Abstract
APETALA1 plays a crucial role in floral transition from vegetative to reproductive phase and in flower development. In this study, a comprehensive analysis of AP1 homologues in poplar was performed by describing the gene structure and chromosomal location. The phylogenetic relationship of the deduced amino acid sequences of Arabidopsis AP1 and AP1 homologues from Populus, to other AP1-like proteins was analyzed. The expression of PtAP1-1 and PtAP1-2 in Populus tomentosa was examined by RT-qPCR. Expression profiles were similar and both genes exhibited a high expression level in the reproductive phase. Seasonal expression profiles in floral buds indicated that the pattern of PtAP1-1 and PtAP1-2 expression in male and female floral buds was different. The trends of the PtAP1-1 and PtAP1-2 transcript levels in both sex floral buds were similar, but the peak of expression of the two genes in male buds was earlier than in female buds. This work would be of value to future functional analysis of AP1 homologues in poplar.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In high plants, flowering is an important developmental process in response to endogenous and environmental signals. During the past two decades, many genes related to flower initiation and development have been isolated and their functions have been well studied (Wellmer and Riechmann 2010). Particularly, APETALA1 (AP1) orchestrates floral initiation by integrating growth, patterning, and hormonal pathways (Kaufmann et al. 2010). In Arabidopsis thaliana, flowering cues gather in FLOWERING LOCUS T (FT), a flowering time integrator, and FT protein interacts physically with FLOWERING LOCUS D (FD) and activates the floral meristem identity genes LEAFY (LFY) and AP1 to specify floral meristems on the flanks of the shoot apical meristem (Abe et al. 2005; Corbesier et al. 2007). These two genes play central roles in the transition from the inflorescence meristems into floral meristems. LFY activates AP1 and they also form a positive feedback loop. AP1 is first observed throughout emerging floral primordia (Mandel et al. 1992; Wagner et al. 1999). TERMINAL FLOWER1 (TFL1) is a shoot identity gene and is referred to as a floral repressor. TFL1 is necessary for indeterminate shoot fate, since tfl1 mutants show early flowering and conversion of inflorescence meristems to floral meristems (Bradley et al. 1997; Ratcliffe et al. 1998). LFY and AP1 antagonize the activity of TFL1 to establish floral meristems. In turn, TFL1 acts to repress LFY and AP1 in inflorescence meristems (Ratcliffe et al. 1998). The balance between these three genes determines the rate of shoot apical phase transition and flowering time (Bradley et al. 1997; Parcy et al. 2002; Ratcliffe et al. 1999). Three MADS-box transcription factors, SHORT VEGETATIVE PHASE (SVP, a floral repressor), AGAMOUS-LIKE24 (AGL24, a floral activator) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1, a floral activator), are identified early as flowering time genes. In floral meristems before late stage 2, class B and C homeotic genes are not expressed because SEP3 is repressed by SVP, SOC1 and AGL24. As floral meristems proceed to late stage 2, direct repression of SVP, SOC1 and AGL24 by AP1 gradually derepresses SEP3. SEP3 interacts physically with LFY to activate downstream other floral organ identity genes in the apical region of early stage 3 floral meristem (Liu et al. 2009). AP1 functions not only as floral meristem identity gene, but also determines floral organ formation, and in later stages of floral development, its expression is confined to the first and second whorls of floral buds, where AP1 is involved in the specification of sepals and petals (Mandel et al. 1992; Weigel and Meyerowitz 1994).
Poplar (Populus spp.) is a perennial woody plant that has been widely grown for landscaping, forestation, timber, pulp and biofuels. Since the release of the Populus trichocarpa draft genome (Tuskan et al. 2006) and subsequent updates in the phytozome database (Goodstein et al. 2012), poplar has emerged as a model species for molecular genetics research in trees. However, the long juvenile phase in poplars presents a substantial obstacle to research and breeding. Additionally, shed flowers, allergenic properties of poplar pollen, and catkin production represent sources of environmental pollution and a hazard to human health (An et al. 2011a, b; Strauss et al. 1995). The ability to control the transition from the juvenile phase to the reproductive phase and identification of the key genes involved in flowering in poplar would represent a significant accomplishment. Recently, the functions of FT (Böhlenius et al. 2006; Hsu et al. 2006, 2011; Shen et al. 2012; Zhang et al. 2010), CO (Hsu et al. 2012), and CEN (Mohamed et al. 2010) homologues in poplar have been well investigated. Although AP1 plays key roles in flowering time and floral organ formation, the mechanisms of controlling flowering in poplar and the patterns of gene expression during poplar floral bud development have not been fully elucidated. In this study, we analyzed the gene structure, chromosomal location and phylogenetic relationship of Populus AP1 homologues and characterized their expression patterns throughout the vegetative and reproductive developmental stages in Chinese white poplar (Populus tomentosa Carr.). This work may help to elucidate the biological functions of AP1 homologues in poplar.
