Abstract
Hippocampal atrophy is often considered as one of the important biomarkers for early diagnosis of Alzheimer’s disease (AD), which is an irreversible neurodegenerative disorder. Traditional methods for hippocampus analysis usually computed the shape and volume features from structural Magnetic Resonance Image (sMRI) for the computer-aided diagnosis of AD as well as its prodromal stage, i.e., mild cognitive impairment (MCI). Motivated by the success of deep learning, this paper proposes a deep learning method with the multi-channel cascaded convolutional neural networks (CNNs) to gradually learn the combined hierarchical representations of hippocampal shapes and asymmetries from the binary hippocampal masks for AD classification. First, image segmentation is performed to generate the bilateral hippocampus binary masks for each subject and the mask difference is obtained by subtracting them. Second, multi-channel 3D CNNs are individually constructed on the hippocampus masks and mask differences to extract features of hippocampal shapes and asymmetries for classification. Third, a 2D CNN is cascaded on the 3D CNNs to learn high-level correlation features. Finally, the features learned by multi-channel and cascaded CNNs are combined with a fully connected layer followed by a softmax classifier for disease classification. The proposed method can gradually learn the combined hierarchical features of hippocampal shapes and asymmetries to enhance the classification. Our method is verified on the baseline sMRIs from 807 subjects including 194 AD patients, 397 MCI (164 progressive MCI (pMCI) + 233 stable MCI (sMCI)), and 216 normal controls (NC) from Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset. Experimental results demonstrate that the proposed method achieves an AUC (Area Under the ROC Curve) of 88.4%, 74.6% and 71.9% for AD vs. NC, MCI vs. NC and pMCI vs. sMCI classifications, respectively. It proves the promising classification performance and also shows that both hippocampal shape and asymmetry are helpful for AD diagnosis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disorder in the world, characterized by the progressive impairment of cognitive functions and ultimately resulting in irreversible loss of neurons. It is one of the leading causes of dementia affecting the late-life of humans. Mild cognitive impairment (MCI) is considered as the prodromal stage and clinical precursor of AD when suffered from memory loss. It can be further divided into two types: stable MCI (sMCI) and progressive MCI (pMCI). As the name suggests, pMCI indicates that the subjects have a greater potential for further deterioration and conversion into AD, while sMCI subjects show relatively stable symptoms (Minati et al. 2009; Prince et al. 2015). Currently, there is no effective way found to cure AD. However, some treatments can be taken to delay the progression of AD, which has made the early diagnosis of AD especially important and meaningful. Magnetic resonance imaging (MRI) is a non-invasive and powerful imaging tool to obtain three-dimensional images of the human brain (Herrup 2011; Jacka et al. 2011). MRI of different modalities can characterize the structural and functional brain changes related to various brain development abnormalities, including AD and its progression. Specifically, structural MRI is widely applied for computer-aided diagnosis of AD and MCI, due to its sensitivity to aging and atrophy of human brain.
The fast development of computer vision and machine learning methods has motivated their applications on the computer-aided-diagnosis of AD with MR images (Liu et al. 2013a, b; Suk et al. 2015; Zhang et al. 2011). The early diagnosis of dementia through MR image computing and analyisis can be performed on the whole brain or regions of interest (ROIs). Compared with the whole brain, ROIs can provide morphological measurements of brain regions for AD diagnosis (Herrup 2011; Suk et al. 2015; Zhang et al. 2011). Suk et al. (2015) proposed to build a deep-stacked auto-encoder to combine the latent information with the original features of ROIs, achieving high diagnostic accuracy of AD classification. Liu et al. (2015) proposed a novel diagnostic framework with the deep-stacked autoencoder to learn high-level features of ROIs and with a zero-masking strategy for fusion of features from multiple image modalities.
Among all ROIs in the brain, the hippocampus is considered to be the earliest region that suffered damage from dementia and is strongly correlated to AD. Many studies have extracted features from the hippocampus and its affiliated areas for AD diagnosis based on sMRI (Chupin et al. 2010; Cui and Liu 2018; Ho et al. 2011; Jyrki et al. 2011; Shen et al. 2012). Since hippocampal atrophy is often found in the early stage of AD, the shape and volume analysis of the bilateral hippocampi based on MRIs are usually investigated in most of the existing methods. Chupin et al. (2010) proposed a fully automatic framework to segment the hippocampus based on probabilistic and anatomical priors, and the volumes of the segmented hippocampus were proved to be significantly associated with AD. However, the volume calculation only represents the global variation of the hippocampus, which is limited by individual differences of hippocampus volumes on different subjects. Therefore, shape analysis was then proposed to focus on the morphological differences and changes of the hippocampus, further improving the efficiency of AD diagnosis (Beg et al. 2013; Emilie et al. 2009; Lindberg et al. 2012). Local atrophy of the hippocampus, which is relevant to AD diagnosis, can be captured more precisely by shape analysis methods. Lindberg et al. (2012) proposed a shape analysis method to map the hippocampus segmentation mask into the spherical harmonic shape description (SPHARM-PDM) and found that most atrophic deformation was mainly distributed in CA1 and subiculum areas in AD. Compared with the volume analysis, this method can realize more comprehensive morphological and structural analysis of the hippocampus. A hippocampus analysis method was proposed to build a 3D Densely Connected Convolutional Networks (DenseNet) to learn the features of hippocampal regions which were further combined with the SPHARM-PDM for AD diagnosis (Cui and Liu 2018). In this method, the DenseNet was built on the local MR image patches of the hippocampus to learn the visual features of MR image while the SPHARM-PDM was applied on the binary hippocampal masks to extract the shape features. These two types of features were combined to enhance AD classification. Moreover, relevant studies found that there are also internal differences and asymmetry in the atrophy between left and right hippocampus in AD patients (Barnes et al. 2005; Hou et al. 2013; Maruszak and Thuret 2014; Yue et al. 2018), which further provided an important reference for early diagnosis and analysis of AD.
