Abstract
Diabetes mellitus, often known as hyperglycemia, is a serious worldwide disease now. In clinical pharmacology, the dipeptidyl peptidase IV (DPP-4) enzyme is important for glucose homeostasis. The clinical DPP-4 blockers are essential oral antidiabetic medications used as alternate treatment following metformin inability as insulinotropic drugs with no inherent risk of hypoglycemia. The objective of this study is to create novel and potent DPP-4 inhibitors by molecular hybridization of eight clinically licensed DPP-4 inhibitors. Molecular hybridization process led to the creation of five novel hybridized DPP-4 inhibitors, which preliminary computational studies suggest may exhibit improved selectivity compared to authorized DPP-4 inhibitors. The pharmacokinetic features of the hybridized inhibitors, including their solubility and potential to pass through biological tissues, were evaluated using Lipinski’s rule of five and other druglikeness filters, indicating favorable properties for reaching the DPP-4 active site. Furthermore, the possible toxicity of suggested inhibitors was investigated using basic toxicity filters and PASS, indicating no immediate red flags regarding their potential toxicity and metabolism. In addition, a mechanism for synthesizing the proposed compounds has been developed via machine learning and artificial intelligence algorithms. At the biomolecular level, using the Gromacs package, molecular dynamics simulations (100 ns) were performed for all the studied systems. Following analyzing the molecular dynamics trajectories and evaluating the dynamic shifts of DPP-4 after its molecular interactions with the designed compounds via dynamic cross-correlation matrix, free energy landscape and MM-PBSA calculations, all data show that the proposed DPP-4 inhibitors create extremely stable complexes when compared to the clinical DPP-4 inhibitor (alogliptin). Finally, the findings of this study might greatly contribute to the development of novel and potent DPP-4 inhibitors and assist in the search for new medications for diabetes type 2.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is critical to regulate insulin secretion in order to maintain euglycemia. Hyperglycemia develops in type 2 diabetes due to a decrease in insulin production and the development of peripheral insulin resistance. Throughout the fasting state, insulin is biologically generated to a limited level with the objective of increasing glucose absorption via peripheral tissues. Following eating, insulin production is rapidly and significantly increased in order to keep plasma glucose quantities within a restricted physiological limit (Gerich 2003). In addition to the rise in glucose quantities, the intestinal hormones gastric.
inhibitory polypeptide (GIP) and glucagon-like peptide-1 (GLP-1) also contribute to the post-prandial regulation of glucose. Under hyperglycemic circumstances, both of these hormones enhance insulin production and account for 70% of post-prandial insulin generation. They are known as incretin hormones because of its crucial physiological in nature role in increasing post-prandial secretion of insulin (Creutzfeldt 1979; NaucK et al. 1993; Nauck and Meier 2018). Because the GLP-1 reliant increase of insulin production occurs only under hyperglycemic circumstances, the inherent risk of hypoglycemia is minimal. GLP-1 has additional favorable function in type 2 diabetes that helps to maintain euglycemia: Glucagon secretion is overly elevated in type 2 diabetes, and glucagon boosts hepatic glucose synthesis (Nauck et al. 1993). GLP-1 suppresses glucagon secretion in hyperglycemic situations, lowering blood sugar levels, and also is a peptide hormone with a few minutes' plasma half-life (Drucker and Nauck 2006). In addition, the fast enzymatic breakdown of GLP-1 by the enzyme dipeptidyl peptidase IV (DPP-4) (Figure 1) is responsible for the limited pharmacological half-life (Mentlein et al. 1993). Bioactive small molecules have the ability to inhibit DPP-4, and when DPP-4 inhibitors are taken orally, endogenous GLP-1 content rises 2–3 times (Holst and Deacon 1998).
The existing DPP-4 inhibitors have a high effectiveness in blocking DPP-4, and DPP-4 is suppressed by 80–90% in clinical circumstances. This suppression results in a twofold to threefold increase in post-prandial GLP-1 levels in the blood, which facilitates the glucose-dependent stimulation of insulin production and a decrease in the release of glucagon (Deacon 2019; Sesti et al. 2019). The oral administration of DPP-4 inhibitors is quite potent, and the pharmacologic and pharmacokinetic properties lead to therapeutically adequate DPP-4 inhibition with once-day treatment (vildagliptin is necessary administered twice daily) (Gallwitz 2016). In phase III clinical trial activities, DPP-4 inhibitors had favorable safety as well as tolerability features, with nasopharyngitis and skin lesions being the most common side effects seen (Scheen 2018).
One of the modern drug discovery technologies is the bioinformatics or computer-aided drug design (CADD) strategy. The breed de novo hybridization strategy analyzes structural data and uses the precise positions of two compounds to reconstitute fragments from each one to make a unique molecule (Pierce et al. 2004). The newly discovered compound will most likely become a hybrid of the two scaffolds or just an addition of features from one scaffold to a different one (Patel et al. 2021). Furthermore, they are not limited to integrating two separate scaffolds, numerous molecules produced following only two breed crossing repetitions, combining scaffold and secondary chain elements from a maximum of four of the lead molecules show no resemblance to any of the original ligand configurations (Schmitt et al. 2002). On other hand, the methodology for drug development overall is a challenging task for organic biologists owing to the intricacy of the pharmacophore characteristic that enhances the property and activity of a medication (Usha et al. 2017).
Because DPP-4 quickly cuts and blocks the incretin hormones (GLP-1 and GIP), which are required for glucose control, its inhibition has been studied as a means of regulating glycemia in diabetes by preserving the diminished incretin function. Based on this knowledge, we used the breed de novo hybridization strategy to design and novel inhibitors of dipeptidyl peptidase IV. Following this, we assessed the pharmacokinetics of the suggested compounds and confirmed their potential toxicity. In addition, we investigate the stability of generated complexes by molecular dynamics simulation (MDS), MM-PBSA calculation, dynamic cross-correlation matrix (DCCM) and free energy landscape (FEL).