Materials and methods
Plant materials and growth conditions
One-month-old tissue-cultured plants of Chinese white poplar (P. tomentosa female clone TC1521) were grown and synchronized (using vegetative stem segments containing an axillary bud) on 1/2 MS solid medium supplemented with 0.4 mg l−1 IBA (pH 5.8) in a growth chamber at 25 °C under a 16-h light/8-h dark photoperiod.
Sequence and phylogenetic analyses of AtAP1 and PtrAP1
The amino acid sequences of AP1 homologue genes were retrieved from Phytozome v9.1 (http://www.phytozome.net/) and aligned with ClustalX 1.81 and Bioedit software. Conserved and functional domains sequences were analyzed using Expert Protein Analysis System (ExPASy) software available on the website of the Swiss Institute of Bioinformatics (http://cn.expasy.org) (Gasteiger et al. 2003). The tertiary structure of the AP1 proteins was predicted using SWISS-MODEL (http://swissmodel.expasy.org/workspace/index.php?func=modelling_simple1) software, and viewed with RasMol 2.7.2.1. Genomic sequence data were used to analyze gene structure and determine chromosomal locations. A phylogenetic tree was constructed using the Neighbor-Joining (N-J) method available in MEGA 5.0 software (Tamura et al. 2011).
PtAP1-1 and PtAP1-2 gene expression analysis in P. tomentosa
RT-qPCR was used to obtain a transcriptomic profile of PtAP1-1 and PtAP1-2 genes in P. tomentosa. Total RNA was extracted as previously described (Chen et al. 2013) from (a) roots, stems and leaves of one-month-old tissue-cultured poplar plants, and (b) mature leaves, vegetative buds, female catkins, and female and male floral buds obtained from female and male adult trees located in the Beijing Forestry University nursery. Samples from mature trees were collected from June, 2012 through February, 2013 monthly, to cover major stages of floral development (An et al. 2010). Total RNA was treated with RQ1 DNase I (Promega, Madison, WI, USA) to remove any contaminating genomic DNA. First-strand cDNA was synthesized using 1.0 μg of DNase-treated total RNA, Superscript III (Invitrogen, Carlsbad, CA, USA) and oligo d(T)20 in a total volume of 20 μl. First-strand cDNA was diluted 1:10 with ddH2O, and 2 μl of the diluted cDNA was used as a template for RT-qPCR analysis. The RT-qPCR reaction was performed using SYBR® Premix Ex Taq™ (TaKaRa, Otsu, Japan) on a DNA Engine Opticon™ 2 system (MJ Research™). Specific primers used for RT-qPCR analysis are listed in Table S1. The PCR condition was 95 °C for 10 s, followed by 40 cycles of amplification (95 °C for 5 s, 60 °C for 20 s and 72 °C for 15 s), with a final extension of 7 min at 72 °C. The plates were read every 0.2 °C for 1 s from 70 to 95 °C, to generate the melting curves. The generated melting curve was employed as a significant parameter to check the specificity of the amplified fragment. All reactions were carried out in triplicate for technical and biological repetitions of the three individuals, respectively, and the generated real-time data were analyzed using the Opticon Monitor Analysis Software 3.1 tool. Data were presented as a mean ± SD. The efficiency of the primer sets was calculated by performing real-time PCR on several dilutions of first-strand cDNAs. Efficiencies of the different primer sets were similar. The PCR products were analyzed by agarose gel electrophoresis and sequencing to verify the presence of a gene-specific PCR product. Relative expressions of all transcripts were calculated using the 2−ΔΔCt method (Livak and Schmittgen 2001). The results obtained for the different tissues and stages we analyzed were standardized to the levels of a poplar ACTIN gene, which have stable expression in different tissues and in different developmental stages (An et al. 2011a, b).
