Abstract
Food safety and quality are issues of great concern to food producers and consumers. Key challenges towards achieving these include availability of rapid, user-friendly, economic and reliable techniques to detect problems such as antibiotic residues. Therefore, the present study was conducted to evaluate performance of plastic film test plate (PFTP) as influenced by antibiotic type, test bacterium, inoculum volume and concentration. Comparison was made to the microtiter test plate (MTP) and the National Standard method (SN/T 3979-2014). For this, antibiotic susceptibility testing was conducted on four bacterial types (Micrococcus luteus, Streptococcus thermophillus suspensions, Staphylococcus aureus and Coliform bacteria) against three antibiotics (penicillin G, sulfadiazine and tetracycline). Results showed high susceptibility of Micrococcus luteus to penicillin G with minimum inhibitory concentration of 3 µg L−1 and 1 µg L−1 via PFTP and MTP respectively. Optimum performance was realized at bacterial concentration of 104 CFU ml−1 with detection limit of 1 µg L−1, sensitivity and predictive positive value (PPV) of 81–83% and 97–100% respectively. Detection time was recorded as 6 and 9 h for MTP and PFTP respectively compared to the 18–24 h of the National Standard method (SN/T 3979–2014). The microbiological plastic film test plate under optimized bacterial culture conditions demonstrated tremendous potential for rapid and reliable detection of antibiotics in milk.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Detection of antibiotic residues along the food supply chain is an issue of great concern not only to food producers but also food regulatory bodies in most developing countries. Antibiotics are most commonly used for treatment of livestock diseases such as mastitis. Excessive use of these veterinary drugs without observation of withdrawal periods can result in important economic losses in production of fermented dairy products such as cheese and yoghurt [1, 2]. Furthermore, consumers are exposed to major health risks including allergic reactions, interference with gut microbiota and development of antibiotic resistance which can cause failure of antibiotic therapy [3, 4]. Moreover, health problems such as carcinogenicity, mutagenicity, reproductive disorders and chemical poisoning have also been reported [5, 6]. Withholding period enables reduction of antibiotic residues to negligible concentration in the animal products following their treatment. Approximately 700,000 deaths per year occur globally as a result of antibiotic-resistant infections [7].
In an effort to address food safety and quality challenges, countries have established monitoring programs which determine antibiotic residue levels and establish maximum residual limits (MRLs) [8]. So far a number of highly sophisticated, sensitive and selective analytical tools have been developed including physic-chemical, chromatographic, immuno-chemical, and biosensors methods [9,10,11,12,13,14]. However, most of them are complex, expensive and labour intensive, hence the need for continuous development of new rapid screening techniques and improvement of traditional test methods specifically to meet the needs of developing countries [15, 16].
Due to their non-specificity, high throughput and cost-effectiveness relative to most tests, microbiological screening techniques have been the most commonly used methods. Techniques such as BR-AS Special, CMT-Copan Milk Test, Delvotest SP-NT, Delvotest T, Brilliant Black Reduction Test MRL, Charm Blue Yellow II, Nouws antibiotic test and Screening test for antibiotic residues (STAR) are some of the few commercial screening tools which are used for routine antibiotics analysis in milk [17,18,19,20]. However, accessibility of these tools in developing countries is a challenge due to the cost involved. Moreover, some of them are specific for particular groups of antibiotics. Nevertheless, other microbiological screening techniques which are relatively cheaper and effective have also been developed and successfully tested under various conditions for rapid and sensitive antimicrobial residues detection in many livestock products [21,22,23,24,25,26,27].
Furthermore, complimentary techniques such as plastic film test plate have also been developed. This is a portable and highly cost-effective technique which has been used for rapid detection of viable colony counts, molds, yeasts and antibiotics in food [28,29,30]. In addition to that, the use of cold-water soluble gelling agent which requires no heating for gelation to occur makes it highly energy efficient relatively to the conventional test methods. For antibiotic detection, the technique has only been tested for penicillin G using Micrococcus luteus as a test bacterium. However, a number of factors can influence the performance of microbiological plastic film test plate. These include antimicrobial concentration, volume of test solution, inoculum density, gelling agent thickness, growth medium composition, and incubation temperature [19, 31, 32]. To the best of our knowledge no study has been done on the effect of bacterial culture on the plastic film test plate. It was therefore upon this basis that the present study was conducted to determine the effect of bacterial culture volume and concentration on the microbiological plastic film test plate performance relative to the commonly used microtiter plate and the Chinese National Standard method (SN/T 3979-2014).
