Abstract
Purpose
Henneguya Thélohan, 1892 is one of the most species-rich genera of myxosporean parasites and infects fish around the world. The present study describes a new species infecting the gill filaments, fins, and kidneys of Plagioscion squamosissimus (Heckel, 1840), an economically important freshwater fish distributed in watersheds in the north of South America.
Methods
A total of 108 P. squamosissimus specimens were examined from three geographic localities in the Amazon basin: the Lago Grande do Curuai, a marginal lake of the Amazon River; the Tapajós River, in the state of Pará; and the Solimões River, in the state of Amazonas, Brazil. The analyses were based on the myxospore morphology, ribosomal DNA sequencing, phylogeny, prevalence, and geographic distribution of the host and its parasite.
Results
Parasite prevalences were 50% in both the Tapajós and Solimões rivers, and 35.4% in the Lago Grande do Curuai. In terms of the site of infection, the prevalence total was 23.1% in the gill filament, 29.6% in the fins, and 1.8% in the kidney. Regarding gender, the prevalence was 59.5% for males, 32.5% for females, and 21.7% for undetermined sex. The specimens found here were both morphologically and molecularly identical regardless of the infected organ and geographic locality, but distinct from all other Henneguya species, revealing that the parasite reported represents a novel species named Henneguya longisporoplasma n. sp. Despite the sampling being carried out in three different geographic localities of the Amazon basin, no population-level genetic variation was observed, even in the typically more variable ITS-1 region, revealing a panmictic population of H. longisporoplasma n. sp. in this large watershed. Maximum likelihood and Bayesian analyses showed the novel Henneguya clustered as a sister branch of the subclade formed of Henneguya that infect fish belonging to the family Cichlidae.
Conclusions
A novel Henneguya species was identified parasitizing P. squamosissimus. The parasite presented wide geographic distribution in the Amazon basin and genetic analyses showed it as revealing a panmictic population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myxozoa (Cnidaria: Endocnidozoa) is a large, widespread aquatic group of microscopic, obligate endoparasites that can cause severe ecological and economic damage [1, 2]. More than 2600 myxozoan species have been identified, in both marine and freshwater environments, worldwide [3]. Some of these parasites have been reported to be highly pathogenic, resulting in severe debility and mortality in fish hosts [4].
Only a few investigations determining the population structure of myxozoan parasites are available. Genetic structure in the population of Ceratonova shasta (Noble, 1950) with different host associations, was found in the upper and lower Klamath River basin, Oregon/California in the USA [5, 6]. Oceanographic barriers in areas of the African coast were hypothesized to play important roles in the population structuring of the parasite Ceratomyxa cottoidii Reed, Basson, Van As & Dyková, 2007 as for its fish host [7]. In South America, evidence for panmixia was observed in populations of Ceratomyxa gracillima Zatti, Atkinson, Maia, Bartholomew & Adriano, 2017 from different rivers along the Amazon basin [8].
Henneguya Thélohan, 1892 is the second most species-rich genus from the Myxobolidae family and is represented by more than 250 species. Most members of this genus parasitize freshwater fish, with few species described from marine and brackish habitats [9, 10]. Sixty Henneguya species have been described in Brazil, of which 26 were reported from the Amazon basin [9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24].
Plagioscion squamosissimus (Heckel, 1840) is a species of the family Sciaenidae Cuvier, 1829. This species was included in Order Acanthuriformes by [25] and allocated for the Order incertae sedis Eupercaria [26]. It is a non-migratory fish popularly known as pescada-branca or curvina (Portuguese) and South American silver croaker (English) [27, 28]. The original distribution is limited to the Orinoco and Amazon basins, and the rivers of the Guianas, but that has been more recently introduced into the La Plata and São Francisco Basins, as well as several artificial reservoirs in Brazil, due to its high economic value [27,28,29,30]. Despite its wide distribution and considerable commercial value, few myxosporean surveys have been conducted on this fish, thus far known to host a Kudoa sp. in the musculature [31], and Ellipsomyxa plagioscioni Zatti, Maia & Adriano, 2020 in the gall bladder [32], both reported from the Amazon River, state of Pará [31, 32].
In the present study, we describe a novel Henneguya species, the first reported from Plagioscion squamosissimus (Heckel 1840) and a Sciaenidae fish worldwide. Catches were performed in three rivers within the Amazon basin. Our analyses were based on the morphology of myxospores, SSU rDNA and ITS-1 sequencing, phylogeny, and geographic distribution of the new myxosporean species.
Materials and Methods
Fish Collection
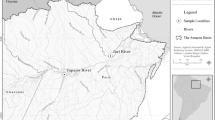
Specimens of P. squamosissimus were collected from three rivers in the Amazon basin: in the Lago Grande do Curuai, a marginal lake of the Amazon River, in the municipal region of Santarém, in the state of Pará (coordinates 2°08′04″S 55°37′54″W), in September 2017 (n = 48); the Tapajós River, in the municipal region of Santarém (coordinates: 2°21′11″S 54°46′11″W) (n = 44) and the Solimões River, near the city of Manaus, in the state of Amazonas (coordinates 3°13′21''S, 59°59′52''W) (n = 16), in March 2018 (Fig. 1). Catches were authorized by the Brazilian Ministry of the Environment (SISBIO nº 44268–9 and 67616–1) and performed using both seine nets and fishing hooks. Live specimens were transported to a field laboratory and were sacrificed by overdose with a benzocaine solution (70 mg/L−1). This methodology was approved by the Ethics Committee on Animal Use of the University of São Paulo (CEUA/FZEA nº 6,885,120,419).
Morphologic and Statistical Analyses
All fish organs (e.g., skin, fins, gills), tissues and fluids (e.g., skin mucous, bile content) were initially observed under a stereomicroscope for the presence of myxozoan. Parasites were fixed in 10% neutral-buffered formalin to measure the myxospores [33] and in 100% ethanol for molecular studies. Myxospores were photographed using differential interference contrast (DIC) and around 30 myxospores were measured using a Carl Zeiss Axio Imager A2 light microscope equipped with an Axio Cam, and the AxioVision AxioVs 40V4.8.2 software package, following the general guidelines of [33]. Measurements were taken from distinct hosts, organs, and localities and then the average of the respective measurements was calculated.
