Abstract
The pivotal role of MRP3 protein in acute leukaemia and the efficacy of natural compounds in cancer treatment have necessitated the current study to identify novel MRP3 inhibitors from natural source. The MRP3 protein was modelled and validated using well-accepted metrics, after which a validated multiple-ligand pharmacophore model (AHHHR_4) was built to screen natural compounds (n = 47,964). The combined pharmacophore screening with molecular docking was conducted to identify the hits drug-like compounds using ADMET profiling. The electronic behaviour of this set of compounds in gas phase was examined using density functional theory. Among the compounds (n = 7) with clean ADMET profile, NPC5486, which possessed the highest binding affinity with MRP3, was further subjected to 50 ns molecular dynamics (MD) simulation to understand its dynamics of binding. Analysis from the resulting MD simulation trajectories of NPC5486 in complex with the model protein alongside that of standard inhibitor (vincristine) showed not only the flexibility and interaction potential of the residues of the MRP3 with NPC5486 as indicated by the RMSF but also stability of the complex as indicated by the RMSD, RoG and number of hydrogen bonds of the ligand–protein complexes. Cluster analysis of the MD simulation trajectory files and dynamics-based MMGBSA computations further revealed that the observed interactions with important residues as well as the free energy contribution per residue were preserved in the dynamic environment. Overall, the current study has revealed drug-like compounds which can serve as potential inhibitors of MRP3 in the treatment of acute leukaemia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute leukaemia stems from a cascade of mutational events (unchecked proliferation and abnormal maturation) that occur during the intricate process of haematopoiesis leading to decreased production of blood cells (Maynadié 2015). Over the years several forms of leukaemia have been described in humans; however, acute myelogenous leukaemia (AML) and acute lymphoblastic leukaemia (ALL) have been the most reported cases (Maynadié 2015). According to the American Cancer Association, an estimated 13,800 cases of AML and 6,000 cases of ALL were reported in the USA in 2012 (Maynadié 2015). The figure was expected to hit a total of 61,090 cases in 2021, with an estimated mortality of 23,660 people (Howlader et al. 2017). In a bid to combat the current morbidity and mortality challenge posed by leukaemia, several studies have been carried out to develop a chemotherapeutic regimen for the treatment of leukaemia. While this was able to achieve major health improvements in the past, there have been several instances of treatment failure in childhood and adult acute leukaemia due to drug resistance (Van Der Kolk et al. 2001).
Drug resistance in acute leukaemia has been linked to overexpression of adenosine triphosphate (ATP)-binding cassette transporters (Van Der Kolk et al. 2001). ATP binding cassette (ABC) proteins consist of families of transporters that drive the movement of different chemical compounds across the biological membrane using energy liberated from ATP hydrolysis (Zhang et al. 2015). Generally, the ABC transport proteins can translocate many substrates, including lipids, ions, nucleotides and drug metabolites, across the membrane. Of particular importance are the subfamily C of the ABC transporters, which includes twelve (12) members, many of which have shown the capacity to efflux cytostatic drugs. Among the subfamily C of the ABC proteins is another group of transporter called multidrug-resistant proteins (MRPs) reported in previous studies to mediate multidrug resistance via active extrusion of chemotherapeutic substances (Hollenstein et al. 2007; Zhang et al. 2015). About seven (7) different types of MRPs have been discovered in humans (Borst et al. 2000; Scheffer et al. 2002; Zhang et al. 2015).
Among the chemotherapeutic agents used in the treatment of AML are daunorubicin, idarubicin, etoposide and mitoxantrone, which are substrates for MRPs. In spite of Fletcher et al. (2016) findings that some MRPs play significant roles in tumour cell resistance to anti-cancer agents, only a few studies have evaluated their prognostic significance in leukaemia. These studies have however noted that MRP3 among other members of the MRP family is associated with high mortality in acute leukaemia. There is also the fact that current studies have indicated that MRP3 is the only ABC transporter that correlates with poor prognosis of AML and ALL regardless of the patient's age (Steinbach and Legrand 2007). In light of these studies, MRP3 may be worth studying in order to develop drugs aimed at ameliorating acute leukaemia.
Located on chromosome 17q22, the MRP3 gene encodes 1527 amino acids (Uchiumi et al. 1998). To date, the three-dimensional structure of MRP3 protein has not been crystallized in spite of its interesting biological features. However, gene and protein sequencing studies have provided reasonable insights that can assist in elucidating the protein structure. MRP1 shows 58% amino acid identity to MRP3; as such, it can be used as an ideal template for modelling MRP3 protein structure. The resulting membrane proteins have different glycosylation patterns and are 170 and 190 kDa in size (Noroozi-Aghideh et al. 2020). MRP3 is thought to be large and contains two large cytoplasmic domains (nucleotide binding domains 1 and 2) (Fukushima-Uesaka et al. 2007). Based on this structural information, previous studies have attempted to develop potential MRP3 inhibitors for the treatment of acute leukaemia. Despite the significant prospects of MRP3 inhibitors in cancer therapeutics, there is currently no clinically established and approved MRP3 inhibitor. However, studies have shown the significant prospects of therapeutic agents including benzbromarone, vincristine and MK571 as MRP3 inhibitors which may improve cancer cell sensitivity to chemotherapeutic drugs (Ali et al. 2017; Gilibili et al. 2017; Song et al.2010). Yet, most of these drugs are rather non-specific and elicit toxic side effects such as peripheral neuropathy, hepatotoxicity and renal failure (Akioka et al. 2013; van de Velde et al. 2017; Ye et al. 2010). Leveraging on the premise that natural compounds have shown profound potential in modulating the activities of specific ABC transporters, including MRP3, in various living tissues (Bobrowska-Hägerstrand et al. 2006; De Vocht et al. 2021; Wu et al. 2005), the current study attempts to identify potential inhibitors of MRP3 by conducting a high-throughput virtual screening of different natural compounds libraries.
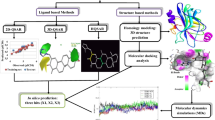
Apparently, one of the drawbacks to the development of MRP3 inhibitors is the lack of sufficient information regarding the morphology of the protein structure. As such, in this study, homology modelling was performed to model the structure of the protein. Using a hybrid of structure-based and ligand-based computational screening (Fig. 1), the current study unveils lead-like compounds with significant prospects as MRP3 inhibitors.
Materials and methods
Software and computational tools
Molecular docking simulation as well as pharmacophore modelling and validation was performed using the Maestro suite available on Schrodinger software version 2017. Molecular dynamics simulation and free energy calculations were performed using NAMD software. Homology modelling, protein structure validation and pharmacokinetic and toxicity properties prediction were achieved using different web servers and discussed in details in subsequent subsections.