Materials and methods
Breed de novo hybridization
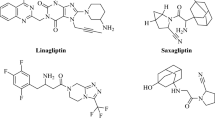
For the present study, we employed Maestro software (version 11.8, Schrödinger, USA). The chemical structures of inhibitors against a certain target must be exploited in breed-based de novo drug development. There are eight DPP-4 inhibitors available in Europe, the USA, Japan, and Korea, including vildagliptin, saxagliptin, alogliptin, linagliptin, sitagliptin, gemigliptin, anagliptin, and teneligliptin (Makrilakis 2019; Gallwitz 2019). Figure 2 illustrates the chemical structures of all the clinical DPP-4 inhibitors mentioned. In the first stage, various fragments have been produced from these clinical inhibitors through the Schrodinger power shell command prompt with the run./fragment_molecule.py script and supplying the folders (Patel et al. 2021). SP docking was used to dock the created fragments within DPP-4 active site, and the fragments with the best score were subsequently linked permanently via the breed ligand creation tool (Lotfi et al. 2023a). The breed panel conducted molecular hybridization by taking into account three distinct characteristics. Initially, the two bonds must possess the same level of order (for instance, a single bond and a double bond cannot be regarded as a suitable match.) This criterion is essential for preserving the hybridization and geometry of the atoms that are bound in the newly formed molecule. Furthermore, the atoms located at both ends of the bond must be at a distance of 1 Å from one other. In addition, the angle formed by the bond vectors of the two bonds should not exceed 15˚ (Patel et al. 2021; Lotfi et al. 2023a). If we conceptualize the original molecules as being divided into two halves at the bond that matches, then one newly formed molecule will consist of the first half of molecule one and the second half of molecule two. The other novel molecule is composed of the latter portion of molecule one and the former portion of molecule two. The new molecules consist of atoms that possess the same atom kinds, locations, and bonds as their equivalent atoms (Pierce et al. 2004; Lotfi et al. 2023a). Finally, Figure 3 represents the breed de novo hybridization technique employed in this study.
Docking modeling
Docking modeling was used to investigate the interactions of breed ligands within DPP-4 active site using the docking system GLIDE (Schrodinger Inc., USA, 2018). The atomic configuration of dipeptidyl peptidase IV (DPP-4) in complex with alogliptin was extracted from Protein Data Bank, PDB ID: 3G0B at a resolution of 2.25 Å and no mutations. The protein was utilizing the protein preparation wizard in the Maestro program, and then the receptor was refined for docking experiments (Venkatesan et al. 2018). In addition, the water molecules in the protein were eliminated and hydrogen’s were supplied, and the energy was minimized via OPLS3e (Optimized potential for liquid simulation) force field. By centering the co-crystallized ligand, the grid box was attached to the DDP-4 protein (Schiering et al. 2011). XP docking was executed to analyze the interactions of the new breed inhibitors with the active site of dipeptidyl peptidase IV in order to conduct thorough investigation and confirm the breed hybridization design outcomes. Subsequently, the eight clinical DDP-4 inhibitors were docked in the DDP-4 receptor as extra reference ligands to assess the inhibitory power of the breed ligands.
We created a decoy of the vildagliptin molecules using the DUDE server (https://dude.docking.org/generate) to validate the docking results and de novo hybridization process.
ADMET and pharmacokinetic features
A good medicine requires a precise balance of pharmacological function, pharmacokinetics, and tolerability. A desired absorption, distribution, metabolism, excretion, and toxicity characteristics, in addition to high effectiveness and selection, are crucial to the successful development of a medicine candidate (Cumming et al. 2013; Hou and Wang 2008). ADMETLAB 2.0 was created with the Python web platform Django and deployed on an Aliyun elastic computing service operating an Ubuntu Linux server (Xiong et al. 2021). All designed inhibitors were converted to SMILES form using Chemdraw Ultra, and the SMILES structures were then submitted to the ADMET lab 2.0 website. It additionally allows it to be easy to use the JMSE editor to generate the appropriate configurations. The submit button was utilized for providing the information while establishing the SMI structure, and it provided ADMET characteristics in pdf file and spreadsheet format, which were able to be retrieved after some time.
Biomolecular activity prediction through PASS
The breed inhibitors were tested for antidiabetic activity spectrum employing the free webserver PASS, which can be found with http://www.pharmaexpert.ru/passonline/ (Lagunin et al. 2000). Based on the structural configuration, this website is able to forecast the therapeutic benefits of a chemical, and its forecast may be examined using the proportion of likelihood to be active (Pa) to probability to be inactive (Pi) (Borkotoky et al. 2021). With possibilities, the Pa and Pi values vary from zero to one, and are generally Pa + Pi ≠ 1, as these possible outcomes are widely foreseen. As a result, biological activities with Pa > Pi are only considered plausible for a specific pharmacological compound.
Molecular dynamic simulation
In this stage, Gromacs-2023 package was used to execute molecular dynamics simulations of protein-ligand complexes for 100 ns. According to the results of molecular docking and pharmacokinetic features, the best complexes were selected for this investigation. In addition, the reference complex (DDP-4_Alogliptin) was then simulated in the same conditions to analyze the biomolecular dynamics of the formed complexes. The SwissParam webserver was consulted to generate the topology parameters for the breed ligands (Zoete et al. 2011). Furthermore, the DDP-4 topology files have been extracted using the CHARMM27 all-atom force field (Lotfi et al. 2023b). After the compilation of topology data, each complex was solvated in a cubic box using the TIP3P water model, also the ions (Na+ and Cl-) have been added to neutralize the charge. For each neutralized-biomolecular system, the energy was minimized using a gradient descent approach until the maximal force was less than 10.0 kJ/mol.
The equilibration phase is a vital component of any molecular dynamics simulation. Before collecting data can commence, each system must be re-equilibrated to ensure that it is stable and representative. Equilibration was done in two phases in the simulation, stressing the need to attain both temperature (NVT) and pressure (NPT) equilibration (Ke et al. 2022). The NVT equilibration phase involved coupling each complex using a v-rescale approach at 300 K for 100 ps with a coupling coefficient of 0.1 ps. The NPT was subsequently equilibrated for 100 ps using a Berenson pressure-coupling system with a coupling constant of 2.0 ps (Bourougaa et al. 2023a).