Myxospores were air-dried onto glass slides, stained with Giemsa, and deposited in the Myxozoa collection of the Museu de Diversidade Biológica (the Museum of Biological Diversity) (MDBio) of the State University of Campinas (IB/UNICAMP), São Paulo, Brazil.
χ2 test or G test (when the expected N was less than five) was performed to assess the effect of the geographic localities, sites of infection and host sex on the prevalence of infection of the new parasite, using BioStat 2.0 package [34], with the level of significance set at p < 0.05.
DNA Extraction, Amplification, and Sequencing
Three parasite samples from each site of infection and geographic locality were used for DNA extraction. Plasmodia preserved in 100% ethanol were mechanically ruptured under a coverslip on a microscopic slide, and the content was washed into a 1.5 mL microtube using ATL Buffer from the DNeasy® Blood & Tissue Kit (animal tissue protocol) (QIAGEN Inc., California, USA). Total DNA was extracted by following the manufacturer’s instructions, and the content was then stored at – 20 °C for further applications. Access to the genetic data was authorized by the Brazilian Ministry of the Environment (SisGen No. A33CB83).
PCR amplification of overlapping fragments of the SSU rDNA was obtained through nested PCR using a combination of specific new primers paired with others available in the literature. The first round targeted almost the entire SSU rDNA, with ERIB1 (5’ACCTGGTTGATCCTGCCAG3’ [35]) and ERIB10 (5’CTTCCGCA GGTTCACCTACGG3’; [35]), followed by a semi-nested second round using the primer set ERIB1 and ACT1R (5’AATTTCACCTCTCGCTGCCA3’; [36]), Henn.4f (5’CACGGTCGCTATTAGCCGA3’; this study), and Henn.1r (5’ACGCTGATCGCAGTTCCA3’; this study). Additionality, we used the primers MC5 (5’CCTGAGAAACGGCTACCACATCCA3’; [37]) with MC3 (GATTAGCCTGACAGATCACTCCACGA; [37]), which cover the most central part of the SSU rDNA gene, to ensure all overlapping DNA fragments could be assembled. ITS-1 was amplified using the Henn.8f primer (5’GCGCGCTACAATGACGATG’; this study) with NC13R (GCTGCGTTCTTCATCGAT; [38]). These primers produced fragments that extended from ~ 1950 bp SSU rDNA through ITS-1 and terminated in 5.8S.
PCRs were conducted in 25 μl reaction volumes comprising: 12.5 μl Dream Taq Green PCR Master Mix (Thermo Fisher Scientific, Massachusetts, USA), 0.5 μl of each primer (10 pmol), 1 μl of DNA (10 – 50 ng/μl), and 10.5 μl of ultrapure water. PCR cycling was performed on a Nexus Mastercycler® (Eppendorf, Hamburg, Germany), using a block preheated to 103 °C. The cycling parameters comprised an initial denaturation step at 95 °C for 2 min, followed by 35 denaturation cycles at 94 °C for 30 s, annealing at 60 °C for 30 s (or 58 °C for ITS-1 primers) and extension at 72 °C for 90 s, terminating in an extension step at 72 °C for 5 min. The amplicons were analyzed via 1.5% agarose gel electrophoresis Tris-borate-EDTA (0.045 M Tris - borate, 0.001 M EDTA, pH 8.0) stained with SYBR™ Safe (Thermo Fisher Scientific, Massachusetts, USA) and analyzed on a Compact Digimage System transilluminator (MajorScience™). The presence of appropriately sized bands was confirmed by direct comparison with the molecular weight marker 1 kb Plus DNA Ladder (Thermo Fisher Scientific, Massachusetts, USA). The amplicons were then purified using the QIAquick PCR Purification Kit (QIAGEN, California, EUA) according to the manufacturer’s instructions, and directly sequenced with PCR primers (5 pmol) in both directions using a BigDye 102 Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, California, USA) in an ABI 3730 DNA 103 Analyzer (Applied Biosystems, California, USA) at the Human Genome and Stem Cell Research Center of the University of São Paulo (USP).
Sequencing Assembly, BLASTn and Distance Analyses
The sequences obtained for each sample were assembled and edited using Geneious Prime ® version 2021.1.1. The low-quality ends of each sequence generated were trimmed and aligned to produce consensus sequences. A nucleotide BLASTn [39] search was conducted for each of the generated consensus sequences to confirm the amplification of myxozoan DNA only. Genetic distances were performed using the closest congeners species according to the phylogenetic tree and were determined using the p-distance model matrix in MEGA X [40]. Gaps and missing data were deleted.
Phylogenetic Analyses
Phylogenetic analyses were performed on an alignment of 40 SSU rDNA sequences from the most closely related myxozoans, as determined by BLASTn search, available in the NCBI database (accession numbers are indicated in the phylogenetic tree). Parvicapsula bicornis Køie, Karlsbakk and Nylund 2007 (GenBank: EF429097) and Parvicapsula irregularis Kodóková, Dyková, Tyml, Ditrich and Fiala (2014) (GenBank: KF874229) were used as outgroups. Sequences were aligned using the MAFFT online server [41] with the G-INS-i strategy and default parameters. The optimum evolutionary model (General Time Reversible substitution model and 4 gamma-distributed ration variations among sites) for the dataset was obtained by applying the Akaike Information Criterion using jModelTest 0.1.1 [42]. Aligned sequences were analyzed using maximum likelihood (ML) and Bayesian Inference (BI). ML analyses were conducted in the PhyML 3.0 online server [43], with topology assessed by bootstrapping with 1000 replicates. BI was conducted using MrBayes v.3.0 [44] with posterior probabilities of 10 million generations, via two independent runs of four simultaneous Markov Chain Monte Carlo (MCMC) algorithms, with every 10000th tree saved. Tracer v1.4 was used to ascertain the length of burn-in and the effective sample size (ESS) values [45]. The trees were visualized using Figtree 1.3.1 [46] and edited using Adobe Photoshop (Adobe Systems Inc., California, USA).
