvD14 gene in Hordeum vulgare encodes the SL receptor, which consists of two exons and a 1055-bp coding sequence (Marzec et al. 2016). The HvD14 protein of 303-amino acid possesses the conserved α/β-hydrolase domain between amino acids 57 and 295 and shows significant structural resemblance, high sequence conservation, and comparable secondary domains to the rice D14 ortholog (Kagiyama et al. 2013). In hvd14.d mutants, a substitution of Glu for Gly at position 193 is observed, which is present in the αD2 α-helical domain forming the cap surrounding the active site along with αD1, αD3, and αD4 (Kagiyama et al. 2013)).
According to Zheng et al. (2016), the woody perennial plant Populus trichocarpa has two highly similar homologs, PtD14a and PtD14b, with a 91.7% identity. PtD14a shares 79% identity and 89.1% similarity with AtD14, while PtD14b shares 77.5% identity and 89.1% similarity with AtD14 (Zheng et al. 2016). The catalytic triad Ser-His-Asp, which is crucial for receptor function, is conserved in both PtD14 homologs at positions 96, 246, and 217. The gene expression level of PtD14a in comparison with PtD14b has higher transcript levels and both the homologs show minimum co-expression levels (Zheng et al. 2016). Identifying the potential SL receptors in parasitic weeds posed a challenge due to the difficulty in genetically dissecting the relevant phenotypes (Toh et al. 2015; Tsuchiya et al. 2015). However, subsequent studies revealed that a group of α/β-hydrolases, known as ShKAI2s/ShHTLs (Striga hermonthica) KARRIKIN INSENSITIVE2/HYPOSENSITIVE TO LIGHT), act as SL receptors through SL hydrolysis and SL-induced seed germination, with these hydrolases being D14 paralogs (Conn et al. 2015; Toh et al. 2015; Yao et al. 2017). Among these paralogs, ShHTL7 serves as the most active receptor of SL in Striga (Conn et al. 2015; Yao et al. 2017). Similar to AtD14, during CLIM formation, ShHTL7 goes through a conformational change, which facilitates signal transduction through its interaction with MAX2/ShMAX2 (Yao et al. 2017).
Conclusion and Future Prospective
Strigolactones and their derivatives are a group of phytohormones which are derived from an intermediate molecule of carotenoid biosynthetic pathway. They are composed of a tricyclic lactone ABC-ring which relates to a D-ring butenolide moiety through enol-ether bridge. Their chemical nature is highly diverse. Since their discovery in 1966 as stimulant of Striga seed germination and rediscovery in 2008 as a new class of phytohormone, the biosynthesis of SL remains a hot topic among researchers. Molecular analysis via genetics and transcriptomics mainly on A. thaliana and O. sativa L. helped in the identification of main components. Although a significant knowledge has been gained over the past few years related to SL biosynthesis and signaling cascade, however, there must be several other unidentified genes which are involved as the nature of SLs is diverse. Thus, it will be of importance to identify and characterize them as this will help to create deep and better understanding and further helps to study the bioactive role of SLs in living organisms. The information regarding the mechanism involved in the SCF-mediated polyubiquitination of targeted protein is scarce and requires intensive study. The SCF complex lacks specificity, so how it differentiates between its target proteins is still need detailed investigation. Although catalytic triad domain in receptor of SL is conserved in diverse plant species, whether these receptors in plants are similar to SL receptors in AMF or not is still a question.
