Abstract
Introduction
The relationship between late post-bariatric surgery weight regain and gut microbiota is not completely understood.
Objective
To analyze the profile of gut microbiota among patients with and without late weight regain after post-Roux-en-Y gastric bypass (RYGB) and to compare it with a control group (CG) comprised of obese Brazilian individuals.
Methods
This is a cross-sectional study which enrolled 34 morbidly obese women divided into 3 groups: post-Roux-en-Y gastric bypass without (RYGB_non-regain), and with weight regain (RYGB_regain) at least 5 years after surgery, and a CG of preoperative individuals. Gut microbiota was determined by metagenomic analyses.
Results
The alpha diversity was higher in groups RYGB non-regain and RYGB regain when compared with CG (p < 0.05). Both RYGB non-regain and RYGB regain groups showed a lower abundance of the phylum Bacteroidetes when compared with CG (p < 0.01). The genera Bacteroides and SMB53 were increased in CG (p < 0.05). Group RYGB non-regain showed more abundance of the Akkermansia genus when compared with CG and group RYGB regain (p < 0.05). RYGB non-regain showed a greater abundance of the Phascolarctobacterium genus and lower of the SMB53 genus when compared with CG (p < 0.05). RYGB non-regain showed a greater abundance of the Phascolarctobacterium genus and a lower of the SMB53 genus when compared with CG (p < 0.05).
Conclusion
The gut microbiota of individuals which presented late weight regain after RYGB was significantly different in comparison to individuals with a successful weight loss, a finding that points towards a significant role of gut microbiota on weight loss and maintenance after surgery.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bariatric surgery (BS) leads to a number of specific changes in patients’ metabolism. These changes might be the key to the success of Roux-en-Y gastric bypass (RYGB) in the treatment of obesity, metabolic syndrome, and type 2 diabetes mellitus (T2DM) [1, 2].
A certain weight regain may occur after BS, which is more pronounced between 24 to 60 months after surgery [3, 4]. Approximately 20–30% of this population does not maintain about 20% of their total weight loss 10 years after surgery. The causes and mechanisms related to this condition have been broadly studied [5, 6].
Evidence has shown that gut microbiota is an important environmental factor that contributes to obesity, altering the host’s energy harvest and storage [7, 8]. Recently, gut microbiota has emerged as an important factor associated with changes in the metabolic processes that mediate some of the beneficial effects of BS. Experiments which involved gut microbiota transplantation suggest a causal relationship between this microbiota and the development of obesity. Germ-free mice which received fecal microbiota transplantation (FMT) from patients who underwent RYGB had a greater reduction in weight loss when compared with mice that received FMT from obese individuals [9, 10]. Studies among humans have associated gut microbial dysbiosis with obesity and metabolic disorders. Usually, obese individuals have decreased bacterial diversity and genus richness, with increased Firmicutes and decreased Bacteroidetes phyla [11, 12].
RYGB is based on substantial physiologic and anatomic changes in the gastrointestinal tract, which significantly modify the intestinal environment and consequently alter the gut microbiota. After this operation, there is a previously reported increase in the diversity of bacterial species, and these changes could benefit weight loss and weight maintenance as well as contribute to weight regain. Additionally, patients have an increased abundance of Proteobacteria and Verrucomicrobia (Akkermansia) and a decreased abundance of Firmicutes phylum after RYGB [13,14,15].
Although there are some studies that have tried to correlate weight regain after BS with gut microbiota, these studies are from the USA and Europe [11, 16], and analyzed individuals over short follow-up times. In this regard, it is known that microbiota vary according to geography, cultural habits, age, lifestyle factors, and also ethnic differences.
In addition, there is no study that correlated gut microbiota with weight regain after BS with a follow-up of at least 5 years. Therefore, the aim of this study was to analyze the profile of the microbiota among patients with and without late weight regain after RYGB and to compare with a control group (CG) comprised of Brazilian obese individuals (mixed ethnic background).