In this paper, we intend to propose a novel hippocampus analysis method based on multi-channel and cascaded convolutional neural networks (CNN), which can automatically learn and extract the combined hierarchical representations of hippocampal shapes and asymmetries from the binary hippocampal masks for AD diagnosis. First, the hippocampus segmentation is performed on the T1-weighted MR images to generate two binary masks, and the asymmetry mask is obtained by calculating the difference between the left and right hippocampus masks. Second, multi-channel 3D CNNs are individually trained with the hippocampal and asymmetry masks to hierarchically extract both shape and asymmetry features for image classification. An upper 2D CNN is cascaded at the end of 3D CNNs to capture the high-level correlation features. Finally, the multiscale features learned by the multi-channel and cascaded CNNs are combined for AD classification, and the saliency maps are also investigated to gauge the importance of corresponding hippocampal surface locations in AD classification for interpretation. The proposed method can jointly learn the combined hierarchical features of hippocampal shapes and asymmetries and the ensemble classifier has a stronger recognition ability for AD. Our method is evaluated with the baseline T1-weighted structural MR images from 194 AD, 164 pMCI, 233 sMCI, and 216 NC subjects from the ADNI dataset.
Compared with our previous works (Cui and Liu 2018), this paper has great differences in the investigated features and the deep neural network architecture of the proposed method for hippocampus analysis in AD diagnosis. In this paper, we propose to learn the combined features of hippocampal surfaces from the binary hippocampal masks, while our previous works explored the detailed visual features of MR images in the hippocampal regions. The network in this paper consists of three-channel and cascaded convolutional neural networks to hierarchically learn the features, while the network proposed in the previous works consists of the DenseNets with a set of dense blocks.
Materials and method
In this section, we present the proposed hippocampus analysis method using the T1-weighted MR brain images. In the proposed method, a multi-channel cascaded deep CNN model is proposed to gradually learn and combine the features of hippocampal shapes and asymmetries for AD diagnosis. Figure 1 shows the flowchart of our proposed algorithm, which consists of four main steps: 1) image acquisition and pre-processing; 2) segmentation of hippocampus; 3) feature learning by multi-channel cascaded CNNs; 4) final ensemble classification. More details of these steps are provided in the following subsections.
The flowchart of the proposed classification algorithm, where “3D CNNs” denotes the deep 3D CNN models to learn the shape and asymmetry features, “2D CNN” denotes the cascaded CNN model to learn the high-level features, and “Fusion by full connection” denotes a fully connected layer for the fusion of the shape and asymmetry features
Materials
The MR images under this study were collected from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset. ADNI was initially launched in 2003 and funded as a private-public partnership contributed by private companies and the Foundations for the National Institutes of Health and the National Institute on Aging. It is a longitudinal multi-center study of healthy controls, mild cognitive impairment (MCI) and AD. Magnetic resonance imaging (MRI), (18F)-fluorodeoxyglucose positron emission tomography (FDG-PET), genetic and biochemical biomarkers, as well as the clinical and cognitive assessments, are captured for the participants at multiple time points. Their aim is to find out the specific correlations among clinical diagnosis results, cognitive assessments, the brain imaging, genetic and biochemical biomarkers during the entire spectrum of AD. The initial five-year study of ADNI, i.e., ADNI-1 was extended to ADNI-GO in 2009 by a Grand Opportunities grant, and further extended to ADNI-2, and ADNI-3 in 2011 and 2016 by competitive renewals.
In this work, we use the baseline T1-weighted MR brain imaging data from ADNI-1 for training the deep learning model and evaluating the proposed classification method. The baseline ADNI-1 dataset applied here consists of 1.5 T T1-weighted MR images of 807 subjects, including 194 AD, 164 pMCI, 233 sMCI and 216 normal controls (NC). The demographic and clinical information of the studied subjects are shown in Table 1, including the age, gender, education and clinical mental scores. Pre-processing of the images was performed before feature extraction. All MR brain images were preprocessed using a nonparametric nonuniform intensity normalization algorithm for correcting the inhomogeneity of image intensities, followed by skull-stripping and cerