Abstract
To realize the genetic transformation of Cry1Ac, Cry3A, and NTHK1 genes in Populus × euramericana “Neva” and obtain transgenic plants, the plant transformation vector p09687199–Cry1Ac–Cry3A–NTHK1 was transformed in Populus × euramericana “Neva” through Agrobacterium-mediated transformation. The complete regeneration of the transformed plant was screened with kanamycin. Resistant roots of the transformed plants were detected through PCR, fluorescence quantitative PCR, and ELISA analysis of the toxic protein. The insect resistance and salt tolerance of transgenic plants were determined based on the findings of these methods. PCR detection showed that among six lines, five lines indicated the presence of three target genes, Cry1Ac and Cry3A genes were detected in one line. Fluorescence quantitative PCR detection showed that the transcript abundance of the Cry1Ac gene was within 1.13E+3 to 3.17E+4, that of Cry3A was within 4.40E+6 to 1.97E+7, and that of NTHK1 was within 2.48E+3 to 8.17E+3. Bt toxic protein detection showed that the content of the Cry1Ac gene was from 1.08 to 22.99 ng·g−1, and that of Cry3A gene was from 146.12 to 17027.96 ng·g−1. The expression of the Cry3A gene was significantly higher than that of the Cry1Ac gene. The insect resistance test demonstrated that the mortality of transgenic plants on Hyphantria cunea first instar larva reached 68.89 %. The mortality on Plagiodera versicolora first and second instar larvae reached 100 %, whereas that on the third instar larva was as high as 81.71 %. Four lines were selected to detect potting salt tolerance, and only the No. 2 line showed strong salt tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, transgenic technology exhibited much progress in the directional improvement of plant traits and has become one of the most important approaches in contemporary molecular breeding. Many studies have examined transgenosis, especially insect-resistant transgenosis, of various Bt and salt tolerance genes in the poplar genome (Génissel et al. 2003; Fladung and Ewald 2006; Yang et al. 2008; Hu et al. 2010).
However, research on transgenic poplar mainly focused on 1–2 gene transformations to change traits via genetic engineering technology. Despite the continuous development of transgenic technology, changing a single gene of plant traits has not been realized. The transformation of two or more resistant genes in the same plant and the improvement of multiple plant traits have become popular research topics (Bates et al. 2005; Li and Zhu 2005; Christou et al. 2006; Wang et al. 2010). Multi-gene transformation can alter insect resistance, salt tolerance, and other qualities of genes in the same plant genome, change the multiple traits of plant variety, and improve the comprehensiveness of plant stress tolerance (Sun et al. 2011; Lian et al. 2008; Gao et al. 2012). The multi-gene transformation methods of plants include agrobacterium-mediated method and particle bombardment induction. Methods applied on tobacco (Komari et al. 1996), rice (Feng et al. 2001), corn (Zhu et al. 2008), and other plants are reported to be successful.
With the constant improvement of multi-gene vector construction technology, the multi-gene vector co-transformation method has been extensively implemented in gene engineering. Multi-gene vector transformation aims to transform the expression vector containing multiple genes and their regulatory elements in the plant to obtain a multi-gene transformation plant co-expressed with the multivalent gene via screening. This method can overcome the substitute of the selectable marker gene in the single-gene repeated transformation method and reduce the workload of genetic transformation, screening, and identification. As for the high-efficiency expression of multiple exogenous genes in a plant and the interaction of gene expressions, transgenic plant traits still need to be studied. In this study, two insect-resistant genes, namely Cry1Ac and Cry3A, and NTHK1 salt tolerance gene were transformed into Populus × euramericana “Neva.” Molecular and biological detections of the transgenic plants were performed, and the expressions of the three exogenous genes were analyzed to provide a fundamental basis for multi-gene transformation and expression.
Materials and methods
Agrobacterium strains
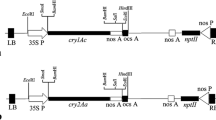
Agrobacterium tumefaciens was GV3101. The plasmid vector p09687199 in Agrobacterium tumefaciens carries the Bt gene (containing Cry1Ac and Cry3A) and the ethylene receptor gene NTHK1. The selectable marker gene was kanamycin resistance gene NPT II (Fig. 1). The Cry1Ac gene is resistant only to Lepidoptera pests, whereas the Cry3A gene is resistant to coleoptera pests. The NTHK1 gene can improve plant salt tolerance (Wang et al. 2008, 2011).
Receptor materials
The transgenic receptor material was Populus × euramericana “Neva”, a hybrid of Populus deltoides and Populus nigra L.; it is a new hybrid variety from Italy introduced by the Chinese Academy of Forestry. The control materials were non-transgenic Populus × euramericana “Neva.”