Methods
Study Design and Population
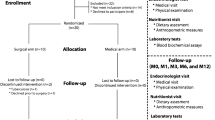
This is a cross-sectional study which enrolled 34 morbidly obese women (body mass index (BMI) 35–49.9 kg/m2), followed at Brasilia University and from a private healthcare facility from January 2016 through December 2017. These individuals were divided into three independent groups according to the period evaluated. Between January 2005 and December 2008, 26 morbidly obese women underwent BS (RYGB). Among them, 12 patients were identified without weight regain (RYGB non-regain), and 14 patients with weight regain (RYGB regain). The third group was comprised of preoperative patients (CG). The inclusion criteria were age between 18 and 65 years, both genders and the ability to understand the study protocol. The exclusion criteria were pregnancy, T2DM, and vulnerable groups (mentally ill institutionalized, or age below 18 years or above 65 years).
Anthropometric measurements of patients were obtained during nutrition evaluation as well as demographic and clinical data. Blood samples were collected. During routine visits, patients who met the study criteria signed the informed consent form. All the bariatric procedures were performed by the same surgical team and with the same technique.
The study protocol was evaluated and approved by the local institutional review board under the reference numbers UNB 2.826.509/2014 and 2.826.509/2018.
Surgical Procedure
The main characteristics of the laparoscopic RYGB were a 30-ml gastric pouch, a 50-cm biliopancreatic limb, and a 100-cm alimentary limb. All proceedings were performed by the same surgical team.
Measurements
BMI was measured in all groups in all preoperative groups and after surgery in the RYGB groups. The percentage excess weight loss (%EWL) was calculated using the formula: %EWL = ((preoperative BMI – current BMI) ÷ (preoperative BMI − 25) × 100). The percentage total weight loss (%TWL) was calculated using the formula %TWL = ((preoperative weight–current weight) ÷ preoperative weight) × 100). The occurrence of weight regain after BS was defined as a minimal 15.0% increase after the lowest weight achieved following surgery [16].
For glucose and insulin analysis, blood samples were collected in tubes with EDTA3 plus Sigma diprotin. Serum samples were stored in the freezer at − 80 °C for later analysis (ELISA, Bayer Corp.). Homeostasis model assessment (HOMA) was calculated by means of the formula of Matthews [17].
Metagenome Profile
Fecal samples were collected and immediately stored at − 80 °C until analysis. After surgery, the microbiota was determined at approximately 55 months in the RYGB non-regain and at 84 months in the RYGB regain. The total DNA of fecal samples was extracted with the Stool PSP Spin DNA kit (STRATEC Biomedical AG, Germany).
To profile microbiota composition, the hyper-variable region (V3–V4) of the bacterial 16S rRNA gene was amplified by following the Illumina 16S Metagenomic Sequencing Library Preparation guide [18] which uses the following sequence: 338F-5′-TCGTCGGCAGCGTCAG ATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3 and 785R-5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′ (2 × 300 bp paired-end and insert size of ~ 550 bp).
Bioinformatics and Statistical Data Analysis
The analysis of the obtained sequences was performed with MiSeq Reporter software (Illumina), which includes demultiplexing, FASTQ file generation, alignment, and variant calling. The bioinformatic analysis of the sequences was performed using the QIIME2 package [19, 20]. This includes quality filtering, OTU picking, taxonomic assignment, phylogenetic reconstruction, diversity analyses, and visualizations For the subsequent data analysis, MicrobiomeAnalyst was used [21]. The different taxonomies were also identified with the linear discriminant analysis (LDA) effect size (LEfSe) [22]. LDA is a method used to find a linear combination of features that characterizes or separates two or more classes of objects or events. The measure of species diversity was performed according to the Shannon and Simpson diversity indices.
The metagenomics of the gut microbiome was realized in the Laboratory of Clinical Investigation in Insulin Resistence – LICRI - University of Campinas-UNICAMP.
Statistics and Analysis
Data are presented as mean values ± standard deviation. For continuous variables, parametric statistics (t tests and ANOVA) were used when necessary. The significance level adopted was 5% for all statistical tests (p < 0.05). SPSS 24.0 and GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA) were used for the analysis.
Results
There were no differences in regard to BMI before and after surgery, postoperative follow-up time, and preoperative age and weight, among groups. RYGB regain presented a significantly higher frequency of weight regain (p < 0.001), but there was no difference in %EWL and %TWL. Homa-IR was significantly higher in CG when compared with groups RYGB non-regain and RYGB regain (p < 0.001). Table 1 summarizes the main anthropometric, demographic, and surgical data, as well as the complete values for HOMA-IR.