References
Al-Babili S, Bouwmeester HJ (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol 66:161–186. https://doi.org/10.1146/annurev-arplant-043014-114759
Alvi AF, Sehar Z, Fatma M, Masood A, Khan NA (2022) Strigolactone: an emerging growth regulator for developing resilience in plants. Plants 11(19):2604. https://doi.org/10.3390/plants11192604
Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50:1416–1424. https://doi.org/10.1093/pcp/pcp091
Banerjee A, Roychoudhury A (2018) Strigolactones: multi-level regulation of biosynthesis and diverse responses in plant abiotic stresses. Acta Physiol Plant 40:1–10
Baz L, Mori N, Mi J, Jamil M, Kountche BA, Guo X et al (2018) 3-Hydroxycarlactone, a novel product of the strigolactone biosynthesis core pathway. Mol Plant 11(10):1312–1314. https://doi.org/10.1016/j.molp.2018.06.008
Bennett T, Leyser O (2014) Strigolactone signaling: standing on the shoulders of DWARFs. Curr Opin Plant Biol 22:7–13. https://doi.org/10.1016/j.pbi.2014.08.001
Bennett T, Liang Y, Seale M, Ward S, Müller D et al (2016) Strigolactone regulates shoot development through a core signalling pathway. Biology Open 5(12):1806–1820. https://doi.org/10.1242/bio.021402
Bouwmeester HJ, Fonne-Pfister R, Screpanti C, De Mesmaeker A (2019) Strigolactones: plant hormones with promising features. Angew Chem Int Ed 58(37):12778–12786. https://doi.org/10.1002/anie.201901626
Brewer PB, Koltai H, Beveridge CA (2013) Diverse roles of strigolactones in plant development. Mol Plant 6(1):18–28. https://doi.org/10.1093/mp/sss130
Brewer PB, Yoneyama K, Filardo F, Meyers E, Scaffidi A, Frickey T et al (2016) LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 113(22):6301–6306. https://doi.org/10.1073/pnas.1601729113
Brun G, Braem L, Thoiron S, Gevaert K, Goormachtig S, Delavault P (2018) Seed germination in parasitic plants: what insights can we expect from strigolactone research? J Exp Bot 69(9):2265–2280. https://doi.org/10.1093/jxb/erx472
Bürger M, Chory J (2020) The many models of strigolactone signaling. Trends Plant Sci 25(4):395–405. https://doi.org/10.1016/j.tplants.2019.12.009
Butt H, Jamil M, Wang JY, Al-Babili S, Mahfouz M (2018) Engineering plant architecture via CRISPR/Cas9-mediated alteration of strigolactone biosynthesis. BMC Plant Biol 18(1):1–9. https://doi.org/10.1186/s12870-018-1387-1
Bythell-Douglas R, Rothfels CJ, Stevenson DW, Graham SW, Wong GKS, Nelson DC, Bennett T (2017) Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues. BMC Biol 15(1):1–21. https://doi.org/10.1186/s12915-017-0397-z
Chen J, Shukla D (2022) Multiple modes of substrate hydrolysis-induced covalent modification of strigolactone receptors. bioRxiv, 2022-04. https://doi.org/10.1101/2022.04.22.489200
Chesterfield RJ, Whitfield JH, Pouvreau B, Cao D, Alexandrov K, Beveridge CA, Vickers CE (2020) Rational design of novel fluorescent enzyme biosensors for direct detection of strigolactones. ACS Synth Biol 9(8):2107–2118. https://doi.org/10.1021/acssynbio.0c00192
Conn CE, Nelson DC (2015) Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front Plant Sci 6:1219. https://doi.org/10.3389/fpls.2015.01219
Conn CE, Bythell-Douglas R, Neumann D, Yoshida S, Whittington B, Westwood JH, Nelson DC (2015) Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349:540–543. https://doi.org/10.1126/science.aab1140
de Saint GA, Clavé G, Badet-Denisot MA, Pillot JP, Cornu D, Le Caer JP, Bonhomme S (2016) A histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol 12:787. https://doi.org/10.1038/nchembio.2147