Test insects
Hyphantria cunea belongs to Insecta Lepidoptera Arctiidae. Larvae egg masses were collected from Baoding. Plagiodera versicolora Laicharting belongs to Insecta Coleoptera Chrysomelidae. Adult worm and egg masses were also collected from Baoding.
Genetic transformation of Populus × euramericana “Neva”
Revival of Agrobacterium tumefaciens
Agrobacterium tumefaciens GV3101 containing plasmid p09687199 preserved at low temperature was utilized for lineation on a YEB solid medium through the three-level dilution method. It was cultured at 28 °C in a thermostat incubator for 16–24 h. After the bacterial colonies grew, a single colony was selected, dissolved in YEB liquid medium, placed on a shaker at 160 rpm (away from light), and cultured to the logarithmic growth phase (OD600 = 0.4–0.6) for explant dip dyeing.
Genetic transformation mediated by Agrobacterium tumefaciens
Under sterile conditions, fully extended and dark green tender leaves of Populus × euramericana “Neva” tissue culture seedling were selected and transected into 3–5 vertical incisions deep into the main vein. The leaves were immersed for 10 min with Agrobacterium. The bacterial suspension was gently shaken to allow the leaves to fully touch the Agrobacterium. The leaves were then removed. The excess bacterial suspension was emptied using sterile filter paper, and the leaves were placed in a co-culture medium (MS + 2.0 mg·L−16-BA + 0.5 mg·L−1NAA + 10 mg·L−1 acetosyringone) and co-cultured for 3 days at 28 °C in dark conditions. The sample was then transferred to a differentiation screening medium (MS + 2.0 mg·L−16-BA + 0.5 mg·L−1NAA + 30 mg·L−1Kan + 400 mg·L−1 Cefotaxime Sodium) every 15 d to 20 days. When the resistance buds grew to 1 cm, they were cut off and transferred into a rooting screening medium (1/2MS + IBA0.3 mg·L−1 + 0.5 mg·L−1NAA + 30 mg·L−1Kan + 400 mg·L−1 Cefotaxime Sodium). The adventitious roots grew within a week and regenerated into a complete plant. Their stem tips were removed and cultured in a differentiation screening culture. When the adventitious buds grew to approximately 2 cm, they were transferred to a rooting screening medium. The rooting seedlings were transplanted into small pots and placed in an isolated garden after domestication. The 30 mg·L−1Kan + 400 mg·L−1 Cefotaxime Sodium were selected for screening referring Yang’s research (Yang 2012).
PCR detection of transgenic plants
Transgenic seedling leaves were collected from the garden in the same year. The genomic DNA of transgenic Populus × euramericana “Neva” and control plants were extracted with improved CTAB method (Wang et al. 2002). The forward and reverse primers of Cry1Ac, Cry3A, NTHK1, and NPTII genes were utilized in PCR detection with genomic DNA as the template (Table 1). A 20 μL reaction system was prepared (ddH20: 7 μl, 2 × Mix: 10 μl, primer R (10 pM): 1 μl, primer F (10 pM): 1 μl, and DNA: 1 μl). The plasmid DNA was considered the positive control, and the non-transgenic Populus × euramericana “Neva” was regarded as the negative control. PCR was conducted as follows: pre-degeneration at 95 °C for 5 min, degeneration at 94 °C for 45 s, renaturation at 50 °C for 60 s, extension at 72 °C for 50 s (30 cycles), extension for 7 min at 72 °C, and preservation for 1 min at 25 °C.
Fluorescence quantitative PCR detection of transgenic plants
The transgenic seedlings’ annual top leaves that had just fully expanded were acquired at the end of July 2014. About 0.1 g of leaves was placed into a mortar. Liquid nitrogen was added, and the leaves were rapidly ground into powder. Total RNA was extracted with EASYEx PLUS Plant RNA Kit (Cyrus, China). Reverse transcription of the first cDNA chain was performed with a reverse transcription kit (Kangwei, China). Fluorescence quantitative PCR primers were designed according to the sequence information of the target genes. cDNA synthesized by reverse transcription was utilized as the template, and 2* Sybr Green qPCR Mix was used for fluorescence quantitative PCR.
The fluorescence quantitative PCR primer is shown below.
Cry1Ac gene F: 5′-GAATTTTTGGTCCCTCTCAAT-3′
R: 5′-AGGATCTGCTTCCCACTCTCT-3′
Cry3A gene F: 5′-TGGGGATACGAGAAGGAGGAT-3′
R: 5′-AGTGGGAACAGTGCGATGAGA-3′
NTHK1 gene F: 5′-CCCTTATCACCCTCTTTCCTAT-3′
R: 5′-AGTGTCTTTGCTAACTCCACCA-3′
The reaction system (20 μL) comprised 10 μL 2* SYBR qPCR Mix, 0.5 μL forward primer (10 μM), 0.5 μL reverse primer (10 μM), 1 μL template, and 8 μL ddH2O. The reaction procedure was as follows: pre-degeneration at 95 °C for 5 min, degeneration at 95 °C for 10 s, and renaturation at 55 °C for 30 s. The amplified products of the known fragment were considered the standard and diluted 10 times for use in real-time fluorescence quantitative PCR. The standard curve was then obtained. According to the cycle threshold (Ct) that indicates whether the fluorescence signal reached the setting threshold in each PCR tube and the standard curve, the transcript abundances of Cry1Ac, Cry3A, and NTHK1 genes in cDNA were calculated with mRNA as the transcript.