Molecular dynamics simulations using Gromacs software are an important tool for research in a variety of domains since they provide valuable information on the behavior of biomolecules and their dynamic changes. The primary metrics were computed in order to analyze the biomolecular stability of the designed inhibitors within the DDP-4 receptor and their impact on its vital and essential functions. Root mean square deviations (RMSD) were calculated to assess the conformational stability of systems, and the relative fluctuations of protein amino acids were determined using root mean square fluctuations (RMSF). In addition, the radius of gyration (Rg) of any configuration is used to determine the general compactness. Finally, we calculated the solvent-accessible surface area (SASA), which is a measurement of the surface area of a protein that solvent can access.
Dynamic cross-correlation matrix (DCCM)
The dynamic cross-correlation matrix is a crucial tool for investigating the relationship between the motions of specific combinations of atoms or residues in a protein structure. The values in this matrix vary from (-1), indicating complete anticorrelation and (+1) show optimal correlation (Hoang et al. 2024). It is worth noting that the numbers over the diagonal of the matrix remain (+1), owing to the fact that an atom’s movement is fully associated with itself. Between two atoms i and j, a covariance matrix characterizing the correlated features of atomic movements was built. The cross-correlation may be calculated using the equation below (Kumari et al. 2022):
Here, Δri and Δrj are molecular motion vectors denoting atoms, i and j from their average position in terms of period interval.
Free energy landscape (FEL)
An effective sampling strategy was used to assess the free energy landscape of DDP-4's dynamic features. The RMSD and Rg of DDP-4 were selected as two reaction coordinates to generate a two-dimensional energy landscape diagram. The following formula was used to compute the energy landscape across these two reaction coordinates (Al-Khafaji and Tok 2020; Pathak et al. 2023):
where Kb denotes the Boltzmann constant, T is the model's temperature, and ρ(p1,p2) is the standardized joint distribution of probabilities.
Binding free energy calculation
Molecular mechanics–Poisson–Boltzmann surface area provides a complete study of the quantitative evaluation of the interaction pathway between DDP-4 and the developed inhibitors. The binding energy components were computed via the g_mmpbsa package's MM-PBSA method (Kumari and Kumar 2014). Furthermore, the g_mmpbsa system calculates the binding energy of the receptor-ligand complex using the following formula (Mohammad et al. 2020):
where GComplex symbolizes the binding complex’s total free energy and Gprotein and Gligand are the DDP-4 and breed inhibitors' total free energies, respectively.
Results and discussion
De novo hybridization and docking modeling
In this research, alogliptin was selected as a reference dipeptidyl peptidase IV (DPP-4) inhibitor. Initially, all eight clinical inhibitors were docked via extra precision (XP) Glide docking to the enzyme active site to estimate their inhibition power. Sitagliptin and linagliptin have the highest binding affinities for DDP-4, with − 7.994 and − 7.919 kcal/mol, respectively (Table 1). Thus, they form the most stable complexes. The gemigliptin molecule subsequently binds to the enzyme’s active site with a binding affinity of − 6.970 kcal/mol. Furthermore, the reference compound alogliptin has a binding energy of − 6.729 kcal/mol to DPP-4 receptor. Finally, the remaining clinical inhibitors interact to the enzyme with binding affinities ranging from − 5.958 to − 4.324 kcal/mol by forming the least stable complexes.
The breed de novo hybridization methodology was used in the present research to create potent and novel DDP-4 inhibitors. Employing the Schrödinger PowerShell system, 191 distinct fragments were created from the initial clinical DDP-4 inhibitors. In the first stage, we docked all 191 generating fragments in the DPP-4 receptor using standard precision (SP) docking. The top121 fragments (with a standard precision score greater than -5 kcal/mol) were chosen for the breed de novo hybridization strategy in the next step. This hybridization method is currently working to completely computerize the design approach, which might greatly speed up its execution and yield superior outcomes. The chemical formulae (Smiles) of the top 121 fragments with SP docking scores greater than − 5 kcal/mol can be seen in Table S1. Additionally, each unique breed molecule was chosen for a very accurate docking (extra precision) procedure into the DDP-4 receptor. Consequently, the top five breed inhibitors have a breeding score ranging from 8.647 to 15.499 (Fig. 4). The Breed 1 was developed via hybridization of anagliptin (F23) and sitagliptin (F18), Breed 2 was generated through hybridization of anagliptin (F27) and sitagliptin (F18), Breed 3 was obtained via hybridization of gemigliptin (F27) and anagliptin (F26), Breed 4 was obtained through hybridization of sitagliptin (F 18) and anagliptin (F 33), and finally, Breed 5 was obtained via hybridization of gemigliptin (F 9) and anagliptin (F 26).
Molecular docking is a computer approach for automatically identifying the conformation of protein–ligand interactions (Bourougaa et al. 2023b). Docking modeling provides data on the binding affinity and binding area of the designed inhibitors to the DDP-4 protein. The docking scores of the breed compounds were contrasted to those of the clinical inhibitor alogliptin. The docking findings have been verified by extracting the DDP-4 co-crystallized compound and re-docking it into the identical location. Using the DockRMSD website, the docked conformation and co-crystallized ligand have an RMSD value of 1.476 Å (Bell and Zhang 2019). The first finding from the docking study reveals that the five hybrid molecules form highly stable complexes with the DDP-4 receptor with XP scores ranging between − 9.139 and − 10.365 kcal/mol, when compared to all clinical inhibitors including the reference ligand (alogliptin). Table 2 contains all results obtained via molecular docking investigations.
The binding affinity of Breed 1 for the DDP-4 active site forms the most stable complex, with an XP score of − 10.365 kcal/mol. It was observed that this hybridized compound has the capacity block the critical function of DDP-4 enzyme and form H-bonds with Glu205 and Glu206. Furthermore, it interacts with Ser30, His470 via halogen bonds and with Ser630, Tyr662 and Tyr666 via hydrophobic interactions. We conclude that the interaction with a halogen bond (similar to Breed 2 and Breed 4) is necessary to inhibit the biological function of the DDP-4 enzyme by forming very stable complexes with it, as compared to alogliptin, which does not react via a halogen bond. Furthermore, the developed compounds interacted strongly and favorably with the essential amino acid residues in the DDP-4 receptor in a way that was similar to alogliptin, but with additional hydrophobic interactions with Ser630, Tyr662, Tyr666, Val656, Val711, Phe357, and Tyr547 amino acids. Figure 5 depicts the molecular binding interactions of the five hybridized molecules and alogliptin with the active site of DDP-4. The results of docking modeling show that all of the created compounds bonded with the binding pocket. They are expected to be new and potent DPP-4 inhibitors due to their interact inside the receptor region formerly occupied by alogliptin. Finally, despite the fact that all of the ligands occupied an identical binding location, the greater number of halogen bonds in Breeds 1, 2, and 3 allows them to interact more effectively than the other compounds.