Sequence alignment and Homology modelling
The FASTA sequence of multidrug-resistant protein 3 containing 1527 amino acid residues was retrieved from UniProt online database (https://www.uniprot.org/) with UniProt id: 015,438. The fasta file was used for the modelling of 3D structure of MRP3 protein based on sequence similarity to known structures using SWISS-MODEL web server (https://swissmodel.expasy.org/). The homology modelling was performed based on the following step; template search, sequence alignment, model building and evaluation. Template search for the target sequence was carried out with Basic Local Alignment Search Tool (BLAST) and hidden Markov models (HMMs): 'HMM-HMM-based lightning-fast iterative sequence search' (HHblits; http://toolkit.genzentrum.lmu.de/hhblits/) against the updated SWISS-MODEL library (Remmert et al. 2011; Studer et al. 2021). The template with the highest sequence identity and best coverage was used to model the protein structure of the query sequence. Model was built based on the alignment between the target and the template using ProMod3 (Studer et al. 2021). The coordinates that are conserved between the target and template are copied from the template to the model. The fragment library was used to remodel insertions and deletions. Following this, side chains were rebuilt. In the final step, the geometry of the model was regularized by using a force field. The global and per-residue model quality was assessed using the QMEAN scoring function (Benkert et al. 2008). The accuracy of the tertiary structure of the generated model was estimated using Global Model Quality Estimate (GMQE) score (Waterhouse et al. 2018). In addition, torsional angles of the amino acid residues contained in the protein were assessed using Ramachandran plot.
Structure refinement and Model Validation
The generated protein from PDB was imported into Maestro workspace for structural refinement to remove steric clashes and fill in missing side chains and loops. Initially, energy minimization was performed with the Prime tool of Maestro that uses VSGB solvation model and OPLS3 force field. Subsequently, loop refinement was performed on the non-template region of the model protein to refine loops that fall within the disallowed regions of the Ramachandran plot. Loop refinement was achieved using Prime ultra-extended sampling option, which performs Ab-initio loop modelling with an implicit solvent system. The protein built was finally subjected to protein preparation using the protein preparation wizard of Maestro as discussed previously (Olawale et al. 2022). The quality of the modelled protein was initially evaluated by ERRAT using the structure analysis and verification server (SAVES) (https://saves.mbi.ucla.edu/). ERRAT measures the quality of a protein by assessing non-bonded atomic interactions. Error values are plotted as a function of position and the overall quality of the protein is expressed as a percentage, where a higher value indicates a higher quality of protein. Further protein validation was performed using ProTSAV (http://www.scfbio-iitd.res.in/software/proteomics/protsav.jsp) which is a meta-server that performs consensus quality assessment using different validation tools including Verify3D, MolProbity, Procheck, ProSA, ERRAT, ProQ, dDFire, Naccess and D2N. The overall quality of the modelled is given as a ProTSAV score which is a cumulative estimate of the individual protein quality score from the different validation tools. Finally, quantitative analysis of the final model was performed using volume, area, dihedral angle reporter (VADAR) (http://vadar.wishartlab.com/) and RESPROX (Resolution by proxy) (http://www.resprox.ca/) score values (Berjanskii et al. 2012; Willard et al. 2003).
Creation of ligand library, compound preparation and Ligand filtering
An in-house library of natural compounds was created by combining compounds retrieved from NPASS natural compound database (26,240) (http://bidd.group/NPASS/), Chinese medicinal compound database (20,712) (http://tcm.cmu.edu.tw/) and South African medicinal compound database (1012) (https://sancdb.rubi.ru.ac.za/). The entire structures were imported into the workspace of maestro to prepare the ligands for subsequent computational simulations. Creation of in-house database was achieved using the generate phase database panel of Maestro in a three step process. In the first step, the compounds from the different databases were merged and duplicates were skipped by generating a unique canonical SMILES string for each structure and comparing the strings. Since the retrieved compounds might not have chirality information, explicit hydrogen atoms or be in the appropriate ionization state for physiological conditions, the compounds were prepared to obtain structures that are suitable for database searching following the procedure described previously (Olawale et al. 2021a). Finally, the prepared compounds were subjected to QikProp to generate topological descriptors which was consequently used to pre-filter the compounds based on the Lipinski rule.
Pharmacophore modelling, Pharmacophore validation and Phase screening
Multiple-ligand pharmacophore was generated from a set of 12 experimentally active inhibitors of MRP3 protein (Köck et al. 2014). Pharmacophore modelling was carried out in four steps, viz: ligand preparation, creating pharmacophore sites from a set of features, finding common pharmacophore and pharmacophore validation. The 2D structures of the compounds were downloaded from PubChem database, and the compounds were prepared using LigPrep to generate 3D conformers of each structure. In the create sites step, a set of chemical structure patterns was used to identify pharmacophore features in each ligand. The general features include hydrogen bond donor (HBD), hydrogen bond acceptor (HBA), hydrophobic group (HG), negative charge (N), positive charge (P) and aromatic ring (AR). Subsequently, a search for common pharmacophores among the set of high-affinity (active) ligands was performed. The search proceeds by enumerating all pharmacophores of a given variant and partitioning them into successively smaller high-dimensional boxes according to their intersite distances. Boxes that contain pharmacophores from the minimum required number of ligands are said to survive the partitioning process. Each surviving box contains a set of common pharmacophores, one of which is ultimately singled out as a hypothesis. The hypotheses generated were validated using an external set of 10 active compounds and 1000 Glide decoy data sets. This is essential to evaluate the ability of each hypothesis of correctly predicting true positive (actives) and rejecting inactive compounds (Glide decoy set) during phase screening. Different statistical metrics including receiver operating characteristics (ROC), area under the accumulation curve (AUAC), phase hypo score and robustness initial enhancement values were used to validate the generated pharmacophore models. The best model based on the validation parameters was used for screening the in-house library of natural compounds discussed in previous section. The top 1,000 compounds with good fitness scores matching that of the pharmacophore were subsequently used for molecular docking simulation.
Drugable site determination and generation of receptor grid
The location of binding pocket of a protein is commonly identified from the binding site of the cocrystallized ligand. However, in the case of modelled proteins which oftentimes do not contain modelled co-ligand, an accurate computational tool is required to determine the likely binding pocket of the protein. In this study to determine the possible ligand binding site of MRP3, SiteMap tool on Maestro was used. SiteMap generates information on the character of binding sites using novel search analysis facility in a three-stage process. In the first step, a site map calculation was performed to determine one or more regions on or near the protein surface called sites which may be suitable for ligand binding. Subsequently, contour maps (site maps) were generated producing hydrophobic and hydrophilic maps. The latter is divided into donor, acceptor and metal binding regions. Finally, depending on the shape and extent of the hydrophilic and hydrophobic regions, each site is ranked and assigned a site score value which is calculated based on the equation below (Halgren 2009):
where n is the number of site points (capped at 100), e is the enclosure score, and p is the hydrophilic score, and is capped at 1.0 to limit the impact of hydrophilicity in charged and highly polar sites.
Druggability score (Dscore) uses the same properties as Site score but different coefficients:
For Dscore, the hydrophilic score is not capped. This is one of the keys for distinguishing "difficult" and "undruggable” targets from “druggable” ones.
In addition, SiteMap also characterizes the binding site in terms of the size of the druggable site, the degree of the enclosure by the protein and exposure to solvent, the tightness with which the site points interact with the receptor, the hydrophobic and hydrophilic character of the site and the balance between them, the degree to which a ligand might donate or accept hydrogen bonds. The site with the highest Site score and Dscore was used to generate the receptor grid files for molecular docking.