Results
The lengths of the 108 specimens of P. squamosissimus analyzed ranged from 14 to 47 cm, while there were 42 males, 43 females, and 23 of undetermined sex. Plasmodia of a Henneguya species were observed in gill filaments, fins, or kidneys of 47 specimens (43.5%). Of the 108 specimens of P. squamosissimus analyzed, 14 (12.9%) were also infected by E. plagioscioni in the gallbladder of fish from the three sites localities: 2 (1.8%) in the Lago Grande do Curuai; 10 (9.2%) in the Tapajós River; and 1 (0.9%) in the Solimões River. Infection by Kudoa sp. was not observed.
The prevalence was 50% in both the Tapajós and Solimões rivers, and 35.4% in the Lago Grande do Curuai (Table 1). These differences were not significant (χ2 = 2.30, df 2). In terms of the site of infection, the prevalence total in the gill filament was 23.1% while in the fins it was 29.6%, and infections of these organs occurred in all sampling localities. The total prevalence of kidney infection was only 1.8% and was restricted to only two specimens from the Tapajós River. Prevalence was significantly different between sites of infection (χ2 = 30.62, df 2), but not when kidney infections restricted to the Tapajós River were excluded from the analysis (χ2 = 1.16, df 1).
Considering the sampling localities separately, the gill filaments were the most prevalent infection site in the Solimões River (50%). Infections in the fins were the most prevalent in the Lago Grande do Curuai and the Tapajós River, with 28.1% and 38.6% respectively (Table 1). Differences in the prevalence between sites of infection were significant in the Tapajós River (χ2 = 22.99, df 2) and the Solimões River (G = 5.51, df 1), but not significant in the Lago Grande do Curuai (χ2 = 2.27, df 1) and in the Tapajós River, if not considered the kidney infection (χ2 = 1.59, df 1).
The prevalence in the males was 59.5%, females of 32.5%, and undetermined sex was 21.7%. These differences were significantly different (χ2 = 10.77, df 2). Considering each geographic localities separately, the prevalences of the infection were of 47.3% for males, 28% for females, and 25% for undetermined sex in the Lago Grande do Curuai (not significantly different - G = 1.96, df 2); 66.6% for males, 40% for females, and 37.5% for undetermined sex in the Solimões River (not significantly different - G = 0.79, df 2), and of 70% for males, 38.4% for females, and 9.1 for undetermined sex in the Tapajós River (significantly different - G = 12.17, df 2).
Considering the organ of infection in each gender and localities there was no significant difference in the prevalence of the fins, with 42.1% for males, 16% for females, and 25% for undetermined sex in the Lago Grande do Curuai (G = 3.72, df 2), 20% for females, 12.5% for undetermined sex, and none infected for males in the Solimões River (G = 1.02, df 2), and 50% for males, 38.4% for females, and 9.1% for undetermined sex in the Tapajós River (G = 5.93, df 2). Regarding gill filaments, the prevalence was 10% for males, 16% for females, and 25% for undetermined sex in the Lago Grande do Curuai, 66.6% for males, 40% for females, and 50% for undetermined sex in the Solimões River, and 35% for males, 23% for females, and none infected for undetermined sex in the Tapajós River. These prevalences were not significantly different between the genders in the Lago Grande do Curuai (G = 0.61, df 2) and in the Solimões River (G = 0.96, df 2), but it was in the Tapajós River (G = 6.69, df 2).
Morphological Description and Taxonomic Summary
Henneguya longisporoplasma n. sp. Figures 2 and 3
Nomarski differential interference contrasting images of wet mount myxospores of Henneguya longisporoplasma n. sp. from Plagioscion squamosissimus in the Amazon basin. A Myxospores found in the gill filaments. White arrow: sporoplasmossomes nuclei. B Myxospores found in the fins. White arrow: sporoplasmossomes nuclei. C Myxospores found in the kidney. Black arrow: polar tubules. D Myxospore in a sutural view. White arrow: suture line. Bar = 10 μm
Morphologic description: microscopic (< 0.1 mm), whitish, and oval-shaped plasmodia were found in the gill filaments, fins, and kidneys of P. squamosissimus. The mature myxospores had an elongated body in the valvular view, with a wider anterior than the posterior end, and were biconvex in the sutural view (Figs. 2 and 3), measuring 53.4 ± 2.9 (range 48.5–59.2) μm in total length, 12.6 ± 0.6 (11.7–13.4) μm in length and 5.7 ± 0.5 (range 4.7–6.5) μm in width. Each valve with a caudal projection averaging 40.7 ± 2.8 (range 36.5–45.9) μm in length. Two pyriform and symmetric polar capsules at the anterior pole of the myxospore, measuring 3.5 ± 0.3 (range 2.8–4) μm in length and 1.9 ± 0.2 (range 1.8–2.3) μm in width, occupying ~ 30% of the myxospore body length, and displaying 4–5 polar tubule turns. Sporoplasm elongated, containing two nuclei and occupying the rest of the myxospore body (Figs. 2 and 3).
Molecular analysis: a total of 10 sequences consisting of partial SSU, complete ITS-1, and partial 5.8S were obtained for H. longisporoplasma n. sp. samples from the distinct infected organs from each of the three localities sampled (see Table 1 for GenBank accession numbers). Apart from differences in length, all the sequences were identical. According to the p-distance analysis, the species with the lowest SSU rDNA difference with H. longisporoplasma n. sp. was H. paraensis (KU535882), 5.6% and the greater was H. tapariensis (MN2249850), 18.04%
Type-host: Plagioscion squamosissimus (Heckel 1840) (Acanthuriformes: Sciaenidae).
Prevalence: of the total of 108 fish examined in the three localities, 47 (43.5%) had plasmodia in at least one site of infection (Table 1).
Site of infection: histozoic, in the gill filaments, fins and kidney.
Etymology: the name derives from the elongated sporoplasm (from the Latin longis = long), occupying ~ 70% of the myxospore body length.