Cry1Ac and Cry3A detection by ELISA
The transgenic and control seedlings’ top annual shoots with fully extended leaves acquired at the end of July 2014 were used for Bt toxic protein detection. The Cry1Ac and Cry3A toxic proteins of all lines were detected with Bt-Cry1Ab/1Ac and Bt-Cry3A ELISA kits (Agdia, USA). The detection methods were in accordance with the instructions provided by the manufacturers. The positive control was the appendant of the kit, and the negative control was the non-transgenic Populus × euramericana “Neva”. The results were determined with a BioRad 550-type microplate reader. Protein concentration was calculated by the number of toxic protein nanogram (ng) in each gram of fresh leaves.
Insect resistance detection
Hyphantria cunea first instar larvae were used to detect the insect resistance of the Cry1Ac toxin protein. Hyphantria cunea larvae were obtained from egg masses in the nursery of Agricultural University of Hebei in July 2014. The first instar larvae that hatched in the greenhouse were directly utilized for the insect feeding test. The insects used in the Cry3A toxic protein insect resistance test were the first, second, and third instar larvae of Plagiodera versicolora. Adult Plagiodera versicolora was acquired from the nursery of Agricultural University of Hebei at the end of April 2014 and cultured at room temperature. Fresh and tender non-transgenic Populus × euramericana “Neva” was replaced every day. The eggs were collected and cultured daily. Non-transgenic Populus × euramericana “Neva” was utilized to feed the larvae. Plagiodera versicolora first instar larvae were obtained in 3 days and cultured with the second and third instar larvae for testing.
All transgenic lines and non-transgenic control seedling fresh leaves on annual shoots were collected. The petioles were inserted in humid flower mud to maintain the fresh leaves. The test insects were gently and evenly placed onto the leaves. The leaves were then placed in a small beaker with 6.5 cm diameter and 8 cm height. The rim of the cup was covered with double-layer moist gauze and tied up. The small beaker was placed in a plastic bag and bound gently to maintain humidity and ventilation. Each treatment was repeated thrice. One bottle underwent one repetition of the treatment, and 30 larvae were placed in a bottle. The fresh leaves were changed every day. The mortality of the larvae was recorded. The test continued until all larvae stabilized or died.
The corrected mortality equation is as follows:
Corrected mortality = (mortality rate of transgenic lines − mortality of control)/(1 − mortality of control).
Salt tolerance test
Four transgenic lines of Populus × euramericana “Neva” containing ethylene receptor gene NTHK1 were regarded as the materials, and the non-transgenic Populus × euramericana “Neva” was considered the control. Branches that were approximately 15 cm in size, a year old, and with constant diameter were cut and placed in a pot with a depth of 40 cm and a diameter of 30 cm during spring. Each pot had five seedlings, including one line of non-transgenic Populus × euramericana “Neva” and four lines of transgenic seedlings. Fifteen pots of seedlings were set for each line. The water container was covered with a plastic film to preserve moisture. The film was gradually removed as the branch sprouted. The salt tolerance treatment was performed when the seedlings reached a height of approximately 15 cm.
NaCl solution concentration gradients of 0, 3, and 6 ‰ were set. Stress treatment was performed on the transgenic lines and control cutting seedling. Five potted plants were set for each treatment, that is, five treatments were repeated for each line. The saline solution was watered each week at 1 L/pot.
Measurement of seedling height The growth of each line and control plant was observed regularly while under salt stress. Seedling height was measured every two weeks.
Biomass measurement The soil of the cutting seedling root was washed with water after subjecting the plant to salt stress for 40 days. The entire seedling was placed inside an envelope and returned to the laboratory. The seedling was baked at 80 °C until the weight was constant after de-enzyming at 105 °C for 10 min. The dry weight was then measured.
Measurement of photosynthetic rate The photosynthetic rate of each line was determined by Li-6400XT photosynthesis in a sunny morning after subjecting the plant to salt stress for 40 days. The first mature leaf of each line was selected. The dust on the surface of the leaves was removed; the main vein was avoided as the leaves were clamped in the leaf chamber. Data were continuously recorded until the numbers stabilized.
Data analysis
Figures were drawn in Microsoft Excel 2007, and the data were processed with DPS v7.05 statistical analysis software. Multiple comparisons were performed on significant differences with Duncan’s new multiple range method.
Results and analysis
Acquisition of transgenic plants
The green buds were differentiated from the transgenic Populus × euramericana “Neva” in the screening medium. The resistant buds grew in the culture medium containing a certain concentration of kanamycin because of neomycin phosphotransferase NPT II gene. The non-transgenic Populus × euramericana “Neva” etiolated and died. The differentiated buds were transferred to the rooting medium. Six lines rooted, but the control did not root. Six resistant lines were obtained after repeated subculture and selection. When the root length was approximately 2 cm in the rooting medium, it was transplanted to a small pot and placed in the field. The transgenic lines were obtained by kanamycin preliminary screening, as shown in Fig. 2.
Acquisition of transgenic lines. a Agrobacterium infection after co-culture stage. b Leaves with resistant shoots after agrobacterium infection. c Proliferation of resistant shoots. d Rooting of resistant shoots. e Rooting plants transplanted to small pots. f Rooting plants transplanted to the field
PCR detection of transgenic lines
PCR amplification of six pre-acquired transgenic plants was performed with the two Bt genes and NTHK1-specific primers. The p09687199 plasmid and non-transgenic Populus × euramericana “Neva” were considered positive and negative controls. Multiple PCR amplification results showed that NPTII (Fig. 3a), Cry1Ac (Fig. 3b), and Cry3A (Fig. 3c) genes were detected in the six pre-acquired transgenic plants. NTHK1 gene was only detected in five lines (Fig. 3d).
PCR amplification of the exogenous genes of various transgenic lines. a NPTII gene PCR amplification. b Cry1Ac gene PCR amplification. c Cry3A gene PCR amplification. d NTHK1 gene PCR amplification. CK + plasmid, CK − control, M DL5000 DNA marker (from top to bottom: 5000, 3000, 2000, 1000, 750, 500, 250, and 100 bp)
Fluorescence quantitative PCR detection of transgenic lines
The cDNA obtained from mRNA reverse transcription was detected by fluorescence quantitative PCR, that is, exogenous gene expression was detected on the transcriptional level. Real-time monitoring of the entire PCR process by fluorescence signal integration showed that the expression of the Cry1Ac fluorescence signal was detected in six lines and those of Cry3A and NTHK1 were detected in five lines. The transcript abundance of the Cry1Ac gene was within 1.13E+3 to 3.17E+4, that of the Cry3A gene was within 4.40E+6 to 1.97E+7, and that of NTHK1 was within 2.48E+3 to 8.17E+3. The transcript abundance of the Cry3A gene was high and that of Cry1Ac and NTHK1 was low. The expressions of the three genes showed no significant difference (Table 2, Fig. 4).
Detection of Bt toxin protein in transgenic lines
The two Bt toxic proteins detected by ELISA are shown in Table 3. The expression of Cry1Ac toxic protein reached as low as 1.08–22.99 ng·g−1 in the transgenic lines. A significant difference was observed among the transgenic lines. The contents of the toxin protein were high in No. 3 and No. 6 lines. The content of Cry3A toxin protein was as high as 17,027.96 ng·g−1, and a significant difference was evident among the transgenic lines. The contents of No. 3 and No. 4 lines were the highest. The toxic protein was not detected in the control. Comprehensive analysis showed that the content of Cry3A toxin protein in transgenic plants was significantly higher than that of Cry1Ac.
Insect resistance detection of transgenic lines
Lethal effect of transgenic lines on Lepidoptera Hyphantria cunea
The transgenic and control lines were utilized to feed the Hyphantria cunea larvae. The insect resistance effect significantly differed among various transgenic lines. Only No. 3 and No. 6 lines exhibited a highly obvious lethal effect on the larvae and a certain inhibitory effect on the growth of the larvae (Fig. 5). The lethal rate and larvae growth of the other lines did not significantly differ from those of the control lines. Correlation analysis between the lethal rate of Hyphantria cunea larvae and Cry1Ac toxic protein expression was performed. The correlation coefficient was r = 0.9832, which indicates significant correlation at a = 0.01 (r a = 0.8745, n = 7) level, thereby suggesting that the indoor insect feeding result is consistent with the ELISA kit test results. The higher the expression of Cry1Ac toxic protein, the higher the lethal rate of Hyphantria cunea larvae was. In contrast, the lethal rate of Hyphantria cunea larvae was low (Table 4).
Lethal effect of transgenic lines on Coleoptera Plagiodera versicolora
As shown in Table 5, except in the No. 6 line, the insect resistance of transgenic Cry3A gene lines to Plagiodera versicolora larvae was high. The lethal rate of the first instar larvae reached 100 % and that of the second instar larvae reached more than 90 %. However, the lethal rate of the third instar larvae decreased. The insect resistance of the No. 3 line on Plagiodera versicolora larvae was the best. Correlation analysis of the lethal rate of Plagiodera versicolora larvae and the expression of Cry3A toxin protein produced a correlation coefficient of r = 0.8451. Therefore, both are significantly correlated at a = 0.01(r a = 0.7545, n = 7)level, suggesting that the insect test result of Plagiodera versicolora is basically consistent with the ELISA kit detection result. The No. 6 line did not show insect resistance, similar to the control.
Salt tolerance test of transgenic lines
Effects of salt stress on seedling height and biomass
Seedling heights of 3 and 6 ‰ were compared with 0 ‰ seedling height. The proportion of 3 and 6 ‰ seedling heights accounting for 0 ‰ was calculated to determine the extent of decrease in different line seedling heights under salt stress. As shown in Fig. 6, the seedling heights of different lines decreased with the increase in salt concentration. Treated by 3 ‰ saline, the monthly growth heights of the control, No. 1, No. 3, and No. 4 lines decreased to approximately 60–70 %; the No. 2 line decreased to only 90 %. Treated with 6 % saline, the monthly height growth of the control, No. 1, No. 3, and No. 4 lines decreased to about 50–60 %. The height growth of the No. 2 line decreased to 78 %. The increase in seedling height showed no significant difference in No. 1, No. 3, No. 4, and control lines. The extent of the decrease in seedling growth height was large under salt stress. Compared with that of the control line, the extent of the decrease in the No. 2 line was smaller under salt stress, suggesting that the No. 2 line can still maintain high growth and has a level of certain salt tolerance.
Seedling total biomass amounts of 3 and 6 ‰ were compared with 0 ‰ biomass. The percentages of 3 and 6 ‰ total biomass accounting for 0 ‰ were calculated to compare the extent of decrease. The total biomass of each line decreased with the increase in salt concentration (Fig. 7). The decrease in CK (Control check) was most significant when treated by 3 ‰ saline. The total biomass of No. 1, No. 3, and No. 4 lines decreased to about 80–90 %. In contrast, the total biomass of the No. 2 line slightly increased. Treated by 6 ‰ saline, the total biomass of the control and No. 4 lines decreased to 40–50 %. The total biomass of No. 2 and No. 3 lines were 82 and 77 %, respectively, reflecting a small extent of decrease. Overall, compared with the control, the total biomass measures of No. 1, No. 3, and No. 4 lines were not significantly different because of the main effect of salt stress. The decrease in total biomass in the No. 2 line was smaller than that in the control.
Effects of salt stress on net photosynthetic rate
The net photosynthetic rates of the control and transgenic lines decreased after treatment with salt solution (Fig. 8), indicating that salt stress inhibited the photosynthesis of poplar. Treated by 3 ‰ saline solution, the net photosynthetic rates of the control, No. 1, and No. 4 lines decreased by 50–60 % compared with 0 ‰ treatment. The No. 3 line decreased by 40 %, and the No. 2 line declined by only 21 %. Treated by 6 ‰ saline solution, the net photosynthetic rates of the control, No. 1, and No. 4 lines decreased by 60–70 % compared with that treated by 0 ‰ saline solution. The No. 3 line decreased by 52 %, and the No. 2 line declined by only 19 %. The net photosynthetic rate of each line showed a downward trend. No significant difference was observed in the extent of decrease between No. 1 and No. 4 lines. The decrease in the No. 3 line was smaller than that in the control line. The extent of decrease in the No. 2 line was the smallest and indicated the least effect of salt stress.
Discussion
Bt gene is a kind of insect-resistant genes derived from microorganisms, and it can express the insecticidal crystal proteins (ICPs). Insecticidal crystal proteins are hydrolyzed by protease in the intestine after ingested by insect, and they turn into toxic peptide which has resistance on protease. The toxic peptide can bind to the insect midgut epithelial cell-specific binding sites, forming perforation and causing cell expansion and cracking, and finally leading to the death of the insect (Li et al. 2010). Different types of crystal proteins have insecticidal specificity. The Bt gene is an insect-resistant gene obtained from microbes. Current studies on poplar focus on the Cry1 gene having a toxic effect on Lepidoptera and the Cry3 gene having a toxic effect on Coleopteran; various transgenic lines were also observed to exhibit insect resistance (Yang et al. 2008; Zhang et al. 2005; Hjältén et al. 2012; Axelsson et al. 2012). Many studies have proven that transgenic Cry1Ac gene poplar has high insect resistance on Hyphantria cunea, Clostera anachoreta, and Porthetria dispar, and so on. Transgenic Cry3A gene poplar has a lethal effect on Coleoptera Plagiodera versicolora (Yang et al. 2003; Wang et al. 2008; Niu et al. 2011). Yanping Wang et al. (2008) fed Plagiodera versicolora and Clostera anachoreta with seven lines of transgenic Cry3A hybrid poplar 741 and two lines of transgenic Cry1Ac gene hybrid poplar 741. The result showed that transgenic Cry3A gene lines have a high lethal rate on Plagiodera versicolora. No significant difference was observed in the lethal rate between Lepidoptera Clostera anachoreta larvae and the control. Transgenic Cry1Ac gene lines showed a high lethal rate on Clostera anachoreta, and no significant difference was shown in the lethal rate between Coleopteran Plagiodera versicolora larvae and the control. These results suggest that with different transgenic Bt genes, poplars exhibit selective insect resistance. Guiying Wang et al. (2012) compared different transgenic Bt gene lines and lines containing double Bt genes obtained by the twice conversion method. The results showed that with different transgenic Bt genes, poplars exhibit selective insect resistance. The effect on the target insects was significant. No toxicity was inflicted on non-target insects. The transgenic double Bt genes had digenic resistance. ELISA detection showed that the expression of Cry3A toxic protein was more than ten times higher than that of Cry1Ac toxic protein.