Materials and methods
Materials
Guar gum and konjac glucomannan were purchased from West Asia Biochemical Reagent Development Co., Ltd (Beijing, China). Beef heart infusion powder, sodium pyruvate, yeast extract powder, peptone, tryptone and triphenyl-tetrazolium chloride (TTC) were purchased from Aladdin Reagent Co, Ltd (Shanghai, China). Nutrient agar was purchased from Tianjin Xingkai Chemical Factory (Tianjin, China). Micrococcus luteus, Streptococcus thermophillus, Staphylococcus aureus and Coliform bacteria were bought from China Center of Industrial Culture Collection (CICC, Beijing, China). Penicillin G, tetracycline and sulfadiazine were all purchased from Latin Reagent Limited Company (Shanghai, China). Skimmed milk powder was purchased from WALMART supermarket (Changchun, China). All chemicals used were of analytical grade.
Compound gel preparation
Compound gel was prepared in accordance with Ntakatsane et al. [28]. Briefly, powdered gels of guar gum (GG) and konjac glucomannan (KG) were finely ground separately with Taiqi gas crusher and sieved with 120-mesh sieve to give powders with particle size of 0.125 mm. Then they were thoroughly mixed at the ratio of 4:6 (GG:KG).
Preparation of plates
Plastic film test plate (PFTP)
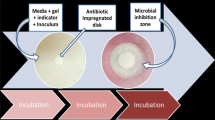
Plastic film disks for antimicrobial detection were prepared according to Xu et al. [30] with slight modifications. The disk structure (8 cm × 7 cm) was made of three-layers; transparent PET silicone film coated with cold water soluble compound gel of guar gum and konjac glucomannan at the ratio of 4:6 (w/w) (upper layer); transparent PP polypropylene with a 6 cm diameter pierced through and a 60 g m−2 non-woven fabrics as culture medium carrier (middle layer); and the bottom layer was a white art paper which held the non-woven fabric in position. Enrichment medium was composed of tryptone (20 mg ml−1), yeast extract powder (15 mg ml−1), sodium chloride (10 mg ml−1), glucose (10 mg ml−1), sodium pyruvate (5 mg ml−1) and beef heart infusion powder (20 mg ml−1). Then 0.2 mg ml−1 triphenyl-tetrazolium chloride (TTC) was added to the enrichment medium. Bacterial inoculation was done according to Ren et al. [29] with slight modifications. Briefly, a volume of 600–1000 µl of bacterial solution with dilutions (103–106 CFU mL−1) was dropped on the center of the PFTP. Then 5–20 µL of milk sample spiked with different concentrations of antibiotics was shortly dropped on the center of the plate after complete dispersion and diffusion of inoculum on the culture region. The plates were then placed in a thermostatic box (Lab Incubator 1925A, VWR International, Hamburg, Germany) at 37 °C for 6–18 h.
Microtiter plates (MTP)
Preparation of the plates was done under aseptic conditions in accordance with Tumini et al. [33] and Sarker et al. [34] with slight modifications. A volume of 100 µL of culture medium was dispensed in each well on the 96-well microtiter plates (Nunclon ™ surface; Nunc, Roskilde, Denmark). Milk sample (100 µL) spiked with antibiotics which were prepared in 0.85% (w/v) sterile saline solution was pipetted into the first row of the plate. Serial dilutions were performed in such a way that 100 µL was transferred from the first row down to the tenth row (100 µL discarded after mixing) leaving a 100 µL solution in each well in serially descending concentrations.
Finally, 5–20 µL of bacterial suspensions (103–6 CFU mL−1, each replicated twice on each plate in three different plates) were added to each well (1st–11th row). The last row was the blank with no antibiotic and bacterial suspension. The plates were covered and sealed to avoid dehydration during incubation (Lab Incubator 1925A, VWR International, Hamburg, Germany) at 37 °C for 6–18 h.