Results
Sequence and phylogenetic analyses of AtAP1 and PtrAP1
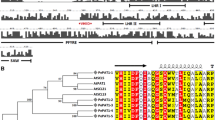
A comprehensive analysis of AP1 homologues in P. trichocarpa is presented in Table 1. Bioinformatics information for each gene includes the length of genomic DNA, length of the transcript, length of the CDS, number of amino acids, theoretical Mw and pI, and the location of the functional domains. Sequence alignment of AtAP1 (AT1G69120.1), PtrAP1-1 (Potri.008G098500.1) and PtrAP1-2 (Potri.010G154100.1) by BLAST indicated a 79.81 % identity to the proteins coded by AP1 homologous genes in Vitis vinifera (VvAP1, GSVIVG01012250001), Glycine max (GmAP1, Glyma16g13070.1) and Malus × domestica (MdAP1, MDP0000013331). The proteins all contain MADS-box, I, K-box, and C-terminal domains, which are typical for MIKC-type MADS-box proteins. The MADS-box domain was located at the N-terminus from 1 to 60 aa. A highly conserved K-box domain was located from 79 to 174 aa. An I domain was located between the MADS-box domain and K-box domain. All of the proteins contain a C-terminal conserved euAP1 motif (Litt and Irish 2003). MADS-box and K-box domains are highly conserved but sequences can be widely variable in the C-terminal domain (Fig. 1a). The euAP1 motif has two short conserved motifs, one LxLT/NLx (where “x” is an acidic residue) type of ERF-associated amphiphilic repression (EAR) motif, which is distinct for negative transcriptional regulation (Kagale et al. 2010), and one CFAA/T (farnesylation/prenylation motif) that terminates the protein (Litt and Irish 2003). The predicted tertiary structures of MADS-box domain in AtAP1 and PtrAP1 are similar, consisting of one α-helix and two β-sheets (Fig. 1b–d). The MADS-box domain is the most highly conserved of the four major MIKC-type MADS-box protein domains and has been widely studied across taxonomic kingdoms (Leseberg et al. 2006).
a Alignment of amino acid sequences of AP1 protein homologues from Arabidopsis, poplar, grapevine, pea and apple. The phytozome v9.1 accession number of each protein is as follows: AtAP1 (A. thaliana, AT1G69120.1), PtrAP1-1 and PtrAP1-2 (P. trichocarpa, Potri.008G098500.1 and Potri.010G154100.1, respectively), VvAP1 (V. vinifera, GSVIVG01012250001), GmAP1 (G. max, Glyma16g13070.1) and MdAP1 (M. × domestica, MDP0000013331). The gaps are attributed to the lack of amino acids. Conserved region, including a MADS-box domain, I domain, K domain, and C-terminal domain, is underlined. The euAP1 motif is double underlined. The first red box indicates an EAR motif within the euAP1 motif. The second red box indicates a prenylation motif. b The predicted tertiary structure of MADS-box domain in AP1 from A. thaliana contains an α-helix (pink) and two β-sheets (yellow). c The predicted tertiary structure of MADS-box domain in PtrAP1-1 from P. tomentosa contains an α-helix (pink) and two β-sheets (yellow). d The predicted tertiary structure of MADS-box domain in PtrAP1-2 from P. tomentosa contains an α-helix (pink) and two β-sheets (yellow)
The genomic sequence of AtAP1 is 3984 bp and consists of eight exons of 185, 79, 65, 100, 42, 42, 155 and 103 bp, encoding a putative protein of 256 amino acids. The genomic sequence length of PtrAP1-1 is 5341 bp and consists of eight exons of 185, 79, 65, 100, 42, 42, 116 and 97 bp, encoding a putative protein of 241 amino acids. The genomic sequence of PtrAP1-2 is 5867 bp and consists of eight exons of 185, 79, 65, 100, 42, 42, 134 and 100 bp, encoding a putative protein of 248 amino acids (Fig. S1a). The lengths of the first six exons in these three genes are the same while the other two are variable. The coding sequence of PtrAP1-1 cDNA exhibited 88.80 % identity to PtrAP1-2 cDNA at the nucleotide level, and the variation is mainly in the seventh and eighth exons (92.43, 92.41, 96.92, 90.00, 92.86, 92.86, 77.61, and 77.67 % for the exons 1–8, respectively). PtrAP1-1 is located on chromosome Scaffold 8, while PtrAP1-2 on Scaffold 10 (Fig. S1b).
In order to clarify the relationship among the homologous AP1/FUL proteins, a phylogenetic tree, based on the deduced amino acid sequences, was constructed using the Neighbor-Joining (N-J) method (Fig. 2). The resulting phylogenetic tree consisted of three major clades, represented by eudicots AP1, eudicots FUL, and monocots AP1/FUL-like. PtrAP1-1 and PtrAP1-2, together with AtAP1, were classified in the eudicots AP1 clade. They were more closely related to AP1-like members in Capsella rubella, Linum usitatissimum, Manihot esculenta, Vitis vinifera and Carica papaya. AP1/FUL-like proteins from the monocots Brachypodium distachyon, Setaria italic, Zea mays, and Oryza sativa were in another clade in the phylogenetic tree.