In the current study, six transgenic lines were obtained with Agrobacterium-mediated method. The expressions of two Bt genes differed. The transcript abundance of Cry3A was 103–104 times higher than that of Cry1Ac at the transcription level. The expression of Cry3A toxic protein was 102–103 higher than that of Cry1Ac toxic protein at the translation level. In the insect resistance test, among the six lines, the lethal rates of five lines to Coleoptera Plagiodera versicolora first instar larvae reached 100 % and that of third instar larvae reached 68.18–81.75 %. However, the lethal effect on Lepidoptera Hyphantria cunea was weak. Only the lethal rates of two lines reached more than 60 %. Compared with the transgenic single Bt poplar line, the expression of the Cry3A gene was maintained at a higher level, and the lethal effect on Plagiodera versicolora larvae was significant. However, the expression of the Cry1Ac gene decreased. Generally, the lethal effect of transgenic single Cry1Ac gene line on Lepidoptera Hyphantria cunea is 80–100 % (Yang et al. 2003; Wang et al. 2008; Tian et al. 2000). This phenomenon may be caused by gene interaction. The two Bt genes were constructed on the same vector and inserted into the plant genome. These genes were closely adjacent and had high homology, resulting in potential interference, namely one gene inhibiting another gene. In addition, the sequence of the target gene may also affect gene expression. Related studies on tobacco showed that when these two Bt genes are constructed on one transformation vector, regardless of whether they are Cry3A and Cry1Ac genes, the toxin protein expression of the Bt gene in the upstream decreases. This observation requires further study and verification.
NTHK1 is an ethylene receptor gene cloned from tobacco; it can be induced by mechanical injury, NaCl, and PEG. Thus, NTHK1 may be involved in the signal transduction process of salt and drought (Zhang et al. 1999, 2001). Tao Feng (2008) injected NTHK1 into soybean through Agrobacterium-mediated method. The result showed that NTHK1 can be effectively expressed and can enhance salt tolerance and drought resistance. Shasha Du et al. (2014) cultured tobacco lines with transgenic NTHK1 gene placed in media containing different NaCl concentrations. The adventitious bud differentiation and callus growth of the explant were observed. The result showed that transgenic NTHK1 tobacco plant growth is superior to that of wild control plants, proving that the NTHK1 gene can improve the salt tolerance of tobacco to a certain degree. In the present study, four transgenic pot seedlings were subjected to a series of NaCl stress. The No. 2 line was proven resistant to salt tolerance. Under NaCl stress, seedling height, the biomass of transgenic lines, and the extent of the decrease in net photosynthetic rate were lower than those of non-transgenic controls. No significant difference in salt tolerance was observed between the three other lines and the control line. However, the salt tolerance test involving short-term pot experiments was insufficient. Comparative tests of the experimental forest should be conducted on salty and alkaline soil to accurately detect the salt tolerance of transgenic lines.
In the six detected transgenic lines, Cry1Ac gene was detected in one line only, and the other two genes were lost. The loss of exogenous gene in transgenic plants had been reported (Jin et al. 2005; Byebier et al. 1987). In general, this phenomenon may be due to the loss in T-DNA integrated into the plant genome or before the integration process (Hua and Huang 1999). Because more repeated sequences existed on the vector, part of target genes was sheared in the T-DNA transformation and integration process, resulting in gene loss. In the study, the missing genes were located near the left border of T-DNA, which was in accordance with the literature reported that T-DNA transformation can cause the loss of the left edge sometimes (Wang et al. 2002; Byebier et al. 1987). But the molecular mechanism of exogenous gene loss is not clear.