Determination of antibiotic susceptibility
Three different antibiotics, namely penicillin G (PG), tetracycline (TC) and sulfadiazine (SF) were tested on four bacterial cultures: Micrococcus luteus, Streptococcus thermophillus suspensions, Staphylococcus aureus and Coliform bacteria. Antibiotic stock solutions (PG 100 mg mL−1, TC 12.5 mg mL−1 and SF 1 mg mL−1) were prepared in appropriate sterile buffers and serially diluted to prepare working solutions (PG 100 µg L−1, TC 1250 µg L−1, and SF 1000 µg L−1). Then reconstituted non-fat milk was spiked with different antibiotic concentrations in such a way that each antibiotic had 12 samples with a total of 36 for the three antibiotics: 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 and 11 µg L−1 (PG) and 0, 10, 25, 50, 75, 100, 150, 200, 250, 300, 400 and 500 µg L−1 (for both TC and SF). The plates were then prepared as previously explained. Development of microbial inhibition zones (MIZ) on the PFTPs and color change on the microtiter plates were monitored and then visually assessed. The change of color to light yellow/orange on the microtiter plates and MIZ > 2–3 mm on the PFTPs were recorded as positive results (Fig. 1).
Micrococcus luteus susceptibility to various antibiotics under different test plates, [plastic film test plate; a penicillin G (PG), b tetracycline (TC), c sulfadiazine (SF)], d agar plate test; and e microtiter plate test. Zones of inhibition on each plate are developed from the same antibiotic concentration
Determination of minimum inhibitory concentration (MIC)
Establishment of MIC value was done in accordance with standard techniques of CLSI [35]. For PFTP, the lowest antibiotics concentration that led to development of MIZ > 2–3 mm was regarded as MIC for that particular test antibiotic at specified concentration of test bacterial strain. For microtiter plate, the lowest antibiotic concentration at which color change occurred was taken as the MIC value.
Effect of bacterial concentration and volume on MIC
Different bacterial concentrations and volumes were tested to establish optimum conditions for determination of MIC for the three antibiotics (penicillin G, tetracycline and sulfadiazine) using the plastic film test plate and microtiter plate. Bacterial suspensions between 103–6 CFU mL−1 at 600, 700, 800, 900 and 1000 µL were dispensed onto sterile thin plastic film test plates. Same bacterial concentrations at 5, 10, 15 and 20 µL were dispensed onto sterile flat-bottomed 96-well polystyrene microtiter plates. Both plates contained culture medium and antibiotic spiked milk samples at serially decreasing concentrations. The two plates were then incubated at 37 °C and monitored over a period of 6–18 h.
Analysis of dose–response curves
Dose–response curve for each antibiotic was constructed in accordance with Codex Alimentarius [36]. For this, reconstituted non-fat milk fortified with 12 different antibiotic concentrations was assayed for each antibiotic (penicillin G, PG: 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 and 11 µg L−1 and tetracycline, TC: 0, 10, 25, 50, 75, 100, 150, 200, 250, 300, 400, and 500 µg L−1) in such a way that the lowest two concentrations gave negative results and the two highest concentrations gave positive results. A volume of 50 µL of antibiotic spiked milk samples was added to the wells containing 40 µL of enriched culture medium and 10 µL of bacterial culture (104 CFU mL−1). Then the microplates were sealed and incubated at 37 °C for 6–18 h while closely monitoring the color change. To ensure high accuracy of the calculation of the detection limits, photometric readings of each microplate were taken at 560 nm using a Multimode Plate Reader (VICTOR Nivo ™, PerkinElmer Inc., Waltham, MA, USA). Percent relative absorbance was calculated according to Tumini et al. [33] using the following equation:
where: A = relative absorbance, Ax = absorbance of sample with antibiotic concentration ‘x’; A0 = absorbance of antibiotic free (ATB-free) sample (negative control), and A100 = absorbance of sample that yielded 100% positive results. Subsequently, the relative absorbance values were analyzed through SPSS 20 for windows (SPSS Inc., Chicago, IL, USA). The detection Limits were then calculated as the concentration of ATBs that produced 45% of relative absorbance [37].
Effect of bacterial concentration on bioassay response
The effect of Micrococcus luteus concentration on assay response time and detection limit for penicillin G and tetracycline were evaluated. Analysis was carried out using four Micrococcus luteus (103 to 106 CFU mL−1 at 10 µL) suspensions prepared in a culture medium. Subsequently, the dose–response curves of penicillin G (PG: 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 and 11 µg L−1) and tetracycline (TC: 0, 10, 25, 50, 75, 100, 150, 200, 250, 300, 400 and 500 µg L−1) were analyzed as in the previous section. A logistic regression model was used to plot the dose–response curves and to calculate the detection limits:
where Lij = linear logistic model; Aij = relative absorbance (Eq. (1)); β0 = intercept; β1, β2, β3 = estimated parameters for model; ATBi = antibiotic concentrations (i: 1, 2…12 levels); logBCj = logarithmic transformation of bacterial concentration; logBCj2 = square of the logarithmic transformation of the bacterial concentration and εij = residual error.