Type-locality: Lago Grande do Curuai, a marginal lake of the Amazon River, in the municipal region of Santarém, in the state of Pará (coordinates: 2°08′04″S 55°37′54″W). Other localities: the Solimões River, near the city of Manaus, in the state of Amazonas, Brazil (coordinates 3°13′21''S, 59°59′52''W); the Tapajós River, near the city of Santarém, in the state of Pará, Brazil (coordinates: 2°21′11″S 54°46′11″W).
Material deposited: Giemsa-stained slide (accession number: ZUEC MYX 112) was deposited in the Myxozoa collection of the Museu de Diversidade Biológica (the Museum of Biological Diversity) (MDBio) of the State University of Campinas (IB/UNICAMP), São Paulo, Brazil. The consensus of the rDNA sequences containing partial SSU, complete ITS-1, and partial 5.8S were deposited in GenBank (accession numbers OM158489-OM158499) (see Table 1 for the GenBank accession numbers of all the sequences).
Phylogenetic Analyses
ML and BI phylogenetic trees showed two main clades, A and B (Fig. 4). Clade A was exclusively composed of Myxobolus parasites of South American characiform hosts. Clade B had several subclades formed of Myxobolus and Henneguya parasites of different fish orders around the world. In this clade, Henneguya longisporoplasma n. sp. was located within clade B, forming a sister branch to a subclade of Henneguya species reported from South American Cichlidae (Henneguya tucunarei Zatti, Atkinson, Maia, Bartholomew & Adriano, 2018); Henneguya jariensis Zatti, Atkinson, Maia, Bartholomew & Adriano 2018; Henneguya tapajoensis Zatti, Atkinson, Maia, Bartholomew & Adriano 2018; Henneguya paraensis Velasco, Videira, Nascimento, Matos, Gonçalves & Matos 2016; and Henneguya sacacaensis Ferreira, Silva, Araújo, Hamoy, Matos & Videira 2020.
Consensus ML phylogenetic tree using SSU rDNA sequences of selected Henneguya and Myxobolus species. GenBank accession numbers are given following the species name. Nodal supports are indicated for ML with a bootstrap of 1000 replicates, and BI with posterior probabilities, respectively. Values for weakly supported nodes (< 70) are not shown
Discussion
Henneguya longisporoplasma n. sp. represents the third myxozoan and the first Henneguya species described from P. squamosissimus. The morphology/morphometry, host, organ/tissue affinities, and DNA sequences of H. longisporoplasma n. sp. were compared with other described species of Henneguya [9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24].
When considering Amazonian species, the myxospores of H. longisporoplasma n. sp. morphologically resembled those of H. tapariensis and H. tapajoensis (Table 2). However, compared to H. tapariensis, the new species had a slightly smaller body length (12.6 vs 13.5 μm), thicker (5.3 vs 2.5 μm) and wider myxospores (5.7 vs 3.6 μm). H. longisporoplasma n. sp. has shorter (3.5 vs 5 μm) and wider (1.9 vs 1.3 μm) polar capsules. Those species also differ in their fish hosts: H. longisporoplasma n. sp. infects a Sciaenid host and H. tapariensis is described as infecting a Serrasalmidae host. In terms of the SSU rDNA sequence, H. longisporoplasma n. sp. and H. tapariensis differed by 18.4% (Table 2).
When compared to H. tapajoensis, H. longisporoplasma n. sp. had a shorter total length (53.4 vs 54.6 μm), shorter (12.6 vs 16.4 μm) and narrower myxospores bodies length (5.7 vs 7 μm), and shorter (3.5 vs 4.2 μm) and narrower (1.9 vs 2.1) polar capsules (Table 2). H. longisporoplasma n. sp. and H. tapajoensis differed by 9.7% of their SSU rDNA.
In comparison with other Henneguya species from South America and other continents, H. longisporoplasma n. sp. exhibited morphological and morphometric differences in at least one of the following characteristics: shape and size of myxospores, number of polar tubule coils, site of infection, fish host, and region of occurrence [5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20]. Henneguya longisporoplasma n. sp. also differed from all other SSU rDNA sequences according to the BLASTn search in the NCBI. Thus, based on morphology/morphometry data, biological trail, and comparison with other available SSU rDNA sequences, we concluded that the myxosporean species isolated in the present study is a novel species.
Henneguya longisporoplasma n. sp. was found parasitizing P. squamosissimus in three localizations in the Amazon basin, and no significant difference was observed in prevalence in the sampled localities, revealing a wide distribution of the parasite in the watershed. Regarding the sites of infection, there was no significant difference between the prevalence of the gill filaments and fins, but the kidney infections were restricted to two specimens from the Tapajós River. This result shows restricted infection in this internal organ in comparison with the wide distribution of the infections in the external organs, but there are no still clear biological/ecological biases to explain these differences. When comparing the prevalence per site of infection in each locality, significant differences were observed for both the Tapajós and Solimões rivers, with the lowest prevalence in kidney and fins, respectively. However, the difference is not significant in the Tapajós River even disregarding the uncommon kidney infection.
Concerning the host gender, there was a significant difference between the total prevalence of males, females, and of undetermined sex, showing a wide distribution regarding the genders. However, when considering the prevalence between the genders in each one of the three localizations, it was significatively different in the Tapajós River, with a lower prevalence for undetermined sex. The comparison of the prevalence per site of infection in each gender in the three geographic regions showed no significant difference for the fins. Gill filament infections also did not differ significantly in the Lago Grande do Curuai and in the Solimões River, but were different in the Tapajós River, with no infection for undetermined sex.
Despite the significant differences in prevalence observed in some aspects of the observed infection, in an overview, our data show a wide distribution of H. longisporoplasma n. sp. infection in the population of P. squamosissimus of the Amazon basin. The wide distribution of myxosporeans in the immensity Amazon basin has also been reported by other authors [8, 47, 48].