Delaux PM, Xie X, Timme RE, Puech-Pages V, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Bécard G, Séjalon-Delmas N (2012) Origin of strigolactones in the green lineage. New Phytol 195:857–871. https://doi.org/10.1111/j.1469-8137.2012.04209.x
Desta B, Amare G (2021) Paclobutrazol as a plant growth regulator. Chem Biol Technol Agric 8(1):1–15. https://doi.org/10.1186/s40538-020-00199-z
Faizan M, Faraz A, Sami F, Siddiqui H, Yusuf M, Gruszka D, Hayat S (2020) Role of strigolactones: signalling and crosstalk with other phytohormones. Open Life Sci 15(1):217–228. https://doi.org/10.1515/biol-2020-0022
Faizan M, Cheng SH, Tonny SH, Robab MI (2022) Specific roles of strigolactones in plant physiology and remediation of heavy metals from contaminated soil. Plant Physiol Biochem. https://doi.org/10.1016/j.plaphy.2022.10.004
Guercio AM, Palayam M, Shabek N (2023) Strigolactones: diversity, perception, and hydrolysis. Phytochem Rev. https://doi.org/10.1007/s11101-023-09853-4
Haider I, Yunmeng Z, White F, Li C, Incitti R, Alam I et al (2023) Transcriptome analysis of the phosphate starvation response sheds light on strigolactone biosynthesis in rice. Plant J. https://doi.org/10.1111/tpj.16140
Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22:2032–2036. https://doi.org/10.1016/j.cub.2012.08.007
Harrison PJ, Newgas SA, Descombes F, Shepherd SA, Thompson AJ, Bugg TD (2015) Biochemical characterization and selective inhibition of β-carotene cis–trans isomerase D27 and carotenoid cleavage dioxygenase CCD 8 on the strigolactone biosynthetic pathway. FEBS J 282(20):3986–4000. https://doi.org/10.1111/febs.13400
Holmquist M (2000) Alpha beta-hydrolase fold enzymes structures, functions and mechanisms. Curr Protein Pept Sci 1:209–235. https://doi.org/10.2174/1389203003381405
Hu Q, He Y, Wang L, Liu S, Meng X, Liu G et al (2017) DWARF14, a receptor covalently linked with the active form of strigolactones, undergoes strigolactone-dependent degradation in rice. Front Plant Sci 8:1935. https://doi.org/10.3389/fpls.2017.01935
Ito S, Kitahata N, Umehara M, Hanada A, Kato A, Ueno K et al (2010) A new lead chemical for strigolactone biosynthesis inhibitors. Plant Cell Physiol 51(7):1143–1150. https://doi.org/10.1093/pcp/pcq077
Ito S, Braguy J, Wang JY, Yoda A, Fiorilli V, Takahashi I et al (2022) Canonical strigolactones are not the major determinant of tillering but important rhizospheric signals in rice. Sci Adv 8(44):eadd1278
Jia KP, Baz L, Al-Babili S (2018) From carotenoids to strigolactones. J Exp Bot 69(9):2189–2204. https://doi.org/10.1093/jxb/erx476
Jia KP, Li C, Bouwmeester HJ, Al-Babili S (2019) Strigolactone biosynthesis and signal transduction. Strigolactones-biology and applications. Springer, New York, pp 1–45
Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L et al (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504(7480):401–405. https://doi.org/10.1038/nature12870
Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C (2006) Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol 142:1014–1026. https://doi.org/10.1104/pp.106.087676
Kagiyama M, Hirano Y, Mori T, Kim SY, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T (2013) Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18:147–160. https://doi.org/10.1111/gtc.12025
Kawada K, Uchida Y, Takahashi I, Nomura T, Sasaki Y, Asami T et al (2020) Triflumizole as a novel lead compound for strigolactone biosynthesis inhibitor. Molecules 25(23):5525. https://doi.org/10.3390/molecules25235525
Kountche BA, Novero M, Jamil M, Asami T, Bonfante P, Al-Babili S (2018) Effect of the strigolactone analogs methyl phenlactonoates on spore germination and root colonization of arbuscular mycorrhizal fungi. Heliyon 4(11):e00936. https://doi.org/10.1016/j.heliyon.2018.e00936