Molecular docking simulation
The top fit 1000 compounds from pharmacophore screening were subjected to grid-based ligand docking and energetics (Glide) docking to estimate the inhibitory prowess of the compound based on receptor binding affinity. Initially, Glide standard precision (SP) docking was carried out to identify the top binding hit compounds. Using SP docking, ligands of unknown quality can be screened in large numbers without imposing limitations on the number of intermediate conformations or reducing the quality of torsional refinement and sampling required for high-throughput virtual screening. The Glide docking process searches the active site for possible ligand sites using a series of hierarchical filters. The top-scoring compounds with docking scores exceeding −6.0 kcal/mol from SP docking analysis were further screened with extra precision (XP) docking as a scoring function. XP employs a more sophisticated scoring function that puts a greater emphasis on the complementarity between receptor binding pockets and ligand shapes to avoid false positives in standard precision docking, penalizing ligands that bind poorly to receptors. Glide XP provides improved hydrogen bond scoring, detection of buried polar groups and detection of pi-cation and pi-pi stack interactions.
Prime MMGBSA post-docking analysis
As a further method of determining the inhibitory potential of the hit compounds, the binding free energy of the ligand-bound protein was measured. In order to calculate binding free energy for the ligand–receptor complex, the Prime MM-GBSA panel of Maestro was utilized (Iwaloye et al. 2020). MMGSA quantifies the energy difference between the ligand and the protein in the unbound state and in the complex state after energy minimization. OPLS3 was selected as the force field for the MM-GBA, and VSGB was used as the continuum solvent model. All other options were left in the default state. The MMGBSA binding free energy was calculated based on the following equations.
Ecomplex, Eprotein and Eligand represent the minimized energies for protein–inhibitor complexes, proteins and inhibitors, respectively.
In this case, ∆GSA is the non-polar contribution to the solvation energy due to the surface area. Surface energies of complexes, proteins and ligands are represented by GSA (complex), GSA (protein) and GSA (ligand).
ADME/Tox Profiling
Following molecular docking analysis, the top hits were subjected to absorption, distribution, metabolism and toxicity profiling to ascertain their potential of being developed into standard drugs. Pharmacokinetics, drug-likeness and medicinal chemistry friendliness of the hit compounds were predicted using SwissADME (http://www.swissadme.ch/index.php) and ADMETlab (https://admetmesh.scbdd.com/service/screening/cal) web servers. The hits compounds were also subjected to in silico toxicological analysis using ProTox-II web server (https://tox-new.charite.de/protox_II/index.php?site=home). The ProTox-II web server incorporates molecular similarity, fragment propensity, most frequent features and (fragment-based CLUSTER cross-validation) machine learning, based on a total of 33 models for the prediction of various toxicity endpoints such as acute toxicity, hepatotoxicity, cytotoxicity, carcinogenicity, mutagenicity, immunotoxicity, adverse outcomes pathways and toxicity targets (Banerjee et al. 2018).
DFT Calculations
Density functional theory was used to estimate the stability of the compounds and their tendency to undergo reactions required for the inhibition of MRP3 protein. By using Becke's three-parameter exchange potential and Lee–Yang–Parr correlation (B3LYP) density functional theory approach with 6-31G** as the basis set, the molecular orbital structure and atomic energy features of the top hit compounds were determined (Becke 1988; Lee et al. 1988). The Schrodinger Materials science (version 3.9) software that accommodates Jaguar fast engine was used to determine the different quantum mechanical parameters of the initially optimized compound (Bochevarov et al. 2013). Frontier molecular orbital parameters were first determined, which included the highest occupied molecular orbital (HOMO), lowest unoccupied molecular orbital (LUMO) and bandgap. HOMO energy describes the electron-donating capability of the compound, while LUMO energy describes the electron-accepting quality of the compound (Olawale et al. 2021b). The space between the HOMO and LUMO orbital is called the bandgap, and it is an indication of a compound's ability to undergo electronic excitation during a chemical reaction. The parameters of global reactivity, such as hardness, electronegativity, softness, electron affinity, chemical potential, electrophilicity and ionization potential, were evaluated.
Molecular dynamics simulation
The complexes of protein with NPC5486 from the docking experiment alongside the reference compound were selected for Molecular Dynamic (MD) simulation using NAMD 2.13. The backbone and protein–ligand complexes were subjected to a 50-ns full atomistic MD simulation production run. The necessary MD simulation files were prepared using CHARMM-GUI (Brooks et al. 2009; Jo et al. 2008; J. Lee et al. 2016). The salt concentration and temperature of the biomolecular systems were set to 0.154 NaCl and 310 K, respectively, in order to mimic physiological conditions. In preparation for the production run, the system was minimized for 10,000 steps using a constant number of atoms, constant volume and constant temperature ensemble (NVT) utilizing a conjugate gradient algorithm, followed by an equilibration in a constant number of atoms, constant pressure and constant temperature ensemble (NPT) for 1 ns. The simulation pressure was set to 1.01325 bar and controlled by the Nose–Hoover Langevin piston, while the temperature was controlled by Langevin dynamics. The CHARMM36 force field was used in the dynamic simulations (Huang and MacKerell 2013). Charmm General Force Field (CGenFF) was used for the phytocompounds (Yu et al. 2012). TIP3P water model was used to resemble the added water box, with 10 Å padding, for the periodic boundary condition to be applied. The simulation box was set to cubic with a length of 158 Å for the three protein–compound complex systems, while for the apo protein the size was 156 Å. The length of the box was determined automatically using a 10 Å as padding from the protein. The numbers of water molecules added during the preparation of the systems using the CHARMM-GUI web server were 338,676, 351,870 and 351,825 for the apo protein, protein–vincristine complex and protein–NPC5486 complex, respectively. All three systems were neutralized using a concentration of 0.154 M of Na+ and Cl− ions which produced 339 Cl− and 342 Na+ ion for the three protein–compound complex systems, while the apo protein system contained 329 Cl− and 332 Na+. The computation of the thermodynamic parameters (RMSD, RMSF SASA, RoG and number of H-bonds) from the MD simulation trajectories was done using VMD Tk console scripts.
Cluster analysis of MD simulation trajectories
TTClust version 4.7.2 was used to cluster the trajectory automatically according to the elbow method, and a representative structure for each cluster was produced (Tubiana et al. 2018). These representative conformations were analysed using Protein–Ligand Interaction Profiler (PLIP) for ligands (vincristine and NPC5486) atom-amino acid residue interactive analysis (Salentin et al. 2015). The images were created using PyMol V2.2.2 (DeLano 2002).
MMGBSA computation
Molecular Mechanics Generalized Born Surface Area (MM-GBSA) was used to compute the binding affinity of the ligands with the proteins using the MMPBSA.py script implemented in Amber tools 17 (Case et al. 2015; Miller et al. 2012). All frames (250 frames) were used in the computation with salt concentration set to 0.154 Mol and other parameters set as default.