To enhance the robustness and reliability of our findings, we conducted a study on a set of decoy compounds to serve as a control group for our fragment-based and docking analyses. This approach will help to validate the specificity and accuracy of the identified active compounds, ensuring that the observed interactions are not merely random but rather indicative of true biological relevance.
Using the vildagliptin molecule, we produced ten decoy inhibitors (Table S2) using the DUDE platform to confirm the de novo procedure and docking process. These decoy inhibitors produced 508 fragments. In addition, all of these fragments were docked to the protein receptor via SP-docking. Table S3 shows the top sixteen generated fragments with an affinity score of greater than − 5 kcal/mol.
Following the completion of the breeding process across the best fragments, 23 new decoy inhibitors were developed. Table S4 represents the docking results of the best-breed decoy inhibitors. In addition, the binding energies of decoy inhibitors ranged from − 6.567 to -5.484 kcal/mol. The binding energies (XP score) of decoy molecules have been demonstrated to be greater when compared to reference molecules, demonstrating the reliability of our approach for de novo hybridization and docking processes.
ADMET and pharmacokinetics analysis
In silico, ADMET and pharmacokinetics analysis helps in understanding how hybridized molecules interact with the human body. Lipinski's rule of five was used to analyze the druglikeness of the hybridized molecules, which comprises metrics such as H-bond donors, H-bond acceptors, molecular weight, topological polar surface area (TPSA) and Nb rotatable bonds.
Firstly, it should be noted that the five discovered inhibitors have molecular weights ranging from 143.110 to 363.130 g/mol, allowing for easier intestine absorption if administered orally. Furthermore, they exhibit LogP (partition coefficient) values ranging from − 0.972 to 2.043, indicating that all designed compounds may traverse biological membranes and are highly soluble in aqueous cellular conditions. About absorption characteristics, all of the molecules had excellent outcomes for Caco-2 permeability and MDCK permeability (varying between 4.6e-06 and 6.9e-05 cm/s), these findings suggest that the new molecules will enter the blood circulation with sufficient quantities.
In terms of distribution, the molecules Breeds 2 and 4 have a greater volume distribution (2.605 and 2.189 L/kg) than the rest of the compounds, implying that these two will arrive in sufficient quantities for the DDP-4 protein. The pharmacokinetic and druglikeness profiles of designed DDP-4 inhibitors are shown in Table 3.
At the liver level, specifically at cytochrome P450 enzymes, the molecules Breeds 1 and breed 2 minimize the enzymatic activity of CYP2C9 and CYP2D6, but the remainder of the molecules is not inhibiting the critical activities of other liver enzymes. As for the excretion process, at the nephron level, all molecules will be eliminated easily and in a short time because of their brief presence in the human body. The half-lives of the proposed molecules range from 0.079 to 0.745 hours.
However, the five discovered DDP-4 inhibitors, on the other hand, are not expected to have any hERG blockers, ames toxicity, carcinogenicity, drug-induced liver injury or toxic features. Finally, all five suggested compounds are in line with Lipinski, Pfizer, and GSK regulations and do not produce pain in the human body.
Retrosynthesis, a powerful organic chemistry approach, is utilized to construct effective synthetic routes for molecule complexes. Synthetic pathway assembler (SPAYA), a recently developed tool, automates the retrosynthesis operation via the use of machine learning and artificial intelligence algorithms. In addition, using SPAYA throughout the synthesis of complex molecules will conserve both money and time. Likewise, SPAYA aids in the identification of potential problems in synthesis strategies, including the formation of undesired by-products, enabling method adjustment and optimization. The molecules Breed 1 was selected for this investigation. Figure 6 shows a complete compilation of the results collected for the predicted ligand (Breed 1). The results show that the identified inhibitors are simple to synthesize in a chemical laboratory, making it easier to evaluate their inhibitory action for dipeptidyl peptidase IV in vitro and in vivo.
In the phase 1, a mixture consisting of 0.724 mole of 1-(2,4,5-trifluorophenyl)propan-2-one, 0.724 mole of paraformaldehyde, 0.724 mole of N-methyl-1-phenylmethanamine, and 7.5 mL of concentrated HCl in 100 mL of ethanol had been refluxed for 2 hours. Following the integration of an additional quantity of paraformaldehyde (0.724 mole), the mixture was refluxed for another 2 hours. 75 ml of acetone was added, agitated for 1 hour at 0 °C. Finally, the produced solid was filtered and washed with acetone. In the next stage (phase 2), a solution of 42.15 g (0.137 mole) of 4-(benzyl(methyl)amino)-1-(2,4,5-trifluorophenyl)butan-2-one dissolved in 221 ml of a MeOH-water (1:1) combination was added, along with 9.68 g of Pd over 5% carbon (56.5% water). At atmospheric pressure, the mixture was hydrogenated for 1 hour. The catalyst was filtered, and the solvent was completely evaporated. The final product was recrystallized from acetone to provide 28.6 g (96%) of the molecule 4-(methylamino)-1-(2,4,5-trifluorophenyl)butan-2-one as a white crystalline solid. Finally, and at the phase 3, the Breed 1 molecule was synthesized via asymmetric hydrogenation catalysts.