Kramna B, Prerostova S, Vankova R (2019) Strigolactones in an experimental context. Plant Growth Regul 88(2):113–128. https://doi.org/10.1007/s10725-019-00502-5
Liang Y, Ward S, Li P, Bennett T, Leyser O (2016) SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 28:1581–1601. https://doi.org/10.1105/tpc.16.00286
Ligerot Y, de Saint GA, Waldie T, Troadec C, Citerne S, Kadakia N, Rameau C (2017) The pea branching RMS2 gene encodes the PsAFB4/5 auxin receptor and is involved in an auxin-strigolactone regulation loop. PLoS Genet 13(12):e1007089. https://doi.org/10.1371/journal.pgen.1007089
Lopez-Obando M, Ligerot Y, Bonhomme S, Boyer FD, Rameau C (2015) Strigolactone biosynthesis and signaling in plant development. Development 142(21):3615–3619. https://doi.org/10.1242/dev.120006
Marzec M (2016) Perception and signaling of strigolactones. Front Plant Sci 7:1260. https://doi.org/10.3389/fpls.2016.01260
Marzec M, Brewer P (2019) Binding or hydrolysis? How does the strigolactone receptor work? Trends Plant Sci 24(7):571–574. https://doi.org/10.1016/j.tplants.2019.05.001
Mashiguchi K, Seto Y, Yamaguchi S (2021) Strigolactone biosynthesis, transport and perception. Plant J 105(2):335–350. https://doi.org/10.1111/tpj.15059
Nakamura H, Asami T (2014) Target sites for chemical regulation of strigolactone signaling. Front Plant Sci 5:623. https://doi.org/10.3389/fpls.2014.00623
Nakamura H, Xue YL, Miyakawa T, Hou F, Qin HM, Fukui K (2013) Molecular mechanism of strigolactone perception by DWARF14. Nat Commun 4(1):2613. https://doi.org/10.1038/ncomms3613
Nardini M, Dijkstra BW (1999) α/β Hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol 9:732–737. https://doi.org/10.1016/S0959-440X(99)00037-8
Nelson DC, Scafdi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Smith SM (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. PNAS 108:8897–8902. https://doi.org/10.1073/pnas.1100987108
Stephen NM, Gayathri R, Niranjana R, Prasad Y, Das AK, Baskaran V, Ganesan P (2016) Carotenoids: types, sources, and biosynthesis. In: Plant secondary metabolites, vol 2, pp 103–132. Apple Academic Press, Washington, DC
Rameau C, Bertheloot J, Leduc N, Andrieu B, Foucher F, Sakr S (2015) Multiple pathways regulate shoot branching. Front Plant Sci 5:741. https://doi.org/10.3389/fpls.2014.00741
Rani V, Sengar RS, Garg SK, Mishra P, Shukla PK (2023) Physiological and molecular role of strigolactones as plant growth regulators: a review. Mol Biotechnol. https://doi.org/10.1007/s12033-023-00694-2
Ruyter-Spira C, Al-Babili S, Van Der Krol S, Bouwmeester H (2013) The biology of strigolactones. Trends Plant Sci 18(2):72–83. https://doi.org/10.1016/j.tplants.2012.10.003
Saeed W, Naseem S, Ali Z (2017) Strigolactones biosynthesis and their role in abiotic stress resilience in plants: a critical review. Front Plant Sci 8:1487. https://doi.org/10.3389/fpls.2017.01487
Seto Y, Yamaguchi S (2014) Strigolactone biosynthesis and perception. Curr Opin Plant Biol 21:1–6. https://doi.org/10.1016/j.pbi.2014.06.001
Seto Y, Kameoka H, Yamaguchi S, Kyozuka J (2012) Recent advances in strigolactone research: chemical and biological aspects. Plant Cell Physiol 53(11):1843–1853. https://doi.org/10.1093/pcp/pcs142
Seto Y, Sado A, Asami K, Hanada A, Umehara M, Akiyama K, Yamaguchi S (2014) Carlactone is an endogenous biosynthetic precursor for strigolactones. PNAS 111:1640–1645. https://doi.org/10.1073/pnas.1314805111
Seto Y, Yasui R, Kameoka H, Tamiru M, Cao M, Terauchi R, Umehara M (2019) Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat Commun 10:191