Results and discussion
Homology modelling and validation
Due to the absence of crystallized 3D structure of MRP3 protein on protein data bank (http: //www. Rcsg.org), homology modelling which is a predictive method based on similar templates was used to model the structure of the protein. Studies have shown that the validity and accuracy of a predicted model is largely dependent on the level of similarity with the template structures as such an intense template search was carried out prior to protein modelling (Dalton and Jackson 2007). A total of 17,318 templates were found by HHblits, while BLAST search algorithm generated 978 templates. The wild-type multidrug-resistant protein 1 with PDB ID: 6uy0 showed the highest amino acid sequence similarity with percentage identity of 57.02% and coverage of 99%, as such was deemed the best fitted template for modelling the 3D structure of MRP3 (Fig. S1 and S2). Ramachandran conformation map was used to evaluate the structural rationality of the modelled MRP3 protein which indicated that most of the amino acid (94.6%) residues were in the favoured region (Fig. 2). As illustrated in Fig. S3b, most of the residues have local quality estimate above 0.7 which is an indication of good model reliability. The GMQE value of 0.60 and QMEAN score of 0.73 further establish that the modelled structure is reliable and has a good quality. Thus, the three-dimensional structure of the protein constructed by homology modelling satisfied quality model reliability and realistic requirements (Fig. S3).
While the previous validation steps have indicated reasonable accuracy of the modelling approach, further structural deformities that may be present in the protein were corrected through a series of loop refinement and protein minimization steps using Glide. Subsequently, the final model developed was validated using different external web servers. As shown in Fig. 3a, the optimized protein showed an RMSD value of about 2.5 Å. Similarly, a protein quality score of 89.8% was obtained using ERRAT (Fig. 3b). ERRAT works by analysing the statistics of non-bonded interactions between different atom types, with higher scores indicating better quality models (Colovos and Yeates 1993). Generally, an ERRAT score exceeding 50% is considered a bench mark for high-quality model. The ERRAT score of 89.9% obtained for the protein suggests that the backbone conformation and non-bonded interactions indicate high-quality model (Saxena et al. 2018). While ERRAT server is arguably one of the best protein quality check server, like other protein quality predictor, it is not individually comprehensive; hence, a protein quality metaserver which combines diverse quality assessment programmes (including Verify3D, MolProbity, Procheck, ProSA, ERRAT, ProQ, dDFire, Naccess and D2N) and outperforms their individual server accuracies was used to give a more robust protein quality estimate. Interestingly, most of the protein structure checks tools gave an RMSD value with the range of 1–3 Å which suggest a good overall quality of the modelled structure.
In addition to the previous qualitative assessments, the VADAR statistics for quantitative evaluation of the predicted model revealed that the model structurally composed of alpha-helix (58%), interspersed beta sheets (12%), coil (28%) and turn (20%) with extensive H-bonding groups (Table 1). The H-bonds distance and energy in the predicted secondary structure protein were similar to the expected value which further corroborates the good quality of the model. Likewise, calculation of the resolution of the protein from coordinate data using ResProx showed a resolution of 1.5 Å (Table 1). Generally, lower-resolution proteins below 2 Å are highly ordered and it allows individual hydrogen atoms to be visualized and heavy atoms (C, O, N) to be very accurately mapped; hence, they are a preferred choice for molecular docking and dynamics simulation (Berjanskii et al. 2012). The validation results therefore indicate that the refined structure of the modelled protein is satisfactory and reliable for the subsequent computational studies.
Active site prediction and analysis
Sequel to MRP3 protein modelling and validation, the potential binding site for small molecule inhibitor was predicted using site map. Five potential ligand binding sites were identified in the protein (Fig. S4). In order to accurately determine the best of the five possible sites which can be used for molecular docking, the values obtained from the site score and D-score were analysed. It was observed that site 1 on the protein with a site score of 1.216 and a D-score of 1.253 had the highest propensity for ligand binding. In addition, site 1 was a preferred active pocket for docking due to the following reasons, viz: large sites with high ligand binding surface and higher number of hydrophobic residues which play important role in the protein structure and activity. As such, site 1 was considered the most pharmacologically relevant site on the protein where lead molecules can bind to modulate its activity. The site contains amino acid residues that include Met584, Pro581, Leu580, Ile577, Lys318, Gln436, Gln321, Asp322, Ser325, Phe326, Glu1200, Asn328, Pro329, Ser360, Gln363, Leu332, Leu1234, Gln1235, Phe1238, Leu367, Trp1242, Leu429, Tyr370, Tyr371 and Ala440 among others.
Pharmacophore modelling, pharmacophore validation and phase screening
Pharmacophore modelling is a ligand-based technique explored in this study to identify pharmacophore features present in the structures of 12 experimentally determined inhibitors of MRP3 protein (Köck et al. 2014) (Fig. S5). The compounds were manually selected based on profound structural diversity and a relatively moderate to high biological activity (Jiang et al. 2016). Ligand-based pharmacophore modelling was necessary since the structures of MRP3 are yet to be fully crystallized. Hence structure-based approach only may result in errors due to the limited knowledge of the 3-dimensional structure of the protein. The pharmacophore model generated was validated using a set of 10 active inhibitors and 1000 decoy sets. While it was observed that a good number of the pharmacophore were able to correctly predict the active compounds, some of them showed low specificity by generating false positive predictions with decoy sets (Tables 2 and 3). As a validation metrics, the ROC curve was calculated by plotting the true positive rates against the false positive rates. From the result, it was observed that the model AHHHR_4 with one hydrogen bond acceptor, three hydrophobic groups and one aromatic group was the best performing model with ROC value of 0.70 and phase score of 0.71 (Fig. 4, 5 and Fig. S6). Similarly, the AUAC measures the probability that an active compound will be ranked ahead of inactive or decoy set. A value exceeding 0.6 can be interpreted as high probability. The robust initial enhancement (RIE) score of 2.09 also confirms the superior robustness of the hypothesis.
Once a pharmacophore has been identified, it can be used for rational drug design processes such as virtual screening, lead optimization and de novo drug design. Based on the pharmacophore validation results obtained here, the model AHHHR_4 was further employed in identifying potential inhibitors of MRP3 from an in-house library of natural compounds. A total of 1,000 hits were identified as drug-like compound with fitness score above 1.5 (Table S1). In addition to the fitness score, several parameters including vector score, volume score and align score were also monitored. The vector score is a parameter that measures how well the vectors for acceptors, donors, hydrophobic groups and aromatic rings are aligned in the structures that contribute to the hypothesis (Lauria et al. 2010). The volume score measures how much volume of the contributing structures overlaps when aligned on the pharmacophore (Lauria et al. 2010). Most of the hit compounds showed vector score, volume score and align score within acceptable limits. The compounds were then used for the subsequent virtual screening analysis.
Receptor grid generation, molecular docking analysis and interaction patterns
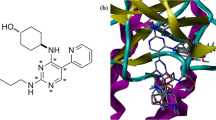
A receptor grid was generated to represent the ligand binding pocket for molecular docking using Site 1 identified from site map (Table 4). Initially, standard precision docking was performed on the compound retrieved from pharmacophore phase screening. A total of 167 compounds had docking score below −6.0 kcal/mol and was used for a more accurate extra precision docking. From XP docking, seven distinct lead compounds (NPC5486, NPC200761, NPC10754, Cyrtophyllone_B, NPC17677, NPC476840 and 33,434) were identified all of which had docking scores below −7.5 kcal/mol (Table 5). Moreover, the lead compounds all had docking scores which was comparable to those of experimentally determined MRP3 inhibitor including vincristine, furosemide and indomethacin. The highly significant docking scores of the hit compounds suggest their potentials in MRP3 inhibitors development. Notably, compound NPC5486 and NPC200761 had the best docking score with the same XP docking score value of −8.877 kcal/mol.