Cooke et al. [49] sampled populations of South American silver croakers from distinct regions with different hydrochemical gradients in the Amazon Basin - blackwater (Negro River), whitewater (Solimões/Amazon Rivers), and clearwater (Tapajós River). Their results suggested that the hydrochemical gradients act as ecological barriers, resulting in populational discontinuities of P. squamosissimus in the Amazon Basin. Measured in a straight line, the Lago Grande do Curuai - a marginal lake of the Amazon River (whitewater) - is ∼100 km from the Tapajós River (clearwater), while the Solimões River (whitewater) is a further 550 km away, approximately. Irrespective of geographic locality, hydrochemical gradient, and organ of tropism, however, the myxospore morphology and SSU rDNA sequences were identical, supporting the belief that these are the same species. Thus, we decided to sequence the ITS-1 marker, which tends to vary more widely, from samples from the different organs and localities, aiming to seek a putative intraspecific variation between those samples. However, in contrast to the populational structure of the South American silver croaker, associated with hydrochemical gradients and observed by Cooke et al. [49], the ITS-1 analysis did not reveal intraspecific variation among the samples of H. longisporoplasma n. sp., despite the wide geographic separation of the collection localities and hydrochemical gradients. Thus, besides the wide distribution of the infection within the Amazon Basin, this genetic mixture among distinct geographical localities and hydrochemical gradients is evidence of high gene flow in the H. longisporoplasma n. sp. population, probably reflecting the passive dispersion of the infective myxospores/actinospores along with the downstream river flow, or natural active dispersion, mainly of vertebrates, but also of invertebrate hosts across the Amazon basin. Similar panmixia was reported for C. gracillima, another Amazonian myxosporean [8].
Generally, histozoic myxosporeans (e.g., Myxobolus, Henneguya, Thelohanelus, Kudoa) exhibit tissue specificity within their hosts [50, 51]. However, some myxozoans have been reported infecting more than one organ in a single host. For example, the plasmodia of Thelohanellus kitauei Egusa & Nakajima 1981 were found in both the skin and intestine of the common carp Cyprinus carpio [52], while those of Henneguya kwangtungensis Chen 1998, developed in the gills and gall bladder of Mylopharyngodon piceus [53]. Histological analyses of Myxobolus Cordeiro Adriano, Arana, Alves, Silva, Ceccarelli, Henrique-Silva & Maia, 2009, which infects the gill arch, skin, serosa of the body cavity, urinary bladder, and eyes of Zungaro jahu, identified infections in the connective tissue of the several organs [54], showing that even in distinct organs, there may be tissue specificity. Thus, the occurrence of H. longisporoplasma n. sp. in multiple organs may not mean there is no tissue specificity. However, as histological analyses were not the focus of the present study, data on the specificity of infection sites will be the subject of further studies.
Phylogenetic analyses placed H. longisporoplasma n. sp. in a clade composed of Henneguya species from South America, as a sister branch of the subclade composed only of parasites of Amazonian cichlids (Fig. 4). Previous studies strongly support that vertebrate host affinity and geographic localization are important phylogenetic signals within Myxobolidae phylogenies 14,15,16,17,18, 54,55,56,57,58,59]. While H. longisporoplasma n. sp. is the first myxobolid species described from this host family worldwide, it is expected that further studies may reveal a lineage of myxobolid parasites of sciaenid hosts.
Besides H. longisporoplasma n. sp., E. plagioscioni, a myxozoa recently described in P. squamosissimus [32], was also reported in the bile of 12.9% of the specimens herein studied. However, Kudoa sp. was noted reported, but this result may be related to the methodology used, which did not focus on the muscle analyses, the methodology used by Oliveira et al. [31].
This study contributes to knowledge of myxosporean biodiversity from South American freshwater rivers and lakes, as well as providing details of the prevalence and remarkable genetic uniformity of a parasite infecting a non-migratory fish, in distant geographical regions within the Amazon basin.
Financial support
Zatti S.A. was supported by a Postdoctoral fellowship from the São Paulo Research Foundation (FAPESP) grant #2018/19285–9. This study was supported by the FAPESP regular project (grant # 2016/22047–7 to Adriano EA.) and partially financed by the Coordination of the Improvement of Higher Education Personnel (CAPES) within the Ministry of Education, Brazil, Finance Code 001. Marinho AMR was supported by a doctoral fellowship provided by CAPES to the University of São Paulo. Adriano E.A. received a research productivity grant from the Brazilian Fostering Agency CNPq (Proc. No. 304687/2020–0).
5. References
Lom J, Dyková I (2006) Myxozoan genera: definition and notes on taxonomy, life-cycle terminology, and pathogenic species. Folia Parasitol 53:1–36. https://doi.org/10.14411/fp.2006.001
Okamura B, Gruhl A, Bartholomew JL (2015) An introduction to myxozoan evolution, ecology, and development. In: Okamura B, Gruhl A, Bartholomew JL (eds) Myxozoan evolution, ecology and development. Springer, Switzerland, pp 69–84