Phylogenetic analysis of AP1/FUL-like proteins. The tree was constructed using the Neighbor-Joining (N-J) method for the deduced amino acid sequence of members of the AP1/FUL from Arabidopsis thaliana, Populus trichocarpa, Malus × domestica, Capsella rubella, Linum usitatissimum, Manihot esculenta, Vitis vinifera, Carica papaya, Brachypodium distachyon, Setaria italic, Zea mays, Oryza sativa, Fragaria vesca, Gossypium raimondii, Medicago truncatula, Phaseolus vulgaris, Prunus persica and Solanum lycopersicum. The protein sequence data were obtained from Phytozome v9.1 database. Numbers on each branch indicate bootstrap values for 1000 replicates
Expression patterns of PtAP1-1 and PtAP1-2 in P. tomentosa
Expression profiles of PtAP1-1 and PtAP1-2 in various tissues of poplar in the juvenile phase and reproductive (adult) phase were obtained using RT-qPCR, with gene-specific primers (Table S1). Overall, the level of PtAP1-1 transcript was relatively higher than that of PtAP1-2. PtAP1-1 transcript accumulated mainly during the reproductive phase in tissues or organs of poplar, such as mature leaves, vegetative buds, male floral buds, female floral buds and female catkins (Fig. 3a). In contrast, the level of PtAP1-1 transcript was low in tissues of one-month-old poplar seedlings (juvenile phase), including roots and leaves, with little detection in stems (Fig. 3a). PtAP1-2 transcript was also mainly detected in the reproductive phase in various tissues and organs, except for mature leaves (Fig. 3b). Similar to PtAP1-1, expression of PtAP1-2 was very low or not detected in tissues, including roots, stems, and leaves, of one-month-old poplar seedlings (Fig. 3b).
Expression profiles of PtAP1-1 (a) and PtAP1-2 (b) in various tissues, as well as the seasonal expression patterns of PtAP1-1 and PtAP1-2 in developing male (c) and female (d) floral buds as determined by RT-qPCR. Samples for (a) and (b) from left to right are as follows: roots (R), stems (S) and leaves (L) from one-month-old tissue-cultured plantlets; mature leaves (ML), vegetative buds (VB), male floral buds (MFB), female floral buds (FFB) and female catkins (FC) from adult trees. Samples for male (c) and female (d) floral buds collected from late June through late January. A native poplar ACTIN gene was used as a reference gene for normalization. Data presented represent the mean ± SD
As indicated in Fig. 3c, d, the trend of PtAP1-1 and PtAP1-2 seasonal expression pattern in male or female floral buds was similar, however, the timing of expression was different. In developing male floral buds, the level of PtAP1-1 transcript was relatively higher than PtAP1-2 during most of the sampling period. The expression of PtAP1-1 and PtAP1-2 was upregulated rapidly during the summer. Expression of both genes peaked in August and decreased thereafter (Fig. 3c). On the other hand, the transcript level of PtAP1-1 was higher than PtAP1-2 in developing female floral buds prior to November, but the pattern subsequently differed from that observed in male floral buds (Fig. 3d). Expressions of PtAP1-1 and PtAP1-2 increased (more slowly than in male floral buds) during the summer and fall and both peaked in November and thereafter decreased. These results indicate that PtAP1-1 and PtAP1-2 are closely associated with the sexual differentiation of P. tomentosa floral organs.
Discussion
Sequence and phylogenetic analyses of AtAP1 and PtrAP1
MADS-box transcription factors play important roles in several aspects of plant growth and development, including the control of flowering time, meristem identity, floral organ identity, and the development of vegetative organs (Arora et al. 2007). AtAP1, PtrAP1-1 and PtrAP1-2 belong to the MADS-box family of transcription factors. The amino acid sequences of AtAP1, PtrAP1-1 and PtrAP1-2 share 81.10 % identity with each other and also have a high degree of identity with AP1 orthologues in Vitis vinifera, Glycine max and Malus × domestica, especially in the MADS-box domain. The MADS-box is a DNA-binding domain and is also involved in dimerization and the functional operation of transcription factors (Riechmann et al. 1996). The K domain is involved in protein–protein interaction and appears to be plant-specific (Davies et al. 1996). The I domain, located between the MADS-box and the K domain, is less conserved (Riechmann et al. 1996). Sequence differences occur mainly in the C-terminus region which is involved in transcriptional activation and ternary complex formation. The information provided in the current study supports the premise that the C-terminus domain of plant MADS-box proteins is the most variable (Egea-Cortines et al. 1999). While the euAP1-like motif in AP1-like proteins is well characterized, neither PtrAP1-1 or PtrAP1-2 contains the farnesylation motif “CFAA” in the C-terminus domain that was found to play an important role in the determining the function and specificity of AtAP1 in Arabidopsis (Litt and Irish 2003). PtrAP1-2 has a C-terminal amino acid motif “GYGA” instead of the farnesylation motif “CFAA” found in AtAP1 and a few other AP1 homologues. The motif “GYGA” is also found in SAP1-1 and SAP1-2 from Salix discolor (Fernando and Zhang 2006). Populus and Salix are both dioecious woody perennials with flowers that lack sepals and petals. The absence of the farnesylation motif “CFAA” is common to many AP1 homologues in several species, such as VvAP1 (Vitis vinifera), MdAP1 (Malus × domestica), GmAP1 (Glycine max), PEAM4 (Pisum sativum), and NtMADS11 (Nicotiana tobacum). These observations support the hypothesis that this kind of modification is not an essential factor in the function of AP1 (Berbel et al. 2001; Chi et al. 2011; Jang et al. 2002).