Computational evaluation of antidiabetic (type 2) activity
A vast number of studies initiatives appear to possess been abandoned since serious adverse effects and toxicity are unidentified and these unfavorable consequences are discovered or appear much too late. In contrast, in contemporary times, it is feasible to anticipate over 3700 pharmacological consequences and other biological features of substances using PASS, a simple internet platform. Table 4 displays the PASS outcomes, which were labeled as Pa and Pi. PASS prediction for antidiabetic (type 2) activity of compounds Breeds 1–5 was found to be 0,102 < Pa < 0,568. In addition, this indicated that the molecules Breeds 1, 2, and 3 had antidiabetic (type 2) features. Based on PASS results and ADMET properties, all five generated compounds were chosen for molecular dynamic simulation to assess and analyze their ability to block the vital function of dipeptidyl peptidase IV as antidiabetic (type 2) drugs.
Molecular dynamic simulation (MDS)
Molecular dynamics examination is a critical tool for understanding the dynamics and molecular interactions of molecules during typical or pathological environments. Several criteria, including root-mean-square deviation, root-mean-square fluctuation, radius of gyration, and solvent-accessible surface area, are useful to evaluate the results of this investigation. RMSD is used to calculate the average shift of molecular configurations throughout simulation, allowing for evaluation of biomolecular structural stability over different times. RMSF estimates the degree of atom changes, offering insight on molecular mobility. Lower RMSF values suggest greater atomic stability. In addition, Rg provides for the observation of structural modifications over the simulation time and assesses molecular compactness. Finally, significant changes in SASA might signal changes in the molecule's interactions with another system, which may impair its pharmacological activity.
In this research, the stability of the five hybridized molecules (Breeds 1–5) and alogliptin within the DDP-4 receptor was analyzed during a 100 ns of simulation. The biomolecular stability of the five breed molecules was evaluated using the data from Table 5, which displayed all of the average values (RMSD, RMSF, Rg and SASA) acquired from this investigation.
Initially, the average RMSD value across all docked complexes, including Alogliptin, remained between 0.172 and 0.319 nm. Nevertheless, in the identical simulation, the DPP4_Breed 5 complex demonstrated substantially more stable conduct with an average RMSD value of 0.172 nm when compared to other complexes (which comprise the reference complex having an average RMSD value of 0.249 nm). The DPP-4_Breed 1 complex had the maximum RMSD value (> 2 nm) at 26 ns, after that, the complex remained stable at 0.3 ns until the simulation ended. In addition, the DPP-4_Breed 2 complex exhibited an upward dynamic from 10 to 35 ns while remaining stable throughout the simulation with an average RMSD of 0.261 ns. In this simulation, the DPP-4_Breed 3 complex displayed fluctuating dynamics. When compared to other RMSD profiles, the behavior from 30 to 85 ns was more stable, with an average RMSD value of 0.372 ns. Finally, the DPP-4_Breed 4 complex exhibited unstable atomic mobility with an average RMSD of 0.391 nm at the period between 0 and 30 ns. The RMSD backbone atoms profiles of the developed inhibitors and alogliptin within the DPP-4 receptor are displayed in Figure 7.
When determining the flexibility of a protein, the root-mean-square fluctuation (RMSF) of each amino acid in a particular frame configuration is compared to the average conformation. RMSF analyses show that the residues in the proposed inhibitors are quite stable, with averages of 0.131, 0.122, 0.127, 0.124, 0.122, and 0.123 nm for DPP-4_Breed 1–5 and alogliptin, respectively. It is important to note that the RMSF profiles of all generated complexes and the reference molecule are very similar. Furthermore, all complexes show significant fluctuations in the molecular level, up to 0.8 nm (near atom 3400).
Some discernible fluctuations were seen in the RMSF data around the 2500, 5500, and 9000 atoms, in Breeds 1, 3, and 4 showing the dynamic changes within 0.5 nm. The RMSF backbone atoms profiles of the designed inhibitors and Alogliptin are presented in Figure 8.
Radius of gyration (Rg) was used to determine the geometric compactness of protein complexes. The results showed that the Rg values of all the docked conformations showed little variation from 2.5 to 2.75 nm, with average values of 2.735, 2.731, 2.721, 2.725, and 2.725 nm for DPP-4_Breed 1–5 complexes and 2.732 nm for the DPP-4_Alogliptin complex. Consequently, the Rg values indicate that, following interaction with hybridized molecules, the protein folding compactness remains mostly unchanged. Nevertheless, there are slight variations in the protein's levels following binding with Breeds 1 and 3.
This implies that all complexes formed between protein and hybridized molecules are structurally stable during 100 ns of simulation. The Rg profiles of the proposed inhibitors and alogliptin are depicted in Figure 9.
SASA signifies the protein region that is highly vulnerable to interaction with neighboring solvent molecules. Thus, the change in solvent-accessible surface area for the six formed complexes is plotted in Figure 10. The DPP-4_Breed 1–5 and alogliptin complexes average SASA values from 100 ns were 330.14, 328.12, 326.49, 326.75, 329.48, and 331.22 nm2, respectively. In addition, all of these findings indicate that none of the complexes appears to have considerable variation. As a result, the data are conclusive evidence of minimal change in the biomolecular structure of DPP-4. More notably, minor shifts were discovered when comparing the clinical inhibitor (alogliptin) to all of the developed compounds.
The findings of molecular dynamics study for the newly hybridized molecules demonstrate a satisfactory RMSD when compared to the clinical DPP-4 Inhibitor. Furthermore, with a few exceptions for residues larger than 0.4 nm, the RMSF remained stable and similar to the clinical inhibitor. The Rg and SASA were stable throughout the trajectory, suggesting that the complexes were stable and compact. According to these findings, the hybridized molecules can block the enzymatic activity of DPP-4 and maintaining high biomolecular stability.
Dynamic cross-correlation matrix (DCCM)
To investigate the modifications in the internal dynamics of DPP-4 caused by the interactions with the designed molecules and the clinical DPP-4 inhibitor (alogliptin), the cross-correlation coefficients were computed employing the atomic coordinates Cα. Figure 11 shows the cross-correlation maps for the six examined complexes. In addition, positive correlation (PC) movements are represented by blue, whereas anticorrelated (AC) movements are depicted by red, with the color indicating the intensity of correlation. Notably, the off-diagonal zones show the relative shifts of the various residues, whereas the diagonal regions represent the dynamics of a specific residue relative to itself (Wang et al. 2023). Following the biomolecular interactions of DPP-4 and the clinical inhibitor alogliptin (Figure 11F), the diagonal zones (Z)1, (Z)2, (Z)3, and the off-diagonal (Z)4 zone exhibit strongly positive correlation movements (PC), whereas (Z)5 and (Z)6 zones cause significantly anticorrelated (AC) movements. As demonstrated in Figure 11A, the presence of Breed 1 within the DPP-4 active site not simply intensifies the PC motion in the diagonal zones but also increases the AC movement in the (Z)5 and (Z)6 zones.