The expressions of the three target genes showed a significant difference in this study. In the mRNA transcription, Bt toxic protein expression and biological resistance detection showed significant differences. However, no obvious difference was observed in the mRNA transcript abundance of different transgenic lines. Bt toxic protein expression and biological resistance showed a significant difference between different transgenic lines, suggesting that lines with a low expression of exogenous gene demonstrate posttranscriptional gene silencing. The expressions of various genes showed no correlation in the six transgenic lines. The expressions of the Cry3A and NTHK1 genes were not ideal. The Cry1Ac gene was expressed well in the No. 6 line. The expression of the Cry3A gene in the No. 2 line was generally acceptable. The expression of the Cry1Ac gene was not ideal, whereas the NTHK1 gene was expressed well. The expressions of Cry1Ac and Cry3A toxic proteins were relatively high in the No. 3 line probably because three exogenous genes had their own independent promoter and reading frame. When the exogenous gene was inserted into the plant genome, many patterns and mechanisms about its integration patterns emerged.
Due to the different T-DNA insertion sites in the genome of poplar, the gene sequences around the exogenous gene insertion sites in host genome may affect the exogenous gene expression, resulting in different expression conditions of exogenous genes in different lines. On the multi-gene vector, three exogenous genes have independent reading frame, but the same promoter and terminator and more repeated sequences of two Bt genes may also lead to genes mutual inhibition at the transcriptional level, resulting in different expressions of different genes. The plant genome may also affect the expression of a certain gene. For example, after the methylation balance of the plant genome was destroyed, the defense occurring on the plant genome allowed different degrees of methylation to occur on the integrated exogenous gene, thereby causing gene silencing (Nan et al. 1998). Therefore, to improve the expression effect, the exogenous gene expression and interaction mechanism must be further studied in polygenic transformation.
Author contribution statement
Dongyan Liu performed most of the experiments and wrote the first draft of the manuscript; Jun Zhang and Yan Dong performed part of the experiments, analyzed part of the data and revised the manuscript; Xu Zhang performed part of the experiments and analyzed part of the data; Minsheng Yang and Baojia Gao designed the work and revised the final version of the text.
References
Axelsson EP, Hjältén J, LeRoy CJ (2012) Performance of insect-resistant Bacillus thuringiensis (Bt)-expressing aspens under semi-natural field conditions including natural herbivory in Sweden. Forest Ecol Manag 264:167–171
Bates SL, Zhao JZ, Roush RT, Shelton A (2005) Insect resistance management in GM crops: past, present and future. Nat Biotechnol 23(1):57–62
Byebier B, Deboeck F, Greve HD, Montagu MV, Hernalsteens JP (1987) T-DNA organization in tumor cultures and transgenic plants of the monocotyledon Asparagus officinaolis. Proc Natl Acad Sci USA 84:5345–5349
Christou P, Capell T, Kohli A, Gatehouse JA, Gatehouse AMR (2006) Recent developments and future prospects in insect pest control in transgenic crops. Trends Plant Sci 11(6):302–308
Du SS, Zhang X, Wang JM, Dong Y, Yang MS (2014) Tobacco transformation with bivalent gene BtCry1Ac and NTHK1 mediated by agrobacterium. J Henan Agric Sci 43(7):31–37
Feng T (2008) Identify the Function of the NTHK1 Gene and Transformation with Agrobacterium-mediated in Soybean. Dissertation, Northeast Agricultural University
Feng DR, Xu XP, Li BJ (2001) Production of rice plants containing multiple transgenes. Acta Phytophysiol Sin 27(4):331–336
Fladung M, Ewald D (2006) Tree transgenesis: recent developments. Springer, Berlin-Heidelberg
Gao C, Lang ZH, Zhu L, Song FP, Zhang YJ, Zhang J, Huang DF (2012) Insecticidal effect of transgenic tobacco harboring cry2Ab4 or vip3Aa11 gene on Agrotis ypsilon. J Agr Sci Tech-Iran 14:42–48
Génissel A, Leplé JC, Millet N, Augustin S, Jouanin L, Pilate G (2003) High tolerance against Chrysomela tremulae of transgenic poplar plants expressing a synthetic cry3Aa gene from Bacillus thuringiensis ssp tenebrionis. Mol Breeding 11:103–110
Hjältén J, Axelsson EP, Whitham TG, LeRoy CJ, Julkunen-Tiitto R, Wennstoem A, Pilate G (2012) Increased resistance of Bt aspens to Phratora vitellinae (Coleoptera) leads to increased plant growth under experimental conditions. PLoS ONE 7(1):30640