Aside from determining the affinity of the lead compound for the target protein, molecular docking is also crucial for exploring the interactions between critical amino acid residues of the protein and targeted small molecule inhibitors. A good lead compound requires the following for ligand bonding: presence of hydrogen bond donors and acceptors that can interact with polar amino acids, an hydrophobic moiety that spans through the binding site to interact with hydrophobic amino acids within the inner pocket of the binding site and an aromatic ring that interacts with phenylalanine residues in the binding site (Le et al. 2021). The 3D binding poses of the top hits and their corresponding 2D protein ligand interaction schematic diagrams are illustrated in Figs. 6, 7, 8, 9, 10 and Fig. S7-10. NPC5486 interacts with amino acid residue Ser325, Tyr371 and Ser1196 via hydrogen bond linkages. Similar interaction fingerprints was observed in NPC200761 which formed hydrogen bonding with Tyr371 and Ser1196 and pi-pi bonds with Arg1245. NPC10754 forms hydrogen bonding interaction with Gln587, Asn583, Leu580 and Arg579, while Cyrtophyllone_B interacts with amino acid residues Asp322 and Ser325. The highest number of hydrogen bonding interaction was observed in NPC476840 which forms five hydrogen bonding network with Ser325, Lys318, Gln326, Ser432 and Trp1242. Although the exact functions of these amino acid residues are yet to be demystified, based on conserved evolutionary relationship with multidrug-resistant protein1, residues like Trp1242, SER 1196 and ASN 1241 might likely function in nucleotide binding. Since ATP is critical for the functions of all MRPs, by binding to these residues, the activity of MRP3 protein can be inhibited by the identified lead compounds.
Equally worth emphasizing is the fact that among the standard inhibitor that was screened vincristine showed the highest binding affinity by forming a single hydrogen bonding interaction with Gln363. Although this was contrary to previous studies which reported interaction with the cytoplasmic domain residues such as Ser752, Gly753, Ser779, Ala780 and Val 781 using blind docking, the authors noted that the standard drug vincristine had the highest docking scores within two cytoplasmic domain of the modelled MRP3 protein (Noroozi-Aghideh et al. 2020). However, since binding site prediction has been reported has a better approach to determining binding site of a protein, the interaction fingerprints reported in this study may appear to hold significant promise.
Binding free energy calculations from rigid docking
In Glide docking, extra precision docking poses were obtained to perform MM-GBSA calculation using surface area energy, solvation energy and energy of the minimization of the protein–ligand complexes. Lyne et al. validated the ability of Glide MMGBSA post-docking scoring protocol to correctly rank cogeneric inhibitors as such can be used as a bench mark suitable for guiding structure-based design of inhibitors (Lyne et al. 2006). As illustrated in Table 6, Prime MM/GBSA (DGbind) ranged from −57.56 to kcal/mol (NPC5486) to −41.571 kcal/mol (NPC10754). Interestingly, the compound NPC5486 which had the best docking scores also showed the most significant binding free energy value. The most remarkable free energy contributions to the ligand binding were from van der Waals (∆G vdw), non-polar solvation energy (∆G Lipo) and Coulombs energy (∆G Coulomb). Overall the entire lead compounds showed strong binding affinity which suggests outstanding potential to form stable interaction with MRP3 protein.
Pharmacokinetics and Toxicological risks assessment
ADMET profiling of the lead compounds was conducted in order to evaluate the pharmacokinetic and toxicity properties of the compounds. More than 90% failure rate has been reported during the process of drug development largely due to toxicity and poor pharmacokinetics which have significantly contributed to the cost of drug development. If the lead compounds identified in this study would stand a chance to be developed as MRP3 inhibitor, then a basic understanding of the pharmacokinetics and toxicological properties is indispensable. In the current study, following pre-filtering of the compound using Lipinski rule, other druglikeness rules including Ghose, Veber, Egan and Muegge were done. Based on the number of hydrogen bond donor, hydrogen bond acceptor, molecular weight (MW) and octanol/water partition coefficient (Log P) (Fig. S11), Lipinski rule predicts the ability of a drug to be orally active in humans. From Table 7, it was evident that none of the lead compounds showed any Lipinski violations which suggest a good oral bioavailability. Veber et al. (2002) proposed that rotatable bonds below 10 and TPSA value less than 140 can better discriminate compounds that are orally active from other large data sets. Ghose rule considers molar refractivity range of 40 and 130, number of atoms between 20 and 70, Log P value between -0.4 and 5.6 and MW range of 160 and 480 have criteria for druglikeness (Ghose et al. 1998). Interestingly, none of the compounds showed any violations for Veber and Ghose rule. Furthermore, the lead compounds did not show more than one violation for Egan rule and Muegge rule which falls within the acceptable limits for good bioavailability of compounds. The Caco-2 permeability scores between 4.0 and 6.0 imply moderate to high intestinal absorption.
In addition to the good drug-like properties, none of the lead compounds showed beyond more than one PAIN and BRENK alert which suggest that the results obtained from the virtual screening are less likely to be false positives that could stem from ligand promiscuity. In addition, due to the role of CYPs inhibition in drug–drug interaction which has been implicated in adverse drug interactions and reactions (Beck et al. 2021), the ability of the lead compounds to inhibit CYP 1A2, 2C9, 2C19, 2D6 and 3A4 was predicted. Although most of the lead compound did not show tendencies to inhibit CYP 1A2, 2C9, 2C19 and 2D6, a higher proportion were predicted as CYP3A4 inhibitor. While CYP inhibition might present a challenge, studies have shown that by controlling the dosing and performing lead optimization the consequent effect of drug–drug interaction can be abated.
As shown in Table 8, the lead compounds are less likely to be hERG blockers with probability values below 0.1. Also, with the exception of NPC17677 which showed AMES toxicity probability of 0.508, most of the compounds show less low tendencies to pose any mutagenic threats. A skin sensitivity probability value below 0.3 in NPC5486, NPC10754, NPC476840 and 33,434 signifies less skin irritation following topical administration. The lead compounds were not predicted to induce any major toxicological targets and pathways. Furthermore, the compounds showed a high LD50 value which means a very high concentration has to be taken before an oral toxicity can be observed. The predicted pharmacokinetic and toxicological studies have thus far shown significant drug-likeness potential of the lead compounds.
Frontier molecular orbital and analysis of chemical reactivity descriptors
Frontier molecular orbital analysis of the fully optimized structures was used to analyse the vital chemical properties of the lead compounds as given in Table 9. FMO theory has shown that the energy level of the HOMO and LUMO orbital is a primary factor that significantly impacts the biological activities of small drug-like molecules (Hagar et al. 2020). The HOMO energy is a measure of the electron-donating ability of the compound, while the LUMO energy quantifies the electron-accepting properties of the compounds. NPC5486 showed the most significant HOMO energy with values of −0.22919 which aligns with the molecular docking result (Fig. 11). The orbital represents probability of finding an electron. The red and blue colour indicates the positive and negative ( ±) sign of the wavefunction, respectively, and the nodal nature of the orbital. HOMO and LUMO cloud were centralized majorly on the aromatic rings. The HOMO of the lead compounds and the LUMO of adjacent residues of MRP3 could share orbital interaction during the ligand binding process. The energy difference between the HOMO and LUMO energy assessed the chemical reactivity tendencies of the lead compounds. Compound NPC17677 was observed as the most chemically reactive with band gap energy of −0.14554 eV, while NPC5486 was predicted as the least chemically reactive with band gap of −0.22964 eV.