The binding of Breed 2 to DPP-4 (Figure 11B) increases the positive correlation motions in the (Z)1, (Z)2 and (Z)3, and it adds anticorrelated motions in the (Z)5 and (Z)6 compared to breed 1. At the internal dynamic level, when Breeds 3 and 4 interact with the DPP-4 active site (Figure 11C and D)), the DPP-4 molecular structure stays very comparable to the effects of the clinical inhibitor on the DPP-4 protein. This supports the notion that DPP-4_Breed 3 and 4 complexes have high molecular structural stability. The interaction of Breed 5 with DPP-4 (Figure 11E) not just clearly reduces the correlation motions in the (Z)1, (Z)2, (Z)3, and (Z)4; however it also reduces the anticorrelated motions in the (Z)5 and (Z)6. These findings show that the molecular binding of the discovered compounds to DPP-4 has no effect on the structure of DPP-4 as opposed to the clinical inhibitor. As a result, it is obvious that the molecular hybridization process is useful for creating novel antidiabetic drugs.
Free energy landscape (FEL)
Molecular dynamics simulations are useful for modeling the free energy landscape (FEL) and investigating the molecular folding behavior of a certain protein at an atomic level (Singh et al. 2019). The FEL of the six DPP-4 complexes was constructed for DPP-4 backbone atoms by combining the root mean square deviation (RMSD) and radius of gyration (Rg), both expressed in nanometers. The outcomes of the FEL analysis are shown in Fig. 12. In the DPP-4_Breed 1 configuration, it demonstrated confirmation stability (ΔG = 0) with an RMSD and Rg of 0.2 and 2.72 nm, respectively, whereas the DPP-4_Breed 2 and 3 configurations had the lowest free energy conformations and were centered around (RMSD = 0.17 nm, Rg = 2.7 nm). Moreover, Breed 4 within DPP-4 has shown confirmation stability via an RMSD of 2.7 nm and an Rg of 0.17 nm, respectively. On other hand, it showed confirmation stability (ΔG = 0) in the DPP-4_Breed 5 configuration, with an RMSD and Rg of 0.16 and 2.7 nm. Finally, the DPP-4 clinical inhibitor (Alogliptin) with DPP-4 illustrated confirmed stability, with RMSD of 0.17 and Rg of 2.73 nm. These findings imply that the hybridized molecules can bind to the DPP-4 receptor and inhibit its biological activities without changing or affecting its biomolecular structure. They additionally possess a very similar effect to the clinical inhibitor alogliptin. Notably, our research showed that the five complexes were stable and maintained their energy minima, indicating that the amino acids constituting the active site would interact with inhibitors on a regular basis.
Binding free energy calculations
MM-PBSA is a useful method for assessing covalent bond stability and identifying the more stable compounds (Jin et al. 2020). Through the execution of the molecular mechanics–Poisson–Boltzmann surface area (MM-PBSA) technique, the energy contributions from the developed systems were further examined. The total binding free energy of each formation is calculated as the combination of van der Waals and electrostatic interactions, NP interactions in a solvated system, NP contribution of repulsive solute-solvent interactions to the solvation energy, total gas phase MM energy and total solvation energy. The total binding energies (Table 6) of the five designed inhibitors were calculated as − 60.43, − 60.94, − 48.52, − 36.38, and − 36.38 kcal/mol for Breeds 1–5, respectively. In addition, the total binding energy of alogliptin was -37.45 kcal/mol. The results show that the five hybridized DPP-4 inhibitors have negative binding energies, indicating a better binding selectivity. Notably, the contributions of ΔEEL and ΔEVDW are negative, indicating positive interactions among the developed inhibitors and the DPP-4. The MM-PBSA results show that the proposed inhibitors limit the biological activities of DPP-4 via the biomolecular interactions, generate relatively stable complexes, and offer therapeutic promise for the treatment of type 2 diabetes.
Using MM-PBSA technique, the binding free energy was decomposed into the contributions from every residue in order to provide a better understanding of the mechanism of binding. In all systems, the amino acids that contributed the most to the binding free energy were Asp624, Glu166, and Glu167. Figure 13 depicts their energy contributions. In addition, it was discovered that Glu166 contributed the most to the created systems, with binding energies ranging from − 19.65 to − 10.58 kcal/mol. whereas, in the DPP-4_Alogliptin system, Glu166 has a binding energy of − 5.85 kcal/mol. In the five developed systems, Glu167 amino acid provided the most binding energy, ranging from -18.21 to − 8.27 kcal/mol. While in the DPP-4_Alogliotin system, this amino acid offered − 5.24 kcal/mol for the binding energy. Finally, all of this data shows that the hybridized inhibitors create very stable complexes with the DPP-4 receptor when compared to the clinical complex. This clearly shows that the designed inhibitors can help in the development of novel and potent medications for treating type 2 diabetes.
While the computational studies, including docking scores and molecular dynamics (MD) simulations, provide valuable insights into the potential binding affinity and stability of the newly designed inhibitors, it is important to acknowledge that these results are preliminary. The correlation between calculated and experimental binding energies is not straightforward, as indicated by studies such as Rifai et al. (Rifai et al. 2019). Which reported a correlation coefficient of only 0.64 between MM/PBSA-generated values and experimental data. Therefore, experimental synthesis and testing of these compounds are essential to confirm their actual binding energies, pharmacokinetic properties, and overall efficacy.
Conclusion
DPP-4 blockers are currently supplanting sulfonylureas as insulinotropic medications and are also an acceptable therapeutic substitution for alternative therapies such as glitazones or glucosidase blockers. In this article, we employed a molecular hybridization strategy to develop novel and potent DPP-4 inhibitors with favorable pharmacokinetic characteristics by combining the molecular features of eight clinical DPP-4 inhibitors currently on the market in Europe and the USA. Our research led to the identification of five promising hybridized DPP-4 inhibitors.