Sensitivity of bioassay
Milk samples (12) spiked with antibiotics above the MIC were analyzed in triplicate (total of 36 measurements) with two bioassays (plastic film test plate and microtiter plate) containing Micrococcus luteus. Two of the 12 samples were antibiotic free. For this purpose, 10 and 700 µL of the bacterial suspension (104 CFU mL−1) were dispensed on microtiter plate and plastic film test plate respectively. A volume of 10 and 50 µL of antibiotic spiked milk samples was used on PFTP and MTP respectively. The plates were then incubated at 37 °C for 6–18 h. For the study of specificity, visual interpretations of bioassay responses were performed, allowing a dichotomous classification according to the presence (positive result: “orange/light yellow” color on the microtiter plate; microbial inhibition zone ≥ 2 mm on the PFTP) or absence (negative result: “red” color on the microtiter plate; microbial inhibition zone < 2 mm on the PFTP) of inhibitors. For this purpose, visual interpretations of the microtiter plates and measurements of MIZ were done by three individuals separately. For ease of calculation of this parameter, positive results were considered as those presenting at least two coincident visual interpretations. Then sensitivity was calculated using the following mathematical equation:
Statistical analysis
All experiments were run in triplicate and the data are expressed as means ± standard deviation. Data was subjected to analysis of variance (ANOVA), and mean comparisons were carried out using Duncan's multiple range test. The significant level was set at P < 0.05. Statistical analysis was performed using SPSS 20 for windows (SPSS Inc., Chicago, IL, USA).
Results and discussions
Determination of antibiotic susceptibility
Table 1 shows susceptibility of four bacterial cultures (Micrococcus luteus, Staphylococcus aureus, Streptococcus thermophiles and Coliform bacteria) to three antibiotics (penicillin G, sulfadiazine and tetracycline) using two bioassays (PFTP and MTP). All the bacterial cultures except Coliform bacteria showed susceptibility to penicillin G (PG). Only Micrococcus luteus and Staphylococcus aureus demonstrated susceptibility to tetracycline (TC) while none of the bacterial cultures displayed any susceptibility to sulfadiazine (SF). However susceptibility was higher in MTP than in PFTP which may describe why the test tube or microtiter technique has been used more often than the agar diffusion test. Although Staphylococcus aureus has shown susceptibility to both penicillin and tetracycline, it can resist or be protected from antimicrobial activity due to its tendency to reside in biofilm [38,39,40]. Figure 1 displays Micrococcus luteus susceptibility to the three antibiotics under the PFTP (Fig. 1a–c) in comparison to the Chinese National Standard method (SN/T 3979-2014) (Fig. 1d). Sulfadiazine was not detected by any of the methods including the MTP (Fig. 1e). The red color in wells of the plate indicates absence of antibiotics in the samples as seen with the control and blank (negative samples), while orange/yellow color suggested presence of antibiotics (positive samples) as demonstrated by PG and TC. Intensity of yellow color diminishes with decreasing antibiotic concentrations.
Minimum inhibitory concentration (MIC)
MIC refers to the lowest concentration of antimicrobial agent under defined in vitro conditions which inhibits visible growth of micro-organisms within defined period of time [41]. This growth prevention leads to development of microbial inhibition zone (MIZ) greater than 2–3 mm in PFTP, and change of color in MTP. The MICs for penicillin G and tetracycline were found to be 3 and 150 µg/L, 1 and 75 µg/L under PFTP and MTP respectively as shown in Table 2. The PFTP results were not significantly different (p > 0.05) from those obtained from the National Standard method (SN/T 3979-2014) while MTP showed significantly lower MICs (1 µg L−1). The lowest MIC was displayed when using Micrococcus luteus followed by Staphylococcus aureus, Streptococcus thermophiles and then Coliform bacteria. PFTP displayed MIC of 4 µg L−1 which is at the same level as the maximum residue limit established by EU [36]. MIC of bacterium determines the size of inhibition zone, which in turn depends on the bacterial lag phase, generation time of the bacterial strain and its inoculum size, volume of antibiotic and its diffusability into the gel [42].