Analysis of the Gut Microbiota
Alpha diversity, estimated by the Simpson and Shannon indices, was higher in groups RYGB non-regain and RYGB regain, when compared with CG (Simpson: obese vs RYGB non-regain (p = 0.0208) or RYGB regain (p = 0.0106); Shannon: obese vs RYGB non-regain (p = 0.0052) or RYGB regain (p = 0.0024)) (Fig. 1a, b).
Comparison of α diversity by Simpson and Shannon indices. a Alpha diversity, estimated by Simpson (Simpson: Obese vs RYGB non-regain p = 0.0208 or RYGB regain p = 0.0106) and b Shannon (Shannon: obese vs RYGB non-regain p = 0.0052 or RYGB regain p = 0.0024). Data are presented as means ± S.E.M. from n ≥ 8 per group. #RYGB regain vs obese, *RYGB non-regain vs obese, and &RYGB regain vs RYGB non-regain
The microbiota analysis showed an increase of Proteobacteria phylum after RYGB. However, there was a difference only between CG and RYGB regain (p = 0.0003) (Fig. 2a). Both RYGB non-regain and RYGB regain groups showed a lower abundance of the phylum Bacteroidetes when compared with CG (p = 0.0018) (Fig. 2b). The abundance of phylum Verrucomicrobia was increased in group RYGB non-regain (RYGB non-regain vs CG; p = 0.0117; RYGB non-regain vs RYGB regain; p = 0.0357), but there was no difference between CG and RYGB regain. (Fig. 2c).
The relative abundance of bacterial phyla: a phylum Proteobacteria (RYGB regain vs obese p = 0.0003). b Phylum Bacteroidetes (RYGB regain vs obese p = 0.0018 or RYGB non-regain vs obese p = 0.0155). c Phylum Verrucomicrobia (RYGB non-regain vs obese p = 0.0117 or RYGB non-regain vs RYGB regain p = 0.0357). Data are presented as means ± S.E.M. from n ≥ 8 per group. #RYGB regain vs obese, *RYGB non-regain vs obese, and &RYGB regain vs RYGB non-regain
Linear discriminant analysis (LDA) of the gut microbiota showed a significant change in the proportion of the nine genera among the groups (p < 0.05) (Fig. 3). The genera Bacteroides and SMB53 were increased in CG. Group RYGB regain showed more abundance of Succinivibrio, Coprococcus, Lachnobacterium, and Klebsiella. The genera Akkermansia (Verrucomicrobia phylum) and Phascolarctobacterium were increased in group RYGB non-regain.
In addition, group RYGB non-regain showed a difference for the abundance of genus Akkermansia when compared with CG and group RYGB regain (RYGB non-regain vs CG (p = 0.0117) or groups RYGB non-regain vs RYGB regain (p = 0.0355)) (Fig. 4a).
The relative abundance of bacterial genera: a genus Akkermansia (RYGB non-regain vs obese p = 0.0117 or RYGB non-regain vs RYGB regain (p = 0.0355)); b-c genus Phascolarctobacterium and genus SMB53 (RYGB non-regain vs obese, p = 0.0035 and p = 0.0041, respectively). d–h The genera Streptococcus (obese vs RYGB non-regain (p = 0.0250) or RYGB regain (p = 0.0330)), Enterococcus (obese vs RYGB non-regain (p = 0.0354) or RYGB regain (p = 0.0498)), Succinivibrio (obese vs RYGB non-regain (p = 0.0065) or RYGB regain (p = 0.0065)), Lachnobacterium (obese vs RYGB non-regain (p = 0.0194) or RYGB regain (p = 0.0080)), and Klebsiella (obese vs RYGB non-regain (p = 0.0153) or RYGB regain (p = 0.0279)). i Genus Faecalibacterium (obese vs RYGB non-regain (p = 0.0170) or RYGB regain (p = 0.0044)). Data are presented as means ± S.E.M. from n ≥ 8 per group. #RYGB regain vs obese, *RYGB non-regain vs obese, and &RYGB regain vs RYGB non-regain
Group RYGB non-regain showed a greater abundance of the Phascolarctobacterium genus and lower of the SMB53 genus when compared with CG (p = 0.0035 and p = 0.0041, respectively). Although without significant difference between groups, group RYGB regain showed intermediate abundance in genera Phascolarctobacterium and SMB53 (Fig. 4b and c).
The genera Streptococcus (CG vs group RYGB non-regain (p = 0.0250) or RYGB regain (p = 0.0330)), Enterococcus (CG vs group RYGB non-regain (p = 0.0354) or RYGB regain (p = 0.0498)), Succinivibrio (CG vs group RYGB non-regain (p = 0.0065) or RYGB regain (p = 0.0065)), Lachnobacterium (CG vs group RYGB non-regain (p = 0.0194) or RYGB regain (p = 0.0080)), and Klebsiella (CG vs group RYGB non-regain (p = 0.0153) or RYGB regain (p = 0.0279)) were increased in both RYGB non-regain and RYGB regain groups when compared with CG (Fig. 4d–h). The genus Faecalibacterium was decreased in both RYGB non-regain and RYGB regain groups when compared with CG (CG vs group RYGB non-regain (p = 0.0170) or RYGB regain (p = 0.0044)) (Fig. 4i).
Discussion
The major finding of the present study is that on a long-term course after BS (5-7 years), the individuals without weight regain showed an increase in the relative abundance of the genera Phascolarctobacterium and a reduction in SMB53 compared with obese patients, and also an increase in Verrucomicrobia and a decrease in Proteobacteria compared with those who presented weight regain.
Although our focus is on the microbiota profile of weight regain, the two groups, with or without regain, presented a microbiota composition completely different from the obese group without BS, a finding which deserves some discussion.
Individuals with obesity have decreased microbial gene richness [11], lower proportions of Bacteroidetes, and higher proportions of Firmicutes [23, 24], but some other studies have produced contradictory results [25]. The reduced diversity of their composition is associated with a reduction of metabolic energy consumption in comparison with the microbiota of lean people [26]. In addition, subjects who underwent gastric bypass have increased abundance of the phyla Verrucomicrobia [27], Gammaproteobacteria (including Enterobacteriaceae), and the Fusobacteriaceae family, and decreases in the proportions of the phylum Firmicutes [14] and the genus Clostridium [26]. It is hypothesized that changes in the duration of the exposure of the gut wall to food, and the differences in pH distribution along the gut after RYGB might contribute to changes in the gut microbiota [26]. The composition of the gut microbiota is established by the host’s genetic background and external factors, including the mode of birth, environmental elements, exercise, and nutrition [28, 29]. In fact, the correlation between weight loss and gut microbiota modulation seems to be not unidirectional, but a complex interplay in both directions.
The genus Succinivibrio was increased in both RYGB groups. Nakayama et al. found that among children exposed to a western high-fat diet, this genus is reduced, reinforcing the relationship of healthy dietary habits and its presence [30]. This genus is known as a plant polysaccharide-fermenting bacterium [31] and can help regulate energy balance [32].
In line with our results, the increase of Enterococcus was observed in humans and animals after surgery [33, 34]. This genus is a butyrate producer that induces significant anti-inflammatory effects and prevents colonization of pathogenic bacteria by competing for adherence sites of the intestinal epithelium [35].
Reduction of gastric acid secretion after RYGB causes an increase of incompletely digested proteins into the intestine, resulting in the production of putrescine [36]. Bacteria from the genus Klebsiella, which was increased in both the RYGB groups, can produce putrescine. This polyamine can be metabolized to GABA, stimulating the increase of GLP-1 levels, improving insulin resistance [37, 38]. Likewise, the genus Lachnobacterium, which was increased in both the RYGB groups, has been shown to improve insulin resistance and glucose homeostasis through short-chain fatty acids (SCFAs) production [39, 40], indicating that both genera Klebsiella and Lachnobacteriumare are associated with an overall anti-obesogenic metabolic profile.
In the individuals without regain, there was an increase in the Verrucomicrobia phylum and a decrease in the Proteobacteria phylum. In this regard it is important to mention that Chevalier et al. analyzed the effects of cold exposure on gut microbiota and observed that this stimulus, which is associated with a necessity of maximizing energy uptake from consumed food, was also correlated with almost complete depletion of the Verrucomicrobia phylum. This finding led to the understanding that the absence of this phylum is linked with an evolutive mechanism associated with calorie uptake and thus potentially associated with obesity, which seems to be clearly linked to the findings of the current study [41].