Hua ZH, Huang DN (1999) Genetic Mode of Exogenes in Transgenic Plants. Acta Botanica Sinica 41:1–5
Hu JJ, Yang MS, Lu MZ (2010) Advances in biosafety studies on transgenic insect-resistant poplars in China. Biodiversity Science 18(4):336–345
Jin WM, Pan QH, Yin SP, Dong J, Jiang LJ, Yang L, Chen QH, Zhao JB (2005) Progress of the genetic stability and breeding behavior of foreign gene in genetically modified plants. Mole Plant Breed 3:864–868
Komari T, Hiei Y, Saito Y, Murai N, Kumashiro T (1996) Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformation free from selection markers[J]. Plant J 10(1):165–174
Li BJ, Zhu HC (2005) On the prospects of applying the multi-gene transformation strategy (MTS) to modify the inheritance of organisms. Acta Scientiarum Naturalium Universitatis Sunyatseni 44(4):79–83
Li YP, Liang GM, Wu JX, Chen H, Ma KS, Wu KM, Gu YY (2010) Progress in insecticidal mechanism of Bt and resistance mechanism of pest insect to Bt. Journal of Northwest A&F University 38(9):118–128
Lian Y, Jian ZW, He KL, Liu YJ, Song FP, Wang BW, Wang GY (2008) Transgenic tobacco plants expressing synthetic Cry1Ac and Cry1Ie genes are more toxic to cotton bollworm than those containing one gene. Chinese Sci Bull 53:1381–1387
Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386–389
Niu XY, Huang DZ, Yang MS, Li XF, Fu XS (2011) Temporal and spatial expression of Bt toxic protein in transgenic Btcry3A hybrid poplar 741. Sci Silv Sin 47(12):154–157
Sun LY, Li J, Yuan HW, Yao JH, Li W, Chen XY (2011) Expression analysis of Bt-BADH-GA20ox-rolB multigenes in transgenic tobacco plants. J Beijing For Univ 33:86–90
Tian YC, Zheng JB, Yu HM, Liang HY, Li CQ, Wang JM (2000) Studies of transgenic hybrid poplar 741 carrying two insect-resistant genes. Acta Bot Sin 42(3):263–268
Wang GL, Fang HJ (2002) Plant genetic engineering. Science press, Beijing
Wang CL, Lu BF, Zhang XC, Li GX, Wang JB, Li YJ, Peng YL, Pan SQ (2002) Molecular basis of Agrobacterium—Plant gene transfer. Chin Bull Life Sci 14:1–5
Wang YP, Li J, Yang MS, Liang HY (2008) Insect-resistance selectivity of transgenic hybrid poplar 741. Sci Silv Sin 8:67–71
Wang JL, Liu DQ, Ding YM, Ge F, Li WX, Tian RH (2010) Research advances in Bt transgenic anti-insect plants. J Biol 27(4):75–78
Wang P, Yang YX, Song J, Zhang Y, Chen SY, Wang JA, Liu ZQ (2011) NTHK1 of tobacco improves salt tolerance of roots in poplars. Plant Physiol J 47(12):1155–1160
Wang GY, Yang MS, Huo XM, Liu XJ (2012) Comparison of exogenous gene expression and insect-resistance ability of transgenic 741 poplars with single and double Bt genes. Acta Entomologica Sinica 55(7):798–803
Yang YL (2012) The research on two insect-resistant gene transforming into Populus × euramericana cv.“74/76”. Dissertation, Agricultural University of Hebei
Yang MS, Liang HY, Gao BJ, Wang JM, Zheng JB (2003) Insecticidal activity and transgene expression stability of transgenic hybrid poplar clone 741 carrying two insect-resistant genes. Silvae Genet 52(6):197–201
Yang LY, Sun Y, Xie LQ (2008) Reviews on transgenic poplar and biosafety evaluation. Mole Plant Breed 1:123–127
Zhang JS, Xie C, Liu F, Liu FH, Chen SY (1999) A novel tobacco gene coding for a product similar to bacterial two-component regulators. Chin Sci Bull 44(11):1025–1029
Zhang JS, Xie C, Wu XL, Du BX, Chen SY (2001) Tobacco two-component gene NTHK2. Chin Sci Bull 46:574–577
Zhang BY, Su XH, Li YL, Zhang YA, Qu JL, Wang YZ, Tian YC (2005) Transformation of poplar (Populus alba × P. glandulosa cv. ‘84 K’) with binary insect resistant genes and analysis of insect resistance. Forest Res 18(3):364–368
Zhu CF, Naqvi S, Breitenbach J, Sandmann G, Christou P, Capell T (2008) Combinational genetic transformation generates a library of metabolic phenotypes for the caroteniod pathway in maize. Proc Natl Acad Sci USA 105:18232–18237
Acknowledgments
This research was supported by the National High Technology Research Development Program of China (863 Program) (No. 2013AA102703) and the National Natural Science Foundation of China (No. 31370663).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Wang.
D. Liu, J. Zhang and Y. Dong contributed equally to this study.
Rights and permissions
About this article
Cite this article
Liu, D., Zhang, J., Dong, Y. et al. Genetic transformation and expression of Cry1Ac–Cry3A–NTHK1 genes in Populus × euramericana “Neva”. Acta Physiol Plant 38, 177 (2016). https://doi.org/10.1007/s11738-016-2195-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-016-2195-6