Comparison of the exon–intron structure of AtAP1 with PtrAP1-1 and PtrAP1-2 indicates that it contains the same number of exons and introns. The length of first six exons of these three genes is the same, indicating that exon length is highly conserved. The length of the gDNAs is largely different due to the length of introns which indicates that intron mutations may distinguish different AP1-like genes in different species. The phylogenetic tree analysis conducted in the present study characterized the relationship between AP1/FUL homologues from several eudicots and monocots. The analysis identified three different clades, designated as eudicots AP1, eudicots FUL, and monocots AP1/FUL-like proteins. AtAP1 from A. thaliana and PtrAP1-1 and PtrAP1-2 from P. trichocarpa are all placed in the eudicots AP1 clade. AtAP1 is most closely associated to the AP1 homologue from Capsella rubella which supports the premise that A. thaliana and C. rubella are closely related species. Additionally, the AP1 homologues from eudicots are well separated from those of the monocots. Our results are consistent with previous study that eudicots AP1 and eudicots FUL belong to core eudicot and relative distance is close to each other (Litt and Irish 2003). In the present phylogenetic tree, non-core eudicot FUL-like is not included and monocot clade is separated from core eudicot gene clade.
Populus trichocarpa is the first sequenced perennial woody plant. Since its public release (Tuskan et al. 2006) and subsequent updates in the phytozome (Goodstein et al. 2012), the availability of the Populus genome has spawned researches in plant molecular biology, morphology, ecology, comparative and functional genomics (Wullschleger et al. 2012). During the past decade, tree physiologists have used this resource in identifying candidate genes that underlie physiological and morphological traits of interest, and the structure, chromosomal location, phylogeny and function of these candidate genes in P. trichocarpa were well and deeply studied. In order to make a comprehensive analysis of AP1-like gene in poplar more concisely, we choose the sequence of AP1-like gene from P. trichocarpa. P. trichocarpa is mainly distributed in western North America and difficult to obtain in China. Previous study showed that all Populus species are closely related, and the sequence identity between homologous genes in different Populus species is often 99 % (Ingvarsson 2005). So, in the gene expression experiment, we choose plant samples from an indigenous tree species (P. tomentosa).
PtAP1-1 and PtAP1-2 expression profiles in P. tomentosa
In Arabidopsis, AP1 is initially expressed specifically in young floral primordia but not in inflorescence meristems, and later becomes localized to sepals and petals (Mandel et al. 1992). In the herbaceous plant, Dendranthema grandiflorum, the AP1 homologue, CDM111, is expressed in inflorescence meristems and developing bracts (Shchennikova et al. 2004). In the woody plant, Betula pendula, the AP1 homologue, BpMADS3, is expressed in both male and female inflorescence meristems (Elo et al. 2001). VAP1, an AP1 homologue in Vitis vinifera, is also expressed in inflorescence meristems (Calonje et al. 2004). In apple (Malus × domestica), the AP1 homologue, MdAP1, is expressed in leaf primordia, the upper cell layers of the shoot apex, inflorescence primordia, both young floral primordia and the inner cell layer of sepal primordia. It is also expressed in the outer cell layer of receptacle primordia, the floral axis, and in developing floral organ primordia (Kotoda et al. 2000; Mimida et al. 2011). In the present study, the expression patterns of PtAP1-1 and PtAP1-2 in P. tomentosa were similar in that they were highly expressed during the reproductive phase and low transcript levels were observed during the vegetative phase (Fig. 3).
In other species, expression profiles of AP1 homologues are somewhat variable. For example in Glycine max, the AP1 homologue, GmAP1, is specifically expressed in the flower, especially in sepals and petals, but not in other organs. RNA in situ hybridization analysis revealed that GmAP1 transcript can be detected in both the outer cell layers of both apical inflorescence meristems and lateral floral meristems of soybean (Chi et al. 2011). In contrast, we observed low levels of AP1 expressions in vegetative tissues or organs of P. tomentosa during the vegetative phase of growth (Fig. 3). Sepals and petals are absent in flowers of both Populus and Salix. In S. discolor, the expression profile of SAP1 (AP1 homologue) in various parts of male reproductive buds indicates that this gene is expressed in inflorescence meristems, bracts, and floral meristems (Fernando and Zhang 2006). In Prunus avium, PaAP1 (AP1 homologue) is highly expressed in petals, sepals, styles, and flower buds (Wang et al. 2013). However, AP1 homologue is expressed in all four whorls of floral organs in Prunus serrulata, Magnolia grandiflora and Persea americana while in all floral organs excluding stamens in P. persica (Kim et al. 2005; Zhang et al. 2008). These data indicate that AP1 homologues in different species may vary considerably in their expression and regulatory function.