Akkermansia muciniphila (Verrucomicrobia phylum) has been shown to have an important role both on improved glucose homeostasis and weight loss as well as on gut epithelium heath [42]. Previous data showed that A. muciniphila increases after RYGB [27], and this genus was increased in the individuals without regain. It is also interesting to observe that the gene richness remains even after 5 years of surgery. Considering this finding and since A. muciniphila is the most abundant species of Verrucomicrobia, we may suggest that this species is linked to the stability of weight loss after RYGB [41].
The RYGB group which presented weight regain showed differences in the relative abundance of two genera related to weight loss: Phascolarctobacterium and SMB53. The genus Phascolarctobacterium, an SCFAs producer (butyrate, acetate, and propionate), has a positive correlation with weight loss [43, 44] which seems to be related with the finding that this genus is associated with lower systemic succinate levels; elevated systemic succinate is paralleled with impairment of glucose homeostasis and atherosclerotic disease [44]. This genus was increased in the individuals without regain, but was decreased in those with regain, which may be associated with different dietary habits between groups, or either present a direct causal relationship with the tendency to weight regain due to its association with circulating succinate levels and their negative metabolic consequences.
In contrast, studies suggest that the genus SMB53 contributes to obesity [45, 46] and this genus was decreased in the individuals in the without regain group and increased in those who regained weight. Studies showed that this genus specifically is found in a lesser proportion in populations that consume high-fiber diets with resistant starch, fructooligosaccharides, and inulin [47]. Different types of fiber have different potentials to generate SCFAs, as do the microbiota that ferments the fiber sources [48].
The present study has some limitations. One of the major limitations is the differences in the postoperative time of gut microbiota assessment (Table 1). Minor limitations are as follows: all of the participants were female and these results might not be applied to men. Second, this is a cross-sectional study, which avoids the determination of cause-effect relationships since it was not possible to compare the gut microbiota profile of each subject, before and after surgery. Lastly, future follow-up studies on metabolomics and metagenomics are recommended to elucidate the metabolic role of the intestinal microbiota after RYGB.
In summary, this study showed the difference in the gut microbiota profile of individuals who underwent RYGB with a successful weight loss or not, 5 to 7 years after surgery. Furthermore, some genera were inherent in the RYGB itself such as Succinivibrio, Enterococcus, Klebsiella, and Lachnobacterium and are associated with an overall anti-obesogenic metabolic profile. However, a difference was found for two other genera, Phascolarctobacterium and SMB53, which are specific to eating habits and related to weight loss or weight regain respectively. In addition, individuals without regain presented an increase in Verrucomicrobia and a decrease in Proteobacteria compared with those who regained weight. It is likely that the anatomical barrier due to surgery may prevent the proliferation of bacteria related to obesity. These data indicate weight regain after RYGB correlated with some gut microbiota patterns, suggesting new targets to maintain weight loss after BS (Fig. 5).
Conclusion
The gut microbiota of individuals who presented weight regain after RYGB was significantly different in comparison to individuals with a successful weight loss, a finding that points towards a significant role of gut microbiota on weight loss and maintenance after surgery.
References
Anhê FF, Varin TV, Schertzer JD, et al. The gut microbiota as a mediator of metabolic benefits after bariatric surgery. Can J Diabetes. 2017;41:439–47.
Angrisani L, Santonicola A, Iovino P, et al. IFSO Worldwide Survey 2016: Primary, endoluminal, and revisional procedures. Obes Surg. 2018;28:3783–94.
Magro DO, Geloneze B, Delfini R, et al. Long-term weight regain after gastric bypass: a 5-year prospective study. Obes Surg. 2008;18:648–51.
Magro DO, Ueno M, Coelho-Neto JS, et al. Long-term weight loss outcomes after banded Roux-en-Y gastric bypass: a prospective 10-year follow-up study. Surg Obes Relat Dis. 2018;14:910–7.
King WC, Hinerman AS, Belle SH, et al. Comparison of the performance of common measures of weight regain after bariatric surgery for association with clinical outcomes. JAMA. 2018;320:1560–9.