Okamura B, Hartigan A, Naldoni J (2018) Extensive uncharted biodiversity: the parasite dimension. Integr Comp Biol 58:132–1145. https://doi.org/10.1093/icb/icy039.l
Jones SRM, Bartholomew JL, Zhang JY (2015) Mitigating myxozoan disease impacts on wild fish populations. In: Okamura B, Gruhl A, Bartholomew JL (eds) Myxozoan evolution ecology and development. Springer, Switzerland, pp 397–413
Atkinson SD, Bartholomew JL (2010) Disparate infection patterns of Ceratomyxa shasta (Myxozoa) in rainbow trout (Oncorhynchus mykiss) and Chinook salmon (Oncorhynchus tshawytscha). Int J Parasitol 40:599–604. https://doi.org/10.1016/j.ijpara.2009.10.010
Atkinson SD, Bartholomew JL (2010) Spatial, temporal and host factors structure the Ceratomyxa shasta (Myxozoa) population in the Klamath river basin. Infect Genet and Evol. https://doi.org/10.1016/j.meegid.2010.06.013
Bartošová-Sojková P, Lövy A, Reed CC, Lisnerová M, Tomková T, Holzer AS et al (2018) Life in a rock pool: radiation and population genetics of myxozoan parasites in hosts inhabiting restricted spaces. PLoS ONE 13(3):e0194042. https://doi.org/10.1371/journal.pone.0194042
Zatti SA, Atkinson SD, Maia AAM, Bartholomew JL, Adriano EA (2017) Ceratomyxa gracillima n. sp. (Cnidaria: Myxosporea) provides evidence of panmixia and ceratomyxid radiation in the Amazon basin. Parasitology 145(9):1137–1146. https://doi.org/10.1017/S0031182017002323
Eiras J (2002) Synopsis of the species of the genus Henneguya Thélohan, 1892 (Myxozoa: Myxosporea: Myxobolidae). Syst Parasitol 52(1):43–54. https://doi.org/10.1023/A:1015016312195
Eiras J, Adriano EA (2012) A checklist of new species of Henneguya Thélohan, 1892 (Myxozoa: Myxosporea, Myxobolidae) described between 2002 and 2012. Syst Parasitol 83(2):95–104. https://doi.org/10.1007/s11230-012-9374-7
Naldoni J, Maia AAM, Correa LL, Silva MRMD, Adriano EA (2018) New myxosporeans parasitizing Phractocephalus hemioliopterus from Brazil: morphology, ultrastructure and SSU-rDNA sequencing. Dis Aquat Organ 128(1):37–49. https://doi.org/10.3354/dao03210
Mathews PD, Maia AA, Adriano EA (2016) Henneguya melini n. sp. (Myxosporea: Myxobolidae), a parasite of Corydoras melini (Teleostei: Siluriformes) in the Amazon region: morphological and ultrastructural aspects. Parasitol Res 115(9):3599–3604. https://doi.org/10.1007/s00436-016-5125-z
Mathews PD, Mertins OM, Espinoza LL, Milanin T, Alama-Bermejo G, Audebert F, Morandini AC (2020) Taxonomy and 18S rDNA-based phylogeny of Henneguya multiradiatus n. sp. (Cnidaria: Myxobolidae) a parasite of Brochis multiradiatus from Peruvian Amazon. Microb Pathog. https://doi.org/10.1016/j.micpath.2020.104372
Videira M, Velasco M, Azevedo R, Silva R, Gonçalves E, Matos P, Matos E (1984) Morphological aspects of Henneguya aequidens n. sp. (Myxozoa: Myxobolidae) in Aequidens plagiozonatus Kullander, 1984 (Teleostei: Cichlidae) in the Amazon region, Brazil. Parasitol Res 114(3):1159–1162
Zatti SA, Atkinson SD, Maia AAM, Bartholomew JL, Adriano EA (2018) Novel Henneguya spp. (Cnidaria: Myxozoa) from cichlid fish in the Amazon basin cluster by geographic origin. Parasitol Res 117(3):849–859. https://doi.org/10.1007/s00436-018-5762-5
Velasco M, Videira M, de Nascimento LC, Matos P, Gonçalves EC, Matos E (2016) Henneguya paraensis n. sp. (Myxozoa; Myxosporea), a new gill parasite of the Amazonian fish Cichla temensis (Teleostei: Cichlidae): morphological and molecular aspects. Parasitol Res 115(5):1779–1787. https://doi.org/10.1007/s00436-016-4916-6
Capodifoglio KRH, Meira CM, Silva MRM, Corrêa LL, Adriano EA, Maia AAM (2020) Morphology and molecular data of two novel cnidarian myxosporean (Myxobolidae) infecting Piaractus brachypomus from the Amazon basin. Acta Trop. https://doi.org/10.1016/j.actatropica.2020.105533
Capodifoglio KRH, Adriano EA, Naldoni J, Meira CM, Silva MRM, Maia AAM (2020) Novel myxosporean species parasitizing an economically important fish from the Amazon basin. Parasitol Res. https://doi.org/10.1007/s00436-020-06641-3
Ferreira RLS, Silva DT, Araújo PG, Hamoy I, Matos E, Videira MN (2020) Henneguya sacacaensis n. sp. (Myxozoa: Myxosporea) parasitizing gills of the acará bicudo Satanoperca jurupari (Osteichthyes: Cichlidae) in eastern Amazon. Braz J Vet Parasitol 29(2):e000620. https://doi.org/10.1590/S1984-29612020030
Azevedo RK, Negrelli DC, Oliveira CP, Abdallah VD, Camara JPS, Matos ER, Vieira DHMD (2021) Morphological and molecular analysis of Henneguya lagunensis n. sp. (Cnidaria Myxosporea) parasitizing the gills of Eugerres brasilianus from Brazil. Parasitol Int. https://doi.org/10.1016/j.parint.2020.102184
Vieira DHMD, Rangel LF, Tagliavini VP, Abdallah VD, Santos MJ, Azevedo RK (2020) A new species, Henneguya lacustris n. sp. (Cnidaria: Myxosporea), infecting the gills of Astyanax lacustris from Brazil. Parasitol Res 119:4259–4265. https://doi.org/10.1007/s00436-020-06871-5
Vieira DHMD, Narciso RB, de Azevedo RK, Silva RJ (2021) Description of two novel Henneguya (Cnidaria: Myxosporea) infecting curimatid fish, using morphological, histological, and molecular analyses. Acta Parasit. https://doi.org/10.1007/s11686-021-00454-9
Vieira DHMD, Rangel LF, Tagliavini VP, Abdallah VD, Santos MJ, Azevedo RK (2021) Morphological and molecular analysis of Henneguya tietensis n. sp. (Cnidaria: Myxosporea), parasitizing the gill filaments of Prochilodus lineatus (Valenciennes, 1837) from Brazil. Parasitol Res 120:27–36. https://doi.org/10.1007/s00436-020-06918-7