RT-qPCR analysis revealed that PtAP1-1 and PtAP1-2 mRNAs were detected in both male and female floral buds at different developmental stages (Fig. 3). Similar observations for AP1 homologues have been reported in other species (Chi et al. 2011; Kotoda et al. 2010). Seasonal expression profiles of PtAP1-1 and PtAP1-2 in male and female floral buds exhibited a similar trend with initial increased expression levels and subsequent reduction. PtAP1-1 exhibited continuous differential expression in male and female floral buds. This indicated that it may play some roles during entire development phase of male and female floral buds. The relative level of PtAP1-1 and PtAP1-2 transcript in developing male floral buds was higher than that in developing female floral buds during June to August. In poplar, more stamen primordia exist in male floral buds than gynoecia primordia in female floral buds. The differences in the level of expression in the male and female floral buds may indicate that higher level of PtAP1 transcript is necessary for male floral buds development and stamen morphogenesis in P. tomentosa from primordial formation stage to enlargement stage. From September to November, the PtAP1-1 and PtAP1-2 mRNA were higher in female floral buds suggesting that a higher level of PtAP1-1 and PtAP1-2 mRNA is necessary to archespore formation in female flowers. A previous study reported that the level of expression of PtLFY (another floral meristem identity gene in P. tomentosa) in male floral buds was higher than that in female floral buds at different developmental stages from floral bud initiation to maturity (An et al. 2011a, b). PtAP1-1 and PtAP1-2 reached a peak in expression three months before in male floral buds than in female floral buds. An anatomical analysis of male and female floral buds in P. tomentosa revealed that initiation and differentiation of male floral buds occur earlier than female floral buds (An et al. 2010). Therefore, the differential expression of PtAP1-1 and PtAP1-2 in male and female floral buds is likely associated with the earlier development of male vs. female floral buds.
Flower induction in P. tomentosa in Beijing, China occurs in June (An et al. 2010). Expression of endogenous PtAP1-1 and PtAP1-2 was low during the period of floral induction in mature trees of P. tomentosa. These data indicate that PtAP1-1 and PtAP1-2 may have little effect on floral initiation. This premise is consistent with previous studies where overexpression of the Populus AP1 orthologue, PTAP1-1 did not induce early flowering in Populus (Strauss et al. 2004). Previous study showed that Like-AP1 (a tree ortholog of Arabidopsis AP1) mediates in photoperiodic control of seasonal growth cessation downstream of the CO/FT module in hybrid aspen and ectopic expression of Like-AP1 fails to induce early flowering in hybrid aspen trees (Azeez et al. 2014). Expressions of both PtAP1-1 and PtAP1-2 increase, however, during floral bud development, indicating that their expression is at least associated with floral bud development. Studies on Citrus plants indicated that the citrus AP1 homologue, CsAP1, has little effect on seasonal flowering of Satsuma mandarin (Citrus unshiu) but may affect floral bud development in trifoliate orange (Poncirus trifoliata) and kumquat (Fortunella crassifolia) (Nishikawa et al. 2007, 2009, 2011). In sweet orange, CsAP1 transcript increased at the end of the floral induction period which suggested that it was involved in floral organ development rather than floral induction (Pillitteri et al. 2004). In addition, seasonal patterns of AP1 homologue expression have been reported in other species such as apple and also in soybean (Chi et al. 2011; Kotoda et al. 2010).
In summary, we analyzed the gene structure, chromosomal location and phylogenetic relationship of Populus AP1 homologues and characterized their temporal and spatial expression profiles. Further studies to investigate the functions of these genes by overexpression, RNAi or using CRISPR-Cas system in transgenic P. tomentosa plants might provide more details about their roles in floral transition and development. To elucidate the molecular mechanism of flowering in poplar would be helpful for shortening the poplar breeding cycle and laying foundation to breed sterility poplar cultivars.
Author contribution statement
Xinmin An and Zhong Chen designed the experiment. Zhong Chen drafted the manuscript. Zhong Chen and Pian Rao performed the experiments. Zhong Chen, Xiong Yang and Xiaoxing Su analyzed the data. Kai Gao and Bingqi Lei helped improve the manuscript. All authors read and approved the final manuscript.