Based on the mathematical equations, descriptors of chemical reactivity parameters were computed (Alyar et al. 2019; Khalid et al. 2019);
where the energy of the HOMO and LUMO orbitals is depicted as E(HOMO) and E(LUMO).
The electrophilicity index is useful for explaining the binding capacity with biomolecules. The higher electrophilicity index of studied compounds will have a higher binding interaction (Parthasarathi et al. 2004). NPC476840 had the highest electrophilicity index when compared with other lead compounds followed by 33,434. The high electrophilicity index of compound 33,434 corroborates the results of the binding free energy calculations which demonstrated significant affinity and stability of the ligand–MRP3 protein complex. While the global hardness is a measure of charge transfer prohibition, the global softness value characterizes the ability of the compound to accept electrons. Generally, soft molecules have a small band gap and are more reactive because they mediate electron transfer easily. NPC17677 was observed to have the highest global softness values. The frontier molecular orbital and chemical reactivity parameters understudied here could significantly impact the degree of binding affinity of these lead compounds to different extents.
Molecular dynamics simulation
The stability and structural integrity of the bound systems as compared to the unbound proteins were studied through the various conformational fluctuations that occurred in the MD-simulated environment. The plots were presented as a function of time frame (Fig. 12). The RMSD plots show the extent of the deviation of each frame from the initial structure and hence used to access the protein stability of the systems. The protein systems were equilibrated at around 30 ns with similar plots of RMSD with average RMSD values of 5.2, 6.2 and 4.7 Å for the unbound protein, vincristine and NPC5486. Vincristine exhibited higher fluctuations with higher mean RMSD values than the NPC5486 bound and unbound protein. The RMSF reveals the flexibility of different regions of the protein. There are spikes occurring at the N terminal end of the proteins corresponding to the terminal motions. The average RMSF values for the protein systems are 1.9 Å for the apoprotein and 1.9 and 1.7 Å for the protein complexed with vincristine and NPC5486, respectively. Some of the amino acid residues in the protein showed significant fluctuations (greater than 2 Å). The two regions of the RMSF plots that showed the highest fluctuations are the VAL51 –ALA101 region as well as, TYR651-GLY726. These regions form a loops and showed greater interaction potentials with the ligands. The RoG measures the compactness of the systems. A stably folded protein structure presents a steady RoG plot. The RoG plot revealed a steady progression of the biomolecular systems during the simulation period with minimal fluctuations. The mean RoG values calculated from the plots for the protein systems are 37.9, 38.4 and 38.2 Å for the apo enzyme, vincristine and NPC5486 complexed systems, respectively. The SASA plots show the degree of solvent accessibility by the surface of the proteins. Both RoG and SASA plots indicate the level of structural unfolding of proteins with reference to its original structure. The SASA plots for the protein system show steady progression throughout the simulation period with average values of 61,809.43, 64,911.86 and 65,256.44 Å2 for the unbound enzyme, vincristine and NPC5486, respectively. The average number of hydrogen bonds for the unbound protein system, vincristine and NPC5486 is 330.2, 328.0 and 324.0, respectively. Overall, the thermodynamic parameters revealed that the stability of the complexes is indicated by the RMSD, RoG and H-bond plots as well as flexibility as shown by the RMSF plot.
Clustering analysis of molecular dynamics simulation trajectories of the protein complexes
In order to gain more insight into the interactions of the ligands with the protein in a more dynamic environment, the MD trajectory files were clustered according to their structure and conformational pattern. From these clusters, representative structures were selected for further analysis. Table 10 shows representative clusters generated, while Fig. 13 shows the representative structures of the first cluster and the last cluster from the MD simulation trajectories. The table and figure further show that the interactions with important catalytic residues were preserved during the course of the dynamic simulations.
Binding free energy from molecular dynamics simulation
Dynamic simulation-based calculations that integrate molecular mechanics with implicit solvation models, such as Molecular Mechanics-Generalized Born Surface Area (MM-GBSA), give substantially accurate estimates of free energies of binding of ligands to proteins. In these calculations, the binding free energy (ΔGbind) measures the affinity of a ligand to its target protein. The free energy difference between the ligand-bound state (complex) and the corresponding unbound states of proteins and ligands is also employed in the calculations (Table 11 and Fig. 14). The result from the MMGBSA calculation further corroborated the docking studies.
Conclusion
In the current study, new sets of inhibitors for MRP3 enzyme were identified from natural compounds libraries using ligand-based drug design approach (pharmacophore modelling and density functional theory calculations) and structure-based approaches (homology modelling, molecular docking, MD simulations and MM-GBSA free energy calculations). Summarily, homology model of MRP3 protein was constructed using bovine MRP1 protein as template. Using a set of experimentally derived MRP3 inhibitors, ligand-based pharmacophore model was generated and used for virtual screening of natural compound libraries. A series of molecular docking steps (standard precision and extra precision docking) retrieved seven potential inhibitors of MRP3 which showed good reactivity parameters following density functional theory analysis. Molecular dynamic simulation of the best binding compound in complex with the protein model was run alongside that of the reference compounds. Analysis from the resulting trajectories as computed from the thermodynamic parameters plots showed not only the increased flexibility and higher interaction potential of the residues of the receptor towards the NPC5486 as indicated by the RMSF but also the higher stability as indicated by the RMSD, RoG and H-bond of the ligand–protein complexes. The cluster analysis of the trajectories further revealed that the observed interactions with important residues were preserved in the dynamic environment; these observations were further verified from MMGBSA computations, which show the free energy of binding of the ligands to the protein as well as the free energy contribution per residue. Thus, the study has revealed possible drug candidates and therapeutic approaches which could help in the development of MRP3 inhibitors in the treatment of acute leukaemia.