Margarido YMM, Adriano EA, Valladão GMR, Naldoni J, Pilarski F (2021) Morphological, molecular, and histopathological characterization of a new species of Henneguya infecting farmed Astyanax lacustris in Brazil. Microb Pathog. https://doi.org/10.1016/j.micpath.2021.104991
Nelson JS, Grande T, Wilson MVH (2016) Fishes of the World, 5th edn. John Wiley & Sons, Hoboken, New Jersey, U.S.A, pp 498–499
Betancur-RR WEO, Arratia G, Acero A, Bailly N, Miya M, Lecointre G, Ortí G (2017) Phylogenetic classification of bony fishes. BMC Evol Biol 17:162. https://doi.org/10.1186/s12862-017-0958-3
Casatti L (2003) Sciaenidae (Drums or croakers). In: Reis RE, Kullander SO, Ferraris CJ Jr (eds) Checklist of the freshwater fishes of south and central America. EDIPUCRS, Porto Alegre, pp 599–602
Casatti L (2005) Revision of the south American freshwater genus Plagioscion (Teleostei, Perciformes, Sciaenidae). Zootaxa 1080:39–64
Sato Y, Godinho HP (1999) Peixes da bacia do rio São Francisco. In: Vazzoler AEAM, Agostinho AA, Cunningham PT. Estudos ecológicos de comunidades de peixes tropicais. EDUSP, São Paulo, pp. 401–412
Queiroz-Souza J, Brambilla EM, Garcia-Ayala JR, Travassos FA, Daga VS, Padial AA, Jean Vitule RS (2018) Biology, ecology and biogeography of the south American silver croaker, an important Neotropical fish species in south America. Rev Fish Biol Fisher 28:693–714. https://doi.org/10.1007/s11160-018-9526-1
Oliveira JC, Velasco M, Santos PFS, Silva JM, São Clemente SC, Matos E (2015) Kudoa spp. (Myxozoa) infection in musculature of Plagioscion squamosissimus (Sciaenidae) in the Amazon region, Brazil. Braz J Vet Parasitol 24:235–240. https://doi.org/10.1590/S1984-29612015023
Zatti SA, Maia AAM, Adriano EA (2020) Growing diversity supports radiation of an Ellipsomyxa lineage into the Amazon freshwater: description of two novel species parasitizing fish from Tapajós and Amazon rivers. Acta Trop 211:105616. https://doi.org/10.1016/j.actatropica.2020.105616
Lom J, Arthur JR (1989) A guideline for the preparation of species descriptions in Myxosporea. J Fish Dis 12(2):151–156. https://doi.org/10.1111/j.1365-2761.1989.tb00287.x
Ayres M, Ayres JRM, Ayres DL, Santos AS (2000) Bio Estat 2.0: aplicações estatísticas nas áreas de ciências biológicas e médicas. Sociedade Civil Mamirauá, Belém. 272p.
Barta JR, Martin DS, Liberator PA, Dashkevicz M, Anderson JW, Feighner SD, Elbrecht A, Perkins-Barrow A, Jenkins MC, Danforth HD, Ruff MD, Profous-Juchelka H (1997) Phylogenetic relationships among eight Eimeria species infecting domestic fowl inferred using complete small subunit ribosomal DNA sequences. J Parasitol 83:262–271. https://doi.org/10.2307/3284453
Hallett SL, Diamant A (2001) Ultrastructure and small-subunit ribosomal DNA sequence of Henneguya lesteri n. sp. (Myxosporea), a parasite of sand whiting Sillago analis (Sillaginidae) from the coast of Queensland, Australia. Dis Aquat Organ 46:197–212
Molnár K, Eszterbauer E, Székely C, Dan A, Harrach B (2002) Morphological and molecular biological studies on intramuscular Myxobolus spp. of cyprinid fish. J Fish Dis 25:643–652. https://doi.org/10.1046/j.1365-2761.2002.00409.x
Gasser RB, Chilton NB, Hoste H, Beveridge I (1993) Rapid sequencing of rDNA from single worms and eggs of parasitic helminths. Nucleic Acids Res 21:2525–2526
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Gapped DJ (1997) BLASTn and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–33902. https://doi.org/10.1093/nar/25.17.3389
Stecher G, Tamura K, Kumar S (2020) Molecular evolutionary genetics analysis (MEGA) for MacOS. Mol Biol Evol 37:1237–1239. https://doi.org/10.1093/molbev/msz312
Katoh K, Rozewicki J, Yamada KD (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:116–1166. https://doi.org/10.1093/bib/bbx108
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. https://doi.org/10.1093/molbev/msn083
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. https://doi.org/10.1093/sysbio/syq010
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:11572–11574
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. https://doi.org/10.1093/sysbio/syy032
Rambaut A (2008) FigTree v1.1.1: Tree figure drawing tool. http://tree.bio.ed.ac.uk/ software/figtree/
Mathews PD, Naldoni J, Maia AA, Adriano EA (2016) Morphology and small subunit rDNA-based phylogeny of Ceratomyxa amazonensis n. sp. parasite of Symphysodon discus, an ornamental freshwater fish from Amazon. Parasitol Res 115(10):4021–4025. https://doi.org/10.1007/s00436-016-5173-4
Sousa FB, Milanin T, Morandini AC, Espinoza LL, Flores-Gonzales A, Gomes ALS, Matoso DA, Mathews PD (2021) Molecular diagnostic based on 18S rDNA and supplemental taxonomic data of the cnidarian coelozoic Ceratomyxa (Cnidaria, Myxosporea) and comments on the intraspecific morphological variation. Zoosystematics Evol 97(2):307–314. https://doi.org/10.3897/zse.97.64769
Cooke GM, Chao NL, Beheregary LB (2011) Marine incursions, cryptic species and ecological diversification in Amazonia: the biogeographic history of the croaker genus Plagioscion (Sciaenidae). J Biogeogr 39:724–738
Molnár K (1994) Comments on the host, organ and tissue specificity of fish myxosporean and on the types of their intrapiscine development. Parasitol Hung 27:5–20
Molnár K, Eszterbauer E (2015) Specificity of infection sites in vertebrate hosts. In Myxozoan evolution, ecology and development, Okamura B, Gruhl A, Bartholomew JL: Springer International Publishing; 2015:295–323
Egusa S, Nakajima K (1981) A new Myxozoa Thelohanellus kitauei, the cause of intestinal giant cystic disease of carp. Fish Pathol 15:213–218