References
Abe M et al (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309:1052–1056
An XM, Wang DM, Wang ZL, Wang JC, Cao GL, Bo WH, Zhang ZY (2010) Expression profile of PtLFY in floral bud development associated with floral bud morphological differentiation in Populus tomentosa. Sci Silvae Sin 46:32–38
An XM, Wang DM, Wang ZL, Li B, Bo WH, Cao GL, Zhang ZY (2011a) Isolation of a LEAFY homolog from Populus tomentosa: expression of PtLFY in P. tomentosa floral buds and PtLFY-IR-mediated gene silencing in tobacco (Nicotiana tabacum). Plant Cell Rep 30:89–100
An X, Ye M, Wang D, Wang Z, Cao G, Zheng H, Zhang Z (2011b) Ectopic expression of a poplar APETALA3-like gene in tobacco causes early flowering and fast growth. Biotechnol Lett 33:1239–1247
Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8:242
Azeez A, Miskolczi P, Tylewicz S, Bhalerao RP (2014) A tree ortholog of APETALA1 mediates photoperiodic control of seasonal growth. Curr Biol 24:717–724
Berbel A, Navarro C, Ferrandiz C, Canas LA, Madueno F, Beltran JP (2001) Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J 25:441–451
Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312:1040–1043
Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275:80–83
Calonje M, Cubas P, Martinez-Zapater JM, Carmona MJ (2004) Floral meristem identity genes are expressed during tendril development in grapevine. Plant Physiol 135:1491–1501
Chen Z et al (2013) A Novel Moderate Constitutive Promoter Derived from Poplar (Populus tomentosa Carriere). Int J Mol Sci 14:6187–6204
Chi Y, Huang F, Liu H, Yang S, Yu D (2011) An APETALA1-like gene of soybean regulates flowering time and specifies floral organs. J Plant Physiol 168:2251–2259
Corbesier L et al (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033
Davies B, Egea-Cortines M, de Andrade Silva E, Saedler H, Sommer H (1996) Multiple interactions amongst floral homeotic MADS box proteins. EMBO J 15:4330–4343
Egea-Cortines M, Saedler H, Sommer H (1999) Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J 18:5370–5379
Elo A, Lemmetyinen J, Turunen ML, Tikka L, Sopanen T (2001) Three MADS-box genes similar to APETALA1 and FRUITFULL from silver birch (Betula pendula). Physiol Plantarum 112:95–103
Fernando DD, Zhang S (2006) Constitutive expression of the SAP1 gene from willow (Salix discolor) causes early flowering in Arabidopsis thaliana. Dev Genes Evol 216:19–28
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Goodstein DM et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186
Hsu CY, Liu Y, Luthe DS, Yuceer C (2006) Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18:1846–1861
Hsu CY et al (2011) FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc Natl Acad Sci USA 108:10756–10761
Hsu CY et al (2012) Overexpression of CONSTANS homologs CO1 and CO2 fails to alter normal reproductive onset and fall bud set in woody perennial poplar. PLoS One 7:e45448
Ingvarsson PK (2005) Nucleotide polymorphism and linkage disequilibrium within and among natural populations of European aspen (Populus tremula L., Salicaceae). Genetics 169:945–953
Jang S, An K, Lee S, An G (2002) Characterization of tobacco MADS-box genes involved in floral initiation. Plant Cell Physiol 43:230–238
Kagale S, Links MG, Rozwadowski K (2010) Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152:1109–1134
Kaufmann K et al (2010) Orchestration of floral initiation by APETALA1. Science 328:85–89
Kim S et al (2005) Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. Plant J 43:724–744
Kotoda N, Wada M, Komori S, S-i Kidou, Abe K, Masuda T, Soejima J (2000) Expression pattern of homologues of floral meristem identity genes LFY and AP1 during flower development in apple. J Am Soc Hortic Sci 125:398–403
Kotoda N et al (2010) Molecular characterization of FLOWERING LOCUS T-like genes of apple (Malus × domestica Borkh.). Plant Cell Physiol 51:561–575
Leseberg CH, Li A, Kang H, Duvall M, Mao L (2006) Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene 378:84–94
Litt A, Irish VF (2003) Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165:821–833
Liu C, Xi W, Shen L, Tan C, Yu H (2009) Regulation of floral patterning by flowering time genes. Dev Cell 16:711–722
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408
Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360:273–277
Mimida N et al (2011) Expression patterns of several floral genes during flower initiation in the apical buds of apple (Malus × domestica Borkh.) revealed by in situ hybridization. Plant Cell Rep 30:1485–1492