References
Akioka K, Masuda K, Harada S, Nakamura T, Okugawa K, Nakano K, Osaka Y, Tsuchiya K, Sako H (2013) Acute renal failure caused by hyperuremic acidemia in ABO-incompatible kidney transplant maintained with cyclosporine and high-dose mizoribine: A case report. Transplant Proc 45(7):2815–2818. https://doi.org/10.1016/j.transproceed.2013.03.052
Ali I, Welch MA, Lu Y, Swaan PW, Brouwer KLR (2017) Identification of novel MRP3 inhibitors based on computational models and validation using an in vitro membrane vesicle assay. Eur J Pharm Sci 103:52–59. https://doi.org/10.1016/j.ejps.2017.02.011
Alyar S, Şen T, Özmen ÜÖ, Alyar H, Adem Ş, Şen C (2019) Synthesis, spectroscopic characterizations, enzyme inhibition, molecular docking study and DFT calculations of new Schiff bases of sulfa drugs. J Mol Struct 1185:416–424
Banerjee P, Eckert AO, Schrey AK, Preissner R (2018) ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res 46(W1):W257–W263. https://doi.org/10.1093/nar/gky318
Beck TC, Beck KR, Morningstar J, Benjamin MM, Norris RA (2021) Descriptors of cytochrome inhibitors and useful machine learning based methods for the design of safer drugs. Pharmaceuticals 14(5):472
Becke AD (1988) Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A Gen Phys 38(6):3098–3100. https://doi.org/10.1103/physreva.38.3098
Benkert P, Tosatto SCE, Schomburg D (2008) QMEAN: A comprehensive scoring function for model quality assessment. Proteins 71(1):261–277. https://doi.org/10.1002/prot.21715
Berjanskii M, Zhou J, Liang Y, Lin G, Wishart DS (2012) Resolution-by-proxy: a simple measure for assessing and comparing the overall quality of NMR protein structures. J Biomol NMR 53(3):167–180
Bobrowska-Hägerstrand M, Lillås M, Mrówczyñska L, Wróbel A, Shirataki Y, Motohashi N, Hägerstrand H (2006) Resveratrol oligomers are potent MRP1 transport inhibitors. Anticancer Res 26(3A):2081–2084
Bochevarov AD, Harder E, Hughes TF, Greenwood JR, Braden DA, Philipp DM, Rinaldo D, Halls MD, Zhang J, Friesner RA (2013) Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int J Quantum Chem 113(18):2110–2142
Borst P, Evers R, Kool M, Wijnholds J (2000) A family of drug transporters: The multidrug resistance-associated proteins. J Natl Cancer Inst 92(16):1295–1302. https://doi.org/10.1093/jnci/92.16.1295
Brooks BR, Brooks CL III, Mackerell AD Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30(10):1545–1614
Case DA, Berryman J, Betz RM, Cerutti DS, Cheatham Iii TE, Darden TA, Duke RE, Giese TJ, Gohlke H, and Goetz AW (2015) AMBER 2015
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2(9):1511–1519
Dalton JAR, Jackson RM (2007) An evaluation of automated homology modelling methods at low target–template sequence similarity. Bioinformatics 23(15):1901–1908
DeLano WL (2002) Pymol: An open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr 40(1):82–92
De Vocht T, Buyck C, Deferm N, Qi B, Van Brantegem P, van Vlijmen H, Snoeys J, Hoeben E, Vermeulen A, Annaert P (2021) Identification of novel inhibitors of rat Mrp3. Eur J Pharm Sci 162:105813. https://doi.org/10.1016/j.ejps.2021.105813
Elekofehinti OO, Iwaloye O, Olawale F, Chukwuemeka PO, Folorunso IM (2021) Newly designed compounds from scaffolds of known actives as inhibitors of survivin: computational analysis from the perspective of fragment-based drug design. Silico Pharmacol 9(1):47. https://doi.org/10.1007/s40203-021-00108-8
Fletcher JI, Williams RT, Henderson MJ, Norris MD, Haber M (2016) ABC transporters as mediators of drug resistance and contributors to cancer cell biology. Drug Resist Updat 26:1–9. https://doi.org/10.1016/j.drup.2016.03.001
Fukushima-Uesaka H, Saito Y, Maekawa K, Hasegawa R, Suzuki K, Yanagawa T, Kajio H, Kuzuya N, Noda M, Yasuda K, Tohkin M, Sawada JI (2007) Genetic variations of the ABC transporter gene ABCC3 in a Japanese population. Drug Metab Pharmacokinet 22(2):129–135. https://doi.org/10.2133/dmpk.22.129
Ghose AK, Viswanadhan VN, Wendoloski JJ (1998) Prediction of hydrophobic (lipophilic) properties of small organic molecules using fragmental methods: an analysis of ALOGP and CLOGP methods. J Phys Chem A 102(21):3762–3772
Gilibili RR, Chatterjee S, Bagul P, Mosure KW, Murali BV, Mariappan TT, Mandlekar S, Lai Y (2017) Coproporphyrin-I: A fluorescent, endogenous optimal probe substrate for ABCC2 (MRP2) suitable for vesicle-based MRP2 inhibition assay. Drug Metab Dispos 45(6):604–611. https://doi.org/10.1124/dmd.116.074740
Hagar M, Ahmed HA, Aljohani G, Alhaddad OA (2020) Investigation of some antiviral N-heterocycles as COVID 19 drug: molecular docking and DFT calculations. Int J Mol Sci 21(11):3922
Halgren TA (2009) Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model 49(2):377–389. https://doi.org/10.1021/ci800324m
Hollenstein K, Dawson RJP, Locher KP (2007) Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol 17(4):412–418
Howlader NNA, Krapcho M, Miller D, Bishop K, Kosary CL, and Yu M (2017) Cronin KA (eds). SEER Cancer Statistics Review, 1975–2014, National Cancer Institute. Bethesda, MD
Huang J, MacKerell ADJ (2013) CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem 34(25):2135–2145. https://doi.org/10.1002/jcc.23354
Iwaloye O, Elekofehinti OO, Momoh AI, Babatomiwa K, Ariyo EO (2020) In silico molecular studies of natural compounds as possible anti-Alzheimer’s agents: ligand-based design. Netw Model Anal Heal Informatics Bioinforma. https://doi.org/10.1007/s13721-020-00262-7
Jiang L, He Y, Luo G, Yang Y, Li G, Zhang Y (2016) Discovery of potential novel microsomal triglyceride transfer protein inhibitors via virtual screening of pharmacophore modelling and molecular docking. Mol Simul 42(15):1223–1232
Jo S, Kim T, Iyer VG, Im W (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29(11):1859–1865
Khalid M, Ullah MA, Adeel M, Khan MU, Tahir MN, Braga AAC (2019) Synthesis, crystal structure analysis, spectral IR, UV–Vis, NMR assessments, electronic and nonlinear optical properties of potent quinoline based derivatives: interplay of experimental and DFT study. J Saudi Chem Soc 23(5):546–560
Köck K, Ferslew BC, Netterberg I, Yang K, Urban TJ, Swaan PW, Stewart PW, Brouwer KLR (2014) Risk factors for development of cholestatic drug-induced liver injury: inhibition of hepatic basolateral bile acid transporters multidrug resistance-associated proteins 3 and 4. Drug Metab Dispos 42(4):665–674
Lauria A, Ippolito M, Fazzari M, Tutone M, Di Blasi F, Mingoia F, Almerico AM (2010) IKK-β inhibitors: An analysis of drug–receptor interaction by using molecular docking and pharmacophore 3D-QSAR approaches. J Mol Graph Model 29(1):72–81
Le M-T, Hoang V-N, Nguyen D-N, Bui T-H-L, Phan T-V, Huynh PN-H, Tran T-D, Thai K-M (2021) Structure-based discovery of ABCG2 Inhibitors: A homology protein-based pharmacophore modeling and molecular docking approach. Molecules 26(11):3115
Lee C, Yang W, Parr RG (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B Condens Matter 37(2):785–789. https://doi.org/10.1103/physrevb.37.785
Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y, Jo S, Pande VS, Case DA, Brooks CL, MacKerell AD, Klauda JB, Im W (2016) CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput 12(1):405–413. https://doi.org/10.1021/acs.jctc.5b00935