Chen CL, Ma CL (1998) Fauna Sinica, Myxozoa, Myxosporea. Science Press, Beijing, p 993
Adriano EA, Arana S, Alves AL, Silva MRM, Ceccarelli PS, Henrique-Silva F, Maia AAM (2009) Myxobolus cordeiroi n. sp., a parasite of Zungaro jahu (Siluriformes: Pimelodidae) from Brazilian pantanal: morphology, phylogeny, and histopathology. Vet Parasitol 162:221–229. https://doi.org/10.1016/j.vetpar.2009.03.030
Naldoni J, Arana S, Maia AA, Silva MR, Carriero MM, Ceccarelli PS, Tavares LER, Adriano EA (2011) Host-parasite-environment relationship, morphology, and molecular analyses of Henneguya eirasi n. sp. parasite of two wild Pseudoplatystoma spp. in Pantanal Wetland, Brazil. Vet Parasitol 177:247–255. https://doi.org/10.1016/j.vetpar.2010.12.008
Adriano EA, Carriero MM, Maia AAM, Silva MRM, Naldoni J, Ceccarelli PS, Arana S (2012) Phylogenetic and host-parasite relationship analysis of Henneguya multiplasmodialis n. sp. infecting Pseudoplatystoma spp. in Brazilian pantanal wetland. Vet Parasitol 185:110–120. https://doi.org/10.1016/j.vetpar.2011.10.008
Carriero MM, Adriano EA, Silva MRM, Ceccarelli PS, Maia AAM (2013) Molecular phylogeny of the Myxobolus and Henneguya genera with several new south American species. PLoS ONE 8(9):e73713. https://doi.org/10.1371/journal.pone.0073713
Azevedo C, Casal G, Matos P, Alves A, Matos E (2011) Henneguya torpedo sp. nov. (Myxozoa), a parasite from the nervous system of the Amazonian teleost Brachyhypopomus pinnicaudatus (Hypopomidae). Dis Aquat Organ 3:235–242. https://doi.org/10.3354/dao02292
Feijó MM, Arana S, Ceccarelli PS, Adriano EA (2008) Light and scanning electron microscopy of Henneguya arapaima n. sp. (Myxozoa: Myxobolidae) and histology of infected sites in pirarucu (Arapaima gigas: Pisces: Arapaimidae) from the Araguaia river, Brazil. Vet Parasitol 157:59–64. https://doi.org/10.1016/j.vetpar.2008.06.009
Azevedo C, Casal G, Matos P, Matos E (2008) A new species of Myxozoa, Henneguya rondoni n. sp. (Myxozoa), from the peripheral nervous system of the Amazonian fish, Gymnorhamphichthys rondoni (Teleostei). J Eukaryot Microbiol 55:229–234. https://doi.org/10.1111/j.1550-7408.2008.00317.x
Matos E, Tajdari J, Azevedo C (2005) Ultrastructural studies of Henneguya rhamdia n. sp. (Myxozoa) a parasite from the Amazon teleost fish, Rhamdia quelen (Pimelodidae). J Eukaryot Microbiol 52:532–537. https://doi.org/10.1111/j.1550-7408.2005.00063.x
Eiras JC, Malta JC, Varela A, Pavanelli GC (2004) Henneguya schizodon n. sp. (Myxozoa, Myxobolidae), a parasite of the Amazonian teleost fish Schizodon fasciatus (Characiformes, Anostomidae). Parasite 11:691–173. https://doi.org/10.1051/parasite/2004112169
Casal G, Matos E, Azevedo C (2003) Light and electron microscopic study of the myxosporean, Henneguya friderici n. sp. from the Amazonian teleostean fish, Leporinus friderici. Parasitology 126:313–319. https://doi.org/10.1017/s0031182003002944
Vital P, Corral L, Matos E, Azevedo C (2003) Ultrastructural aspects of the myxosporean Henneguya astyanax n. sp. (Myxozoa: Myxobolidae), a parasite of the Amazonian teleost Astyanax keithi (Characidae). Dis Aquat Organ 53(1):55–60. https://doi.org/10.3354/dao053055
Azevedo C, Matos E (2002) Fine structure of the myxosporean, Henneguya curimata n. sp., parasite of the Amazonian fish, Curimata inormata (Teleostei, Curimatidae). J Eukaryot Microbiol 49(3):197–200. https://doi.org/10.1111/j.1550-7408.2002.tb00522.x
Azevedo C, Corral L, Matos E (1997) Light and ultrastructural data on Henneguya testicularis n. sp. (Myxozoa, Myxobolidae), a parasite from the testis of the Amazonian fish Moenkhausia oligolepis. Syst Parasitol 37:111–114
Azevedo C, Matos E (1996) Henneguya malabarica sp. nov. (Myxozoa, Myxobolidae) in the Amazonian fish Hoplias malabaricus. Parasitol Res 82(3):222–224. https://doi.org/10.1007/s004360050099
Azevedo C, Matos E (1995) Henneguya adherens n. sp. (Myxozoa, Myxosporea), a parasite of the Amazonian fish. Acestrorhynchus falcatus. J Eukaryot Microbiol 42(5):515–518. https://doi.org/10.1111/j.1550-7408.1995.tb05898.x
Rocha E, Matos E, Azevedo C (1992) Henneguya amazonica n. sp. (Myxozoa, Myxobolidae), parasitizing the gills of Crenicichla lepidota Heckel 1840 (Teleostei, Cichlidae) from Amazon river. Eur J Protistol 28(3):273–278. https://doi.org/10.1016/S0932-4739(11)80233-6
Acknowledgements
The authors would like to thank Dr Lincoln Corrêa for help with fieldwork, and the fishermen of the Amazon and Tapajós rivers, for their local knowledge of fish availability and the provision of biological material for this study.
Author information
Authors and Affiliations
Contributions
SAZ.: research conceptualization, methodology, investigation, data analysis, original draft preparation. AMRM; Molecular analysis, review, and editing. EAA: research conceptualization, funding acquisition, project administration, data analysis, review, and editing. AAMM: project administration, supervision, and funding acquisition. All the authors discussed the results and contributed to the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval.
Ethics approval and consent to participate are not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zatti, S.A., Marinho, A.M.R., Adriano, E.A. et al. Integrative Taxonomy Reveals a Panmictic Population of Henneguya longisporoplasma n. sp. (Cnidaria: Myxozoa) in the Amazon Basin. Acta Parasit. 67, 1644–1656 (2022). https://doi.org/10.1007/s11686-022-00615-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11686-022-00615-4