Montastier E, Chalret du Rieu M, Tuyeras G, et al. Long-term nutritional follow-up post bariatric surgery. Curr Opin Clin Nutr Metab Care. 2018;21:388–93.
Saad MJA, Santos A, Prada PO. Linking gut microbiota and inflammation to obesity and insulin resistance. Physiology. 2016;31:283–93.
Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–5.
Valentina Tremaroli FK, Werling M, Stahlman M, et al. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 2015;22:10.
Liou AP, Paziuk M, Luevano JM, et al. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med. 2013;5:178ra141.
Aron-Wisnewsky J, Prifti E, Belda E, et al. Major microbiota dysbiosis in severe obesity: fate after bariatric surgery. Gut. 2019;68:70–82.
Wabitsch M. Gastrointestinal endocrinology in bariatric surgery. Endocr Dev. 2017;32:124–38.
Koutnikova H, Genser B, Monteiro-Sepulveda M, et al. Impact of bacterial probiotics on obesity, diabetes and non-alcoholic fatty liver disease related variables: a systematic review and meta-analysis of randomised controlled trials. BMJ Open. 2019;9:e017995.
Kamvissi-Lorenz V, Raffaelli M, Bornstein S, et al. Role of the gut on glucose homeostasis: lesson learned from metabolic surgery. Curr Atheroscler Rep. 2017;19:9.
Magouliotis DE, Tasiopoulou VS, Sioka E, et al. Impact of bariatric surgery on metabolic and gut microbiota profile: a systematic review and meta-analysis. Obes Surg. 2017;27:1345–57.
Varma S, Clark JM, Schweitzer M, et al. Weight regain in patients with symptoms of post-bariatric surgery hypoglycemia. Surg Obes Relat Dis. 2017;13:1728–34.
Matthews DR, Hosker JP, Rudenski AS, et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–9.
Illumina 16S metagenomic sequencing library preparation (Illumina Technical Note 15044223)
Lawley B, Tannock GW. Analysis of 16S rRNA gene amplicon sequences using the QIIME software package. Methods Mol Biol. 2017;1537:153–63.
Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol 2012: Chapter 1:Unit 1E.5.
Dhariwal A, Chong J, Habib S, et al. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45:W180–8.
Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60.
Rosenbaum M, Knight R, Leibel RL. The gut microbiota in human energy homeostasis and obesity. Trends Endocrinol Metab. 2015;26:493–501.
Zhu J, Gupta R, Safwa M. The mechanism of metabolic surgery: gastric center hypothesis. Obes Surg. 2016;26:1639–41.
Tseng CH, Wu CY. The gut microbiome in obesity. J Formos Med Assoc. 2019;118(Suppl 1):S3–9.
Muscogiuri G, Cantone E, Cassarano S, et al. On behalf of the obesity programs of nutrition E, R.search and assessment (OPERA) group: gut microbiota: a new path to treat obesity. Int J Obes Suppl. 2019;9:10–9.
Shen N, Caixàs A, Ahlers M, et al. Longitudinal changes of microbiome composition and microbial metabolomics after surgical weight loss in individuals with obesity. Surg Obes Relat Dis. 2019;15:1367–73.
Levin AM, Sitarik AR, Havstad SL, et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci Rep. 2016;6:31775.
Hughes RL. A review of the role of the gut microbiome in personalized sports nutrition. Front Nutr. 2019;6:191.
Nakayama J, Yamamoto A, Palermo-Conde LA, et al. Impact of westernized diet on gut microbiota in children on Leyte Island. Front Microbiol. 2017;8:197.
Ou J, Carbonero F, Zoetendal EG, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. 2013;98:111–20.
Murugesan S, Ulloa-Martínez M, Martínez-Rojano H, et al. Study of the diversity and short-chain fatty acids production by the bacterial community in overweight and obese Mexican children. Eur J Clin Microbiol Infect Dis. 2015;34:1337–46.
Guo Y, Huang ZP, Liu CQ, et al. Modulation of the gut microbiome: a systematic review of the effect of bariatric surgery. Eur J Endocrinol. 2018;178:43–56.
Ulker İ, Yildiran H. The effects of bariatric surgery on gut microbiota in patients with obesity: a review of the literature. Biosci Microbiota Food Health. 2019;38:3–9.