Shabek N, Ticchiarelli F, Mao H, Hinds TR, Leyser O, Zheng N (2018) Structural plasticity of D3–D14 ubiquitin ligase in strigolactone signalling. Nature 563:652–656
Simon L, Levesque RC, Lalonde M (1993) Identification of endomycorrhizal fungi colonizing roots by fluorescent single-strand conformation polymorphism-polymerase chain reaction. Appl Environ Microbiol 59:4211–4215. https://doi.org/10.1128/aem.59.12.4211-4215.1993
Smith SM, Li J (2014) Signalling and responses to strigolactones and karrikins. Curr Opin Plant Biol 21:23–29. https://doi.org/10.1016/j.pbi.2014.06.003
Snowden KC, Janssen BJ (2016) Structural biology: signal locked in. Nature 536:402
Song X, Lu Z, Yu H, Shao G, Xiong J, Meng X, Yao XF (2017) IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res 27:1128
Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A et al (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27(11):3143–3159. https://doi.org/10.1105/tpc.15.00562
Souri Z, Karimi N, Farooq MA, Akhtar J (2020) Phytohormonal signaling under abiotic stress. In: Plant life under changing environment, pp 397–466. Academic Press, Washington, DC
Stanga JP, Smith SM, Briggs WR, Nelson DC (2013) SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol 163:318–330. https://doi.org/10.1104/pp.113.221259
Stirnberg P, van De Sande K, Leyser HO (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129:1131–1141. https://doi.org/10.1242/dev.129.5.1131
Sun T, Tadmor Y, Li L (2020) Pathways for carotenoid biosynthesis, degradation, and storage. Plant and food carotenoids: methods and protocols. Methods Mol Biol 2083:3–23
Sun T, Rao S, Zhou X, Li L (2022) Plant carotenoids: Recent advances and future perspectives. Mol Hortic 2(1):3
Takeuchi J, Fukui K, Seto Y, Takaoka Y, Okamoto M (2021) Ligand–receptor interactions in plant hormone signaling. Plant J 105(2):290–306. https://doi.org/10.1111/tpj.15115
Toh S, Holbrook-Smith D, Stogios PJ, Onopriyenko O, Lumba S, Tsuchiya Y, McCourt P (2015) Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350:203–207
Tsuchiya Y, McCourt P (2009) Strigolactones: a new hormone with a past. Curr Opin Plant Biol 12(5):556–561. https://doi.org/10.1016/j.pbi.2009.07.018
Tsuchiya Y, Yoshimura M, Sato Y, Kuwata K, Toh S, Holbrook Smith D et al (2015) Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349:864–868
Wakabayashi T, Ueno K, Sugimoto Y (2022) Structure elucidation and biosynthesis of orobanchol. Front Plant Sci 13:835160. https://doi.org/10.3389/fpls.2022.835160
Waldie T, McCulloch H, Leyser O (2014) Strigolactones and the control of plant development: lessons from shoot branching. Plant J 79:607–622. https://doi.org/10.1111/tpj.12488
Wang Y, Bouwmeester HJ (2018) Structural diversity in the strigolactones. J Exp Bot 69(9):2219–2230. https://doi.org/10.1093/jxb/ery091
Wang B, Wang Y, Li J (2017) Institute of genetics and developmental biology. Chinese Academy of Sciences, Beijing
Wang L, Xu Q, Yu H, Ma H, Li X, Yang J, Li J (2020) Strigolactone and karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell 32(7):2251–2270. https://doi.org/10.1105/tpc.20.00140
Wang Q, Smith SM, Huang J (2022) Origins of strigolactone and karrikin signaling in plants. Trends Plant Sci 27(5):450–459. https://doi.org/10.1105/tpc.20.00140
Wani KI, Zehra A, Choudhary S, Naeem M, Khan MMA, Castroverde CDM, Aftab T (2020) Mechanistic insights into strigolactone biosynthesis, signaling, and regulation during plant growth and development. J Plant Growth Regul. https://doi.org/10.1007/s00344-020-10234-w
Wani KI, Chaudhary S, Zehra A, Naeem M, Aftab T (2021) Precise role of strigolactones and its crosstalk mechanisms in root development. Rhizobiology: Molecular Physiology of Plant Roots, 253–270.
Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA (2012a) The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol 159(3):1073–1085. https://doi.org/10.1104/pp.112.196253
Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM (2012b) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139(7):1285–1295. https://doi.org/10.1242/dev.074567
Waters MT, Gutjahr C, Bennett T, Nelson D (2017) Strigolactone signalling and evolution. Annu Rev Plant Biol. https://doi.org/10.1146/annurev-arplant-042916-040925
Wu H, Li H, Chen H, Qi Q, Ding Q, Xue J et al (2019) Identification and expression analysis of strigolactone biosynthetic and signaling genes reveal strigolactones are involved in fruit development of the woodland strawberry (Fragaria vesca). BMC Plant Biol 19(1):1–19. https://doi.org/10.1186/s12870-019-1673-6
Xie X (2016) Structural diversity of strigolactones and their distribution in the plant kingdom. J Pestic Sci 41(4):175–180. https://doi.org/10.1584/jpestics.J16-02
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–251. https://doi.org/10.1146/annurev.arplant.59.032607.092804
Yang T, Lian Y, Wang C (2019) Comparing and contrasting the multiple roles of butenolide plant growth regulators: strigolactones and karrikins in plant development and adaptation to abiotic stresses. Int J Mol Sci 20(24):6270. https://doi.org/10.3390/ijms20246270
Yang R, Liu W, Sun Y, Sun Z, Wu Z, Wang Y et al (2022a) LATERAL BRANCHING OXIDOREDUCTASE, one novel target gene of Squamosa Promoter Binding Protein-like 2, regulates tillering in switchgrass. New Phytol 235(2):563. https://doi.org/10.1111/nph.18140
Yang W, Mirbahar AA, Shoaib M, Lou X, Sun L, Sun J et al (2022b) The carotenoid cleavage dioxygenase gene CCD7-B, at large, is associated with tillering in common wheat. Agriculture 12(2):306. https://doi.org/10.3390/agriculture12020306
Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L et al (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536:469–473. https://doi.org/10.3390/agriculture12020306
Yao R, Wang F, Ming Z, Du X, Chen L, Wang Y, Xie D (2017) ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Res 27:838
Yoneyama K (2020) Recent progress in the chemistry and biochemistry of strigolactones. J Pestic Sci 45(2):45–53. https://doi.org/10.1584/jpestics.D19-084
Yoneyama K, Brewer PB (2021) Strigolactones, how are they synthesized to regulate plant growth and development? Curr Opin Plant Biol 63:102072. https://doi.org/10.1016/j.pbi.2021.102072
Yoneyama K, Xie X, Yoneyama K, Kisugi T, Nomura T, Nakatani Y et al (2018) Which are the major players, canonical or non-canonical strigolactones? J Exp Bot 69(9):2231–2239. https://doi.org/10.1093/jxb/ery090
Zhang Y, Van Dijk AD, Scaffidi A, Flematti GR, Hofmann M, Charnikhova T et al (2014) Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat Chem Biol 10(12):1028–1033
Zhao LH, Zhou XE, Yi W, Wu Z, Liu Y, Kang Y, Scafdi A (2015) Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling efector DWARF3. Cell Res 25:1219
Zheng K, Wang X, Weighill DA, Guo HB, Xie M, Yang Y, Muchero W (2016) Characterization of DWARF14 genes in Populus. Sci Rep 6:21593
Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N et al (2013) D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 504(7480):406–410
Zorrilla JG, Rial C, Varela RM, Molinillo JM, Macias FA (2022) Strategies for the synthesis of canonical, non-canonical and analogues of strigolactones, and evaluation of their parasitic weed germination activity. Phytochem Rev 21(5):1627–1659. https://doi.org/10.1007/s11101-022-09801-8
Zwanenburg B, Pospíšil T (2013) Structure and activity of strigolactones: new plant hormones with a rich future. Mol Plant 6(1):38–62. https://doi.org/10.1093/mp/sss141
Acknowledgements
Author gratefully acknowledges the financial assistance rendered by the University Grant Commission (UGC), New Delhi, India, in the form of a non-net fellowship.
Author information
Authors and Affiliations
Contributions
SH gave the idea for the article. The first draft of the manuscript was written by SZ, YA, HI, and MS, and SH revised critically for important intellectual content and gave substantive support. All authors read and approved the final manuscript. All authors contributed to the study’s conception and design.
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Handling editor: Anket Sharma.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zaidi, S., Arif, Y., Imtiaz, H. et al. Structural Chemistry, Biosynthesis, and Signaling of Multifaceted Plant Growth Regulator: Strigolactone. J Plant Growth Regul 43, 2489–2502 (2024). https://doi.org/10.1007/s00344-024-11285-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-024-11285-z