Mohamed R et al (2010) Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J 62:674–688
Nishikawa F, Endo T, Shimada T, Fujii H, Shimizu T, Omura M, Ikoma Y (2007) Increased CiFT abundance in the stem correlates with floral induction by low temperature in Satsuma mandarin (Citrus unshiu Marc.). J Exp Bot 58:3915–3927
Nishikawa F, Endo T, Shimada T, Fujii H, Shimizu T, Omura M (2009) Differences in seasonal expression of flowering genes between deciduous trifoliate orange and evergreen Satsuma mandarin. Tree Physiol 29:921–926
Nishikawa F, Iwasaki M, Fukamachi H, Nonaka K, Imai A, Endo T (2011) Seasonal changes of citrus Flowering Locus T gene expression in kumquat. Bull Natl Inst Fruit Tree Sci 12:27–32
Parcy F, Bomblies K, Weigel D (2002) Interaction of LEAFY, AGAMOUS and TERMINAL FLOWER1 in maintaining floral meristem identity in Arabidopsis. Development 129:2519–2527
Pillitteri LJ, Lovatt CJ, Walling LL (2004) Isolation and Characterization of LEAFY and APETALA1 Homologues from Citrus sinensis L. Osbeck ‘Washington’. J Am Soc Hortic Sci 129:846–856
Ratcliffe OJ, Amaya I, Vincent CA, Rothstein S, Carpenter R, Coen ES, Bradley DJ (1998) A common mechanism controls the life cycle and architecture of plants. Development 125:1609–1615
Ratcliffe OJ, Bradley DJ, Coen ES (1999) Separation of shoot and floral identity in Arabidopsis. Development 126:1109–1120
Riechmann JL, Krizek BA, Meyerowitz EM (1996) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA 93:4793–4798
Shchennikova AV, Shulga OA, Immink R, Skryabin KG, Angenent GC (2004) Identification and characterization of four chrysanthemum MADS-box genes, belonging to the APETALA1/FRUITFULL and SEPALLATA3 subfamilies. Plant Physiol 134:1632–1641
Shen L, Chen Y, Su X, Zhang S, Pan H, Huang M (2012) Two FT orthologs from Populus simonii Carrière induce early flowering in Arabidopsis and poplar trees. Plant Cell Tiss Organ Cult 108:371–379
Strauss SH, Rottmann WH, Brunner AM, Sheppard LA (1995) Genetic engineering of reproductive sterility in forest trees. Mol Breed 1:5–26
Strauss SH, Brunner AM, Busov VB, Ma C, Meilan R (2004) Ten lessons from 15 years of transgenic Populus research. Forestry 77:455–465
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tuskan GA et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Wagner D, Sablowski RW, Meyerowitz EM (1999) Transcriptional activation of APETALA1 by LEAFY. Science 285:582–584
Wang J, Zhang X, Yan G, Zhou Y, Zhang K (2013) Over-expression of the PaAP1 gene from sweet cherry (Prunus avium L.) causes early flowering in Arabidopsis thaliana. J Plant Physiol 170:315–320
Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78:203–209
Wellmer F, Riechmann JL (2010) Gene networks controlling the initiation of flower development. Trends Genet 26:519–527
Wullschleger SD, Weston DJ, DiFazio SP, Tuskan GA (2012) Revisiting the sequencing of the first tree genome: Populus trichocarpa. Tree Physiol 33:357–364
Zhang L, Xu Y, Ma R (2008) Molecular cloning, identification, and chromosomal localization of two MADS box genes in peach (Prunus persica). J Genet Genomics 35:365–372
Zhang HL et al (2010) Precocious flowering in trees: the FLOWERING LOCUS T gene as a research and breeding tool in Populus. J Exp Bot 61:2549–2560
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (2011AA100201, 2013AA102703), the Project of National Natural Science Foundation of China (31170631, J1103516), the Changjiang Scholars and Innovative Research Team Program of China (IRT13047) and the Fundamental Research Funds for the Central Universities (BLYJ201410).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Wang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11738_2015_1805_MOESM1_ESM.tif
Fig. S1 Schematic representation and physical location of the structure of AtAP1, PtrAP1-1 and PtrAP1-2 genes. a Schematic representation of the structure of AtAP1, PtrAP1-1 and PtrAP1-2 genes in Arabidopsis and poplar. Exons and untranslated regions (UTRs) are indicated as black and gray boxes, respectively, and lines between boxes indicate introns. b Physical locations of PtrAP1-1 and PtrAP1-2 genes on the genomic scaffolds (chromosomes). The scale bar represents 2.0 Mb. Scaffold numbers and sizes (Mb) are indicated below each scaffold (TIFF 425 kb)
Rights and permissions
About this article
Cite this article
Chen, Z., Yang, X., Su, X. et al. Identification and expression analysis of APETALA1 homologues in poplar. Acta Physiol Plant 37, 50 (2015). https://doi.org/10.1007/s11738-015-1805-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-015-1805-z