Avram-Hananel L, Stock J, Parlesak A, et al. E durans strain M4-5 isolated from human colonic flora attenuates intestinal inflammation. Dis Colon Rectum. 2010;53:1676–86.
Palleja A, Kashani A, Allin KH, et al. Roux-en-Y gastric bypass surgery of morbidly obese patients induces swift and persistent changes of the individual gut microbiota. Genome Med. 2016;8:67.
Kurihara S, Kato K, Asada K, et al. A putrescine-inducible pathway comprising PuuE-YneI in which gamma-aminobutyrate is degraded into succinate in Escherichia coli K-12. J Bacteriol. 2010;192:4582–91.
Urbain JL, Penninckx F, Siegel JA, et al. Effect of proximal vagotomy and Roux-en-Y diversion on gastric emptying kinetics in asymptomatic patients. Clin Nucl Med. 1990;15:688–91.
Cornejo-Pareja I, Martín-Núñez GM, Roca-Rodríguez MM, Cardona F, Coin-Aragüez L, Sánchez-Alcoholado L, Gutiérrez-Repiso C, Muñoz-Garach A, Fernández-García JC, Moreno-Indias I, Tinahones FJ. Eradication treatment alters gut microbiota and GLP-1 secretion in humans. J Clin Med 2019: 8.
Zupancic ML, Cantarel BL, Liu Z, et al. Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome. PLoS One. 2012;7:e43052.
Chevalier C, Stojanović O, Colin DJ, et al. Gut microbiota orchestrates energy homeostasis during cold. Cell. 2015;163:1360–74.
Debédat J, Clément K, Aron-Wisnewsky J. Gut microbiota Dysbiosis in human obesity: impact of bariatric surgery. Curr Obes Rep. 2019;8:229–42.
Muñiz Pedrogo DA, Jensen MD, Van Dyke CT, et al. Gut microbial carbohydrate metabolism hinders weight loss in overweight adults undergoing lifestyle intervention with a volumetric diet. Mayo Clin Proc. 2018;93:1104–10.
Serena C, Ceperuelo-Mallafré V, Keiran N, et al. Elevated circulating levels of succinate in human obesity are linked to specific gut microbiota. ISME J. 2018;12:1642–57.
Nirmalkar K, Murugesan S, Pizano-Zárate ML, et al. Gut microbiota and endothelial dysfunction markers in obese Mexican children and adolescents. Nutrients. 2018;10
Ziętak M, Kovatcheva-Datchary P, Markiewicz LH, et al. Altered microbiota contributes to reduced diet-induced obesity upon cold exposure. Cell Metab. 2016;23:1216–23.
Liu TW, Cephas KD, Holscher HD, et al. Nondigestible Fructans alter gastrointestinal barrier function, gene expression, histomorphology, and the microbiota profiles of diet-induced obese C57BL/6J mice. J Nutr. 2016;146:949–56.
Lewis JD, Abreu MT. Diet as a trigger or therapy for inflammatory bowel diseases. Gastroenterology. 2017;152:398–414.e396.
Funding
We also acknowledge the financial support of the INCT (National Institute of Science and Technology for Diabetes and Obesity) 465693/2014-8 and also the financial support of the FAPDF (Research Support Foundation from Federal District) number 0569.56.30088.09042016.
Author information
Authors and Affiliations
Contributions
SLF, AS, DOM, and MKI designed the study; SLF, DOM, AS, FTV, ESD, and HBA collected and elaborated data; AS, DG, and HBA Microbiota profile; AS, DOM, EC, and MJAS critical revision of the manuscript; DOM and AS statistical analysis; DOM, AS, EC, and MJAS drafted the article, and all authors gave final revision and permission for publication.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
During routine visits, subjects who agreed in participating in the study signed up an informed consent form. All methods were performed in accordance with the relevant guidelines and regulations. The study was approved by Institutional Ethics Review Board the reference UNB 2.826.509/2014 and 2.826.509/2018.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Faria, S.L., Santos, A., Magro, D.O. et al. Gut Microbiota Modifications and Weight Regain in Morbidly Obese Women After Roux-en-Y Gastric Bypass. OBES SURG 30, 4958–4966 (2020). https://doi.org/10.1007/s11695-020-04956-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11695-020-04956-9