Bacterial concentration and volume on MIC
Effect of bacterial concentration and volume on MIC of penicillin G was evaluated using Micrococcus luteus and Staphylococcus aureus. Penicillin G was chosen because it is the one used with Chinese National Standard method (SN/T 3979-2014), and it also belongs to a large group of antibiotics (beta-lactams) which are most commonly used in treatment of dairy animal diseases [1, 4]. Bacterial volume and concentration did not show significant effect on MIC (Table 3). This was in agreement with Eloff [43] who found no change in MIC for 1% of Staphylococcus aureus incubated for 1–24 h using a 50% inoculum in the serial microdilution assay. However, a significant effect of inoculum size on MIC has also been reported [44, 45]. The difference in results might be related to the sensitivity of bioassay, test bacteria and type of antibiotics. Eloff [31] hypothesized that with low inoculum, bacterial cells need some time for adaptation to the new environment, which is not the case with large inoculum, hence the significant difference in MIC due to inoculum size. However, the initial microbial population size plays an important role towards the extent of increase in MIC [44, 46]. Efficacy for antibiotics is influenced by bacterial density or inoculum which shows substantial effect such as decrease in the rate and extent of microbial growth inhibition and increases in MIC [47]. For the concentration of Micrococcus luteus, MIC varied between 4–6 and 1–2 µg L−1 in PFTP and MTP respectively. On the other hand, for Staphylococcus aureus concentration, MIC varied from 6–7 to 4–5 µg L−1 in PFTP and MTP respectively. These results were not significantly different (p > 0.05) from those obtained from the Chinese National Standard method, (SN/T 3979-2014), and they were also at or not far from the maximum residue limit of 4 µg L−1 penicillin G in milk established by the European Union [36, 48].
Furthermore, detection time for the bioassays was closely monitored. Bacterial culture volume and concentration did not show significant difference (p > 0.05) in detection time. Development of orange/yellow color and microbial inhibition zones on the MTP and PFTP were observed after 6 and 9 h respectively. Detection time for the PFTP was in line with the study of Ntakatsane et al. [28] who reported the results after 9–15 h. For the MTP, detection time was similar to that of 5.5 and 6 h when using the spores of Bacillus licheniformis & Geobacillus stearothermophilus respectively [33, 49]. The detection time of both bioassays was significantly shorter than the 18–24 h of the conventional disk diffusion test. This can be associated with properties of the gelling agent as previously described [28]. Furthermore, Bidlas et al. [44] also indicated that under optimum environmental conditions, time to detection of bacterial culture depends on the initial inoculum size.
Dose-dependency analysis
Figure 2 shows the dose-dependency curves for penicillin G and tetracycline constructed from antibiotic concentrations and relative absorbance. As antibiotic concentration increases, relative absorbance increased faster for penicillin G (Fig. 2a) than tetracycline curve (Fig. 2b). Detection limits were then calculated as 1 µg L−1 and 300 µg L−1 for penicillin G and tetracycline respectively, and this was in accordance with Althaus et al. [37] who reported detection limit as the concentration of antibiotics that produced 45% of relative absorbance. The rate of increase in relative absorbance could be associated with sensitivity of bacterial culture to a particular antibiotic. Micrococcus luteus was earlier in this study reported to be more sensitive to penicillin G than tetracycline.
Bacterial volume and concentration on assay performance
Assay performance was evaluated using Micrococcus luteus and penicillin G. Relative absorbance for various bacterial volume-concentration combinations are plotted in Fig. 3, and the assay detection limits (DL) for penicillin G and tetracycline are shown in Table 4. Bacterial culture volume and concentration showed significant (p < 0.05) effect on the assay detection limit. At low (103 CFU ml−1) and high (106 CFU ml−1) concentrations (Fig. 3a, d), bioassay performance was negatively affected as shown by high detection limits of 9–10 µg L−1 and 7–10 µg L−1 respectively. This display was ascribed to the low (5 µL) and high (20 µL) bacterial volume. At low bacterial concentration and volume, the cells might be too few to effect change while they also take some time for adaptation to the new growth conditions. Nevertheless, low inoculum size has also been associated with increased number of viable colonies [50]. At the concentration of 104 CFU ml−1, DL was improved to the range of 1–3 µg L−1 at the volume of 5–20 µL (Fig. 3b). However, beyond 104 CFU ml−1 (Fig. 3c), DL increased up to 5 µg L−1. Optimum assay performance was realized at the concentration of 104 CFU ml−1 and volume of 10 µL. These results were in agreement with previous studies which indicated that bacterial concentration of 104–5 CFU ml−1 is ideal for microbiological test methods [28, 29, 45].