Preliminary computational studies, including molecular docking and molecular dynamics simulations (RMSD, RMSF, Rg, and SASA), suggest that these compounds form stable complexes with DPP-4 receptors compared to the clinical inhibitor alogliptin. The DCCM and FEL data indicate that the suggested compounds maintain the initial configurations of the DPP-4 structure, unlike alogliptin, which causes substantial changes upon interaction. MM-PBSA calculations further indicated that all complexes formed between DPP-4 and the new compounds exhibit favorable binding free energies and atomic mobility.
Pharmacokinetic profiles, estimated using Lipinski's rule of five and similar druglikeness filters, suggest these molecules have suitable properties for entering the circulatory system. Basic toxicity filters and PASS predictions indicated no immediate red flags regarding potential toxicity.
Although these computational findings are promising, experimental validation is necessary to confirm the efficacy, safety, and pharmacokinetic properties of these proposed DPP-4 inhibitors. We believe the findings of this study provide a foundation for the development of effective type 2 diabetes drugs.
Data availability
The datasets used in the manuscript are publicly available from the repositories below:
A dataset of eight known DPP-4 inhibitors including vildagliptin, saxagliptin, alogliptin, linagliptin, sitagliptin, gemigliptin, anagliptin, and teneligliptin were used in the manuscript. [link to the repository: https://www.frontiersin.org/journals/endocrinology/articles/https://doi.org/10.3389/fendo.2019.00389/full] and deposited from article https://doi.org/https://doi.org/10.3389/fendo.2019.00389. Repository Name: RCSB Protein Data Bank; Deposited Date: 2009-01-27; Released Date: 2010-02-16; by source author(s): Zhang, Z., Wallace, M.B., Feng, J., Stafford, J.A., Kaldor, S.W., Shi, L., Skene, R.J., Aertgeerts, K., Lee, B., Jennings, A., Xu, R., Kassel, D., Webb, D.R., Gwaltney, S.L. Number: https://doi.org/https://doi.org/10.2210/pdb3G0B/pdb ; Macromolecular structure : 3G0B [link to the repository: https://www.rcsb.org/structure/3G0B ] and originally deposited from article, https://doi.org/https://doi.org/10.1021/jm101016w. All other data generated during the current investigation are described in the manuscript
References
Al-Khafaji K, Tok TT (2020) Molecular dynamics simulation, free energy landscape and binding free energy computations in exploration the anti-invasive activity of amygdalin against metastasis. J Comput Methods Programs Biomed 195:105660
Bell EW, Zhang Y (2019) DockRMSD: an open-source tool for atom mapping and RMSD calculation of symmetric molecules through graph isomorphism. J Cheminform 11(1):40
Borkotoky S, Banerjee M, Modi GP, Dubey VK (2021) Identification of high affinity and low molecular alternatives of boceprevir against SARS-CoV-2 main protease: a virtual screening approach. J Chem Phys Lett 770:138446
Bourougaa L, Ouassaf M, Shtaiwi A (2023a) Discovery of novel potent drugs for influenza by inhibiting the vital function of neuraminidase via fragment-based drug design (FBDD) and molecular dynamics simulation strategies. J Biomol Struct Dyn 28:1–15
Bourougaa L, Ouassaf M, Khan SU (2023b) Comparative molecular field analysis (CoMFA), molecular docking and ADMET study on thiazolidine-4-carboxylic acid derivatives as new neuraminidase inhibitors. J Acta Chim Slov 70:333–344
Creutzfeldt W (1979) The incretin concept today. J Diabetologia 16:75–85
Cumming JG, Davis AM, Muresan S, Haeberlein M, Chen H (2013) Chemical predictive modelling to improve compound qualityn. J Nat Rev Drug Discov 12(12):948–962
Deacon CF (2019) Physiology and pharmacology of DPP-4 in glucose homeostasis and the treatment of type 2 diabetes. J Front Endocrinol 10:80
Drucker DJ, Nauck MA (2006) The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. J Lancet 368(9548):1696–1705
Gallwitz B (2016) Novel therapeutic approaches in diabetes. J Endocr Dev 31:43–56
Gallwitz B (2019) Clinical Use of DPP-4 Inhibitors. J Front Endocrinol 10:389
Gerich JE (2003) Contributions of insulin-resistance and insulin-secretory defects to the pathogenesis of type 2 diabetes mellitus. J Mayo Clin Proc 78(4):447–456
Hoang SH, Lam EM, Dao H (2024) Computational assessment of gut microbiota metabolite enterolactone as a promising Aβ42 inhibitor in Alzheimer’s disease. J D L S 5:9–20
Holst JJ, Deacon CF (1998) Inhibition of the activity of dipeptidyl-peptidase IV as a treatment for type 2 diabetes. J Diabetes 47(11):1663–1670
Hou T, Wang J (2008) Structure-ADME relationship: still a long way to go? J Expert Opin Drug Metab Toxicol 4(6):759–770
Jin Z, Wang Y, Yu X, Tan Q, Liang S, Li T, Zhang H, Shaw P, Wang J, Hu C (2020) Structure-based virtual screening of influenza virus RNA polymerase inhibitors from natural compounds: molecular dynamics simulation and MM-GBSA calculation. J Comput Biol Chem 85:107241
Ke Q, Gong X, Liao S, Duan C, Li L (2022) Effects of thermostats/barostats on physical properties of liquids by molecular dynamics simulations. J Mol Liq 365:120116
Kumari R, Kumar R (2014) g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962
Kumari M, Singh R, Subbarao N (2022) Exploring the interaction mechanism between potential inhibitor and multi-target Mur enzymes of mycobacterium tuberculosis using molecular docking, molecular dynamics simulation, principal component analysis, free energy landscape, dynamic cross-correlation matrices, vector movements, and binding free energy calculation. J Biomol Struct Dyn 40(24):13497–13526