Lyne PD, Lamb ML, Saeh JC (2006) Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. J Med Chem 49(16):4805–4808
Maynadié M (2015) Acute Leukemia. Rev Francoph Des Lab 2015(471):27–28. https://doi.org/10.1016/s1773-035x(15)30070-8
Miller BR, McGee TD, Swails JM, Homeyer N, Gohlke H, Roitberg AE (2012) MMPBSA.py: An efficient program for end-state free energy calculations. J Chem Theory Comput 8(9):3314–3321. https://doi.org/10.1021/ct300418h
Noroozi-Aghideh A, Safavi A, Ghodousi ES, Rajaeinejad M, Moghaddam PT (2020) Homology modeling and molecular docking of hABCC3/MRP3 with chemotherapeutic agents in acute leukemia. Jundishapur J Nat Pharm Prod. https://doi.org/10.5812/jjnpp.69407
Olawale F, Iwaloye O, Elekofehinti OO (2021a) Virtual screening of natural compounds as selective inhibitors of polo-like kinase-1 at C-terminal polo box and N-terminal catalytic domain. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2021.1991476
Olawale F, Olofinsan K, Iwaloye O, and Ologuntere TE (2021b). Phytochemicals from Nigerian medicinal plants modulate therapeutically-relevant diabetes targets: insight from computational direction. Adv Tradit Med 1–15
Olawale F, Olofinsan K, Iwaloye O, Chukwuemeka PO, Elekofehinti OO (2022) Screening of compounds from Nigerian antidiabetic plants as protein tyrosine phosphatase 1B inhibitor. Comput Toxicol 21:100200. https://doi.org/10.1016/j.comtox.2021.100200
Parthasarathi R, Subramanian V, Roy DR, Chattaraj PK (2004) Electrophilicity index as a possible descriptor of biological activity. Bioorg Med Chem 12(21):5533–5543
Remmert M, Biegert A, Hauser A, Söding J (2011) HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9(2):173–175. https://doi.org/10.1038/nmeth.1818
Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M (2015) PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res 43(W1):W443–W447. https://doi.org/10.1093/nar/gkv315
Saxena S, Abdullah M, Sriram D, Guruprasad L (2018) Discovery of novel inhibitors of Mycobacterium tuberculosis MurG: Homology modelling, structure based pharmacophore, molecular docking, and molecular dynamics simulations. J Biomol Struct Dyn 36(12):3184–3198
Scheffer GL, Kool M, de Haas M, de Vree JML, Pijnenborg ACLM, Bosman DK, Oude Elferink RPJ, Van Der Valk P, Borst P, Scheper RJ (2002) Tissue distribution and induction of human multidrug resistant protein 3. Lab Investig 82(2):193–201. https://doi.org/10.1038/labinvest.3780411
Song XR, Zheng Y, He G, Yang L, Luo YF, He ZY, Li SZ, Li JM, Yu S, Luo X, Hou SX, Wei YQ (2010) Development of PLGA nanoparticles simultaneously loaded with vincristine and verapamil for treatment of hepatocellular carcinoma. J Pharm Sci 99(12):4874–4879. https://doi.org/10.1002/jps.22200
Steinbach D, Legrand O (2007) ABC transporters and drug resistance in leukemia: Was P-gp nothing but the first head of the Hydra? Leukemia 21(6):1172–1176. https://doi.org/10.1038/sj.leu.2404692
Studer G, Tauriello G, Bienert S, Biasini M, Johner N, Schwede T (2021) ProMod3 - A versatile homology modelling toolbox. PLoS Comput Biol 17(1):e1008667. https://doi.org/10.1371/JOURNAL.PCBI.1008667
Tubiana T, Carvaillo J-C, Boulard Y, Bressanelli S (2018) TTClust: a versatile molecular simulation trajectory clustering program with graphical summaries. J Chem Inf Model 58(11):2178–2182
Uchiumi T, Hinoshita E, Haga S, Nakamura T, Tanaka T, Toh S, Furukawa M, Kawabe T, Wada M, Kagotani K, Okumura K, Kohno K, Akiyama SI, Kuwano M (1998) Isolation of a novel human canalicular multispecific organic anion transporter, cMOAT2/MRP3, and its expression in cisplatin-resistant cancer cells with decreased ATP-dependent drug transport. Biochem Biophys Res Commun 252(1):103–110. https://doi.org/10.1006/bbrc.1998.9546
van de Velde ME, Kaspers GL, Abbink FCH, Wilhelm AJ, Ket JCF, van den Berg MH (2017) Vincristine-induced peripheral neuropathy in children with cancer: A systematic review. Crit Rev Oncol Hematol 114:114–130. https://doi.org/10.1016/j.critrevonc.2017.04.004
Van Der Kolk DM, De Vries EGE, Noordhoek L, Van Den Berg E, Van Der Pol MA, Müller M, Vellenga E (2001) Activity and expression of the multidrug resistance proteins P-glycoprotein, MRP1, MRP2, MRP3 and MRP5 in de novo and relapsed acute myeloid leukemia. Leukemia 15(10):1544–1553. https://doi.org/10.1038/sj.leu.2402236
Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45(12):2615–2623. https://doi.org/10.1021/jm020017n
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303. https://doi.org/10.1093/nar/gky427
Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS (2003) VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res 31(13):3316–3319
Wu CP, Calcagno AM, Hladky SB, Ambudkar SV, Barrand MA (2005) Modulatory effects of plant phenols on human multidrug-resistance proteins 1, 4 and 5 (ABCC1, 4 and 5). FEBS J 272(18):4725–4740. https://doi.org/10.1111/j.1742-4658.2005.04888.x
Ye ZW, Camus S, Augustijns P, Annaert P (2010) Interaction of eight HIV protease inhibitors with the canalicular efflux transporter ABCC2 (MRP2) in sandwich-cultured rat and human hepatocytes. Biopharm Drug Dispos 31(2–3):178–188. https://doi.org/10.1002/bdd.701
Yu W, He X, Vanommeslaeghe K, MacKerell ADJ (2012) Extension of the CHARMM General Force Field to sulfonyl-containing compounds and its utility in biomolecular simulations. J Comput Chem 33(31):2451–2468. https://doi.org/10.1002/jcc.23067
Zhang YK, Wang YJ, Gupta P, Chen ZS (2015) Multidrug Resistance Proteins (MRPs) and Cancer Therapy. AAPS J 17(4):802–812. https://doi.org/10.1208/s12248-015-9757-1
Acknowledgements
The authors acknowledge the technical support received from Computational Biologists at the Bioinformatics and Molecular Biology Unit, Department of Biochemistry, Federal University of Technology, Akure, Ondo State, Nigeria.
Funding
This research received no external financial or non-financial supports.
Author information
Authors and Affiliations
Contributions
FO helped in project design, generation and analysis of data, interpretation of data, preparation of manuscript draft and approval of manuscript for submission. OI and OO performed data analysis, data interpretation, substantial manuscript revision and approval of manuscript for submission. KO was involved in manuscript draft, substantial manuscript revision and approval of manuscript for submission. II contributed to data acquisition, preparation of manuscript draft and approval of manuscript for submission. GG helped in substantial manuscript revision, project supervision and provision of resources for research and approval of manuscript for submission. All authors have read and approved the manuscript for submission.
Corresponding author
Ethics declarations
Ethical approval
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The authors declare the following financial interests/personal relationships which may be considered as potential competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Olawale, F., Iwaloye, O., Olofinsan, K. et al. Homology modelling, vHTS, pharmacophore, molecular docking and molecular dynamics studies for the identification of natural compound-derived inhibitor of MRP3 in acute leukaemia treatment. Chem. Pap. 76, 3729–3757 (2022). https://doi.org/10.1007/s11696-022-02128-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11696-022-02128-w