The effect of bacterial concentration on the assay detection limit for tetracycline is reflected in Fig. 4. Assay performance improved with increasing concentration from 103 CFU ml−1 to 105 CFU ml−1 (Fig. 4a–c), beyond which it started to decline as displayed in Fig. 4d. The DL improved from the initial minimum concentration of 250 µg L−1 down to 100 µg L−1 at the volume of 10 µL (Table 4). This was in line with the Clinical Lab Standards Institute (CLSI) which recommends the inoculum size of 105 CFU ml−1 for microbiological tests of antibiotics [45]. Therefore, an optimum assay detection conditions for tetracycline are at the bacterial concentration and volume of 105 CFU ml−1 and 10 µL respectively.
Bioassay sensitivity
Sensitivity of the bioassays was calculated for the two antibiotics, penicillin G and tetracycline as shown in Table 5. There was no significant (p > 0.05) difference in the sensitivity between the two techniques (PFTP and MTP). PFTP displayed sensitivity and predictive positive value (PPV) of 81 and 97% respectively for penicillin G while 75 and 90% was found for tetracycline. For both antibiotics, high sensitivity and PPV (83 and 100% respectively) were found when using MTP. Similar high sensitivity of microbiological bioassay has also been reported by Tumini et al. [14] using Bacillus pumilus.
Conclusion
It can be concluded from this study that the performance of plastic film test plate and microtiter plate can be influenced by test bacterium and test conditions such as inoculum concentration and volume. Micrococcus luteus proved to be the most susceptible bacterium to penicillin G and tetracycline. MIC was not significantly affected by the inoculum volume and concentration as was the detection limit. Sensitivity of PFTP was higher for penicillin G than tetracycline, and MTP proved to be more sensitive than the PFTP. Nevertheless, PFTP has shown great potential as a simple portable antimicrobial screening tool with several advantages such as simplicity, cost-effectiveness, energy saving and relative speed compared to the conventional agar plate test. Time to detection was recorded as 6 and 9 h for MTP and PFTP respectively.
References
M.I. Berruga, A. Molina, R.L. Althaus, M.P. Molina, Small Rumin. Res. 142, 38 (2016)
N. Bilandžić, B. Kolanović, I. Varenina et al., Food Control 22, 1941 (2011)
F. Conzuelo, V. Montiel, S. Campuzano et al., Anal. Chim. Acta 820, 32 (2014)
P. Sanders, A. Bousquet-Melou, C. Chauvin, P.L. Toutain, INRA Prod. Anim. 24, 199 (2011)
Z. Khoshbin, A. Verdian, M. Housaindokht, M. Izadyar, Z. Rouhbakhsh, Biosens. Bioelectron. 122, 263 (2018)
W.S. Darwish, E.A. Eldaly, M.T. El-Abbasy et al., Jpn. J. Vet. Res. 61, S13 (2013)
C.Y. Liu, Y.Y. Han, P.H. Shih et al., Science Reports 6, 23375 (2016)
H.H. Chung, J.B. Lee, Y.H. Chung, K.G. Lee, Food Chem. 113, 29 (2009)
Z. Liu, Y. Zhong, Y. Hu et al., Food Chem. 270, 573 (2019)
Q. Wang, W.M. Zhao, Sens. Actuators B 269, 238 (2018)
M. Kamal, R. Karoui, LWT Food Sci. Technol. 79, 586 (2017)
L. Kantiani, M. Farré, D. Barceló, TrAC Trends Anal. Chem. 28, 729 (2009)
S. Aytenfsu, G. Mamo, B. Kebede, J. Nutr. Food Sci. 6, 4 (2016)
M. Tumini, O.G. Nagel, R.L. Althaus, J. Dairy Res. 82, 248 (2015)
M. Kamal, R. Karoui, Trends Food Sci. Technol. 46, 27 (2015)