Lagunin A, Stepanchikova A, Filimonov D, Poroikov V (2000) PASS: prediction of activity spectra for biologically active substances. J Bioinfo 16(8):747–748
Lotfi B, Mebarka O, Alhatlani BY, Abdallah EM, Kawsar SMA (2023a) Pharmacoinformatics and breed-based de novo hybridization studies to develop new neuraminidase inhibitors as potential anti-influenza agents. J Mol 28(18):6678
Lotfi B, Mebarka O, Khan SU, Htar TT (2023b) Pharmacophore-based virtual screening, molecular docking and molecular dynamics studies for the discovery of novel neuraminidase inhibitors. J Biomol Struct Dyn 19:1–13
Makrilakis K (2019) The role of DPP-4 inhibitors in the treatment algorithm of type 2 diabetes mellitus: when to select, what to expect. Int J Environ Res Public Health 16(15):2720
Mentlein R, Gallwitz B, Schmidt WE (1993) Dipeptidyl-peptidase IV hydrolyses gastric inhibitory polypeptide, glucagon-like peptide-1(7–36)amide, peptide histidine methionine and is responsible for their degradation in human serum. J Eur Biochem 214(3):829–835
Mohammad T, Siddiqui S, Shamsi A, Alajmi MF, Hussain A, Islam A, Faizan Ahmad Md, Hassan I (2020) Virtual screening approach to identify high-affinity inhibitors of serum and glucocorticoid-regulated kinase 1 among bioactive natural products: combined molecular docking and simulation studies. Molecules 25(4):823. https://doi.org/10.3390/molecules25040823
Nauck MA, Meier JJ (2018) Incretin hormones: their role in health and disease. J Diabetes Obes Metab 20:5–21
Nauck MA, Kleine N, Orskov C, Holst JJ, Willms B, Creutzfeldt W (1993) Normalization of fasting hyperglycaemia by exogenous glucagon-like peptide 1 (7-36 amide) in Type 2 (non-insulin-dependent) diabetic patients. J Diabetologia 36(8):741–744
NaucK MA, Bartels E, Orskov C, Ebert R, Creutzfeldt W (1993) Additive insulinotropic effects of exogenous synthetic human gastric inhibitory polypeptide and glucagon-like peptide-1-(7–36) amide infused at near-physiological insulinotropic hormone and glucose concentrations. J Clin Endocrinol Metab 76(4):912–917
Patel HM, Shaikh M, Ahmad I, Lokwani D, Surana SJ (2021) BREED based de novo hybridization approach: generating novel T790M/C797S-EGFR tyrosine kinase inhibitors to overcome the problem of mutation and resistance in non small cell lung cancer (NSCLC). J Biomol Struct Dyn 39(8):2838–2856
Pathak RK, Kim W, Kim J (2023) Targeting the PEDV 3CL protease for identification of small molecule inhibitors: an insight from virtual screening, ADMET prediction, molecular dynamics, free energy landscape, and binding energy calculations. J Biol Eng 17(1):29
Pierce AC, Rao G, Bemis GW (2004) BREED: Generating novel inhibitors through hybridization of known ligands. application to CDK2, p38, and HIV protease. J Med Chem 47(11):2768–2775
Rifai EA, van Dijk M, Vermeulen NPE, Yanuar A, Geerke DP (2019) A comparative linear interaction energy and MM/PBSA Study on SIRT1–ligand binding free energy calculation. J Chem Inf Model 59(9):4018–4033. https://doi.org/10.1021/acs.jcim.9b00609
Scheen AJ (2018) The safety of gliptins : updated data in 2018. J Expert Opin Drug Saf 17(4):387–405
Schiering N, D’Arcy A, Villard F, Simic O, Kamke M, Monnet G, Hassiepen U, Svergun DI, Pulfer R, Eder J, Raman P, Bodendorf U (2011) A macrocyclic HCV NS3/4A protease inhibitor interacts with protease and helicase residues in the complex with its full-length target. J Proc Natl Acad Sci USA 108(52):21052–21056
Schmitt S, Kuhn D, Klebe G, Schmitt S (2002) A new method to detect related function among proteins independent of sequence and fold homology. J Mol Biol 323(2):387
Sesti G, Avogaro A, Belcastro S, Bonora BM, Croci M, Daniele G, Dauriz M, Dotta F, Formichi C, Frontoni S, Invitti C, Orsi E, Picconi F, Resi V, Bonora E, Purrello F (2019) Ten years of experience with DPP-4 inhibitors for the treatment of type 2 diabetes mellitus. J Acta Diabetol 56(6):605–617
Singh A, Somvanshi P, Grover A (2019) Pyrazinamide drug resistance in RpsA mutant (∆438A) of Mycobacterium tuberculosis: dynamics of essential motions and free-energy landscape analysis. J Cell Biochem 120(5):7386–7740
Usha T, Shanmugarajan D, Goyal AK, Kumar CS, Middha SK (2017) Recent updates on computer-aided drug discovery: time for a paradigm shift. J Curr Top Med Chem 17(30):3296–3307
Venkatesan A, Rambabu M, Jayanthi S, Dass JFP (2018) Pharmacophore feature prediction and molecular docking approach to identify novel anti-HCV protease inhibitors. J Cell Biochem 119(1):960–966
Wang L, Lu D, Wang Y, Xu X, Zhong P, Yangb Z (2023) Binding selectivity-dependent molecular mechanism of inhibitors towards CDK2 and CDK6 investigated by multiple short molecular dynamics and free energy landscapes. J Enzyme Inhib Med Chem 38(1):84–99
Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, Yin M, Zeng X, Wu C, Lu A, Chen X, Hou T, Cao D (2021) ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. J Nucleic Acids Res 49(1):5–14
Zoete V, Cuendet MA, Grosdidier A, Michielin O (2011) SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem 32(11):2359–2368
Acknowledgements
The Researchers would like to thank the Deanship of Graduate Studies and Scientific Research at Qassim University for financial support (QU-APC-2024-9/1).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bourougaa, L., Ouassaf, M. & Alhatlani, B.Y. Clinical informatics and molecular hybridization of established clinical DPP-4 inhibitors to generate next-level diabetes type 2 drugs. Chem. Pap. (2024). https://doi.org/10.1007/s11696-024-03697-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11696-024-03697-8