M. Ntakatsane, P. Chen, J. Liu et al., Food Meas. 14, 1892 (2020)
M. Tumini, O.G. Nagel, R.L. Althaus, Rev. Argent Microbiol. 51, 345 (2019)
M.G. Pikkemaat, M.L.B.A. Rapallini, S.O. Van Dijk, J.W.A. Elferink, Anal. Chim. Acta 637, 298 (2009)
R. Romero, S. Van Weyenberg, M.P. Molina, W. Reybroeck, Int. Dairy J. 62, 39 (2016)
M.C. Beltrán, T. Romeo, R.L. Althaus, M.P. Molina, J. Dairy Sci. 96, 2737 (2013)
R. Parthasarathy, C.E. Monette, S. Bracero, M.S. Saha, FEMS Microbiol. Ecol. 94, fiy105 (2018)
Q. Wu, Q. Zhu, Y. Liu et al., J. Dairy Sci. 102, 10825 (2019)
I. Kukurova, B. Hozova, J. Food Nutr. Res. 46, 9 (2007)
R.M. Kalunke, G. Grasso, R. D'Ovidio, R. Dragone, C. Frazzoli, Microchem. J. 136, 128 (2018)
Q. Wu, S. Peng, Q. Liu, et al., Front. Microbiol. 10 (2019)
P. Sun, Y. Liu, J. Sha et al., Biosens. Bioelectron. 26, 1993 (2011)
F. Deiss, M.E. Funes-Huacca, J. Bal, K.F. Tjhung, R. Derda, Lab Chip 14, 167 (2014)
M. Ntakatsane, P. Chen, J. Liu et al., Food Measure 14, 2087 (2020)
D. Ren, P. Chen, Y. Wang et al., LWT Food Sci. Technol. 82, 335 (2017)
L. Xu, P. Chen, T. Liu et al., LWT Food Sci. Technol. 117, 108687 (2020)
J.N. Eloff, BMC Complement. Alter. Med. 19, 106 (2019)
O.G. Nagel, M.P. Molina, J.C. Basίlico et al., Lett. Appl. Microbiol. 48, 744 (2009)
M. Tumini, O. Nagel, M.P. Molina, R. Althaus, Int. Dairy J. 64, 9 (2017)
S.D. Sarker, L. Nahar, Y. Kumarasamy, Methods 42, 321 (2007)
R. Saginur, M. StDenis, W. Ferris et al., Antimicrob. Agents Chemother. 50, 55 (2006)
European Commission, OJ 1, L15 (2010)
R.L. Althaus, A. Torres, A. Montero, S. Balach, M.P. Molina, J. Dairy Sci. 86, 457 (2003)
D. Chudobova, S. Dostalova, I. Blazkova et al., Int. J. Environ. Res. Public Health 11, 3233 (2014)
F. Van den Driessche, G. Brackman, R. Swimberghe, P. Rigole, T. Coenye, Int. J. Antimicrob. Agents 49, 315 (2017)
G.A. O'Toole, J. Vis. Exp. 47, 2437 (2011)
EUCAST of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID), (2003) https://doi.org/10.1046/j.1469-0691.2003.00790.x
N. Høiby, K. Henneberg, H. Wang et al., Int. J. Antimicrob. agents 53, 564 (2019)
J.N. Eloff, Planta Med. 64, 711 (1998)
E. Bidlas, T. Du, R.J. Lambert, Int. J. Food Microbiol. 126, 140 (2008)
K.P. Smith, J.E. Kirby, Antimicrob. Agents Chemother. 62, e00433–e518 (2018)
J. Karslake, J. Maltas, P. Brumm, K.B. Wood, PLoS Comput. Biol. 12, e1005098 (2016)
K.I. Udekwu, N. Parrish, P. Ankomah, F. Baquero, B.R. Levin, J. Antimicrob. Chemother. 63, 745 (2009)
O.G. Nagel, M.C. Beltrán, M.P. Molina, R.L. Althaus, Small Ruminant Res. 102, 26 (2012)
O. Nagel, M.P. Molina, R. Althaus, Int. Dairy J. 32, 150 (2013)
K.E.R. Davies, S.J. Joseph, P.H. Janssen, Appl. Environ. Microbiol. 71, 826 (2005)
Funding
We gratefully acknowledge the financial support from Jilin Provincial Science & Technology Development Plan under International Science and Technology Cooperation Project: Research on Rapid Detection Technology of Antibiotic Residues in Milk. (No.: 20200801072GH).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ntakatsane, M., Chen, P., Liu, J. et al. Optimization of microbiological plastic film test plate conditions for rapid detection of antibiotics in milk. Food Measure 14, 3473–3482 (2020). https://doi.org/10.1007/s11694-020-00576-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11694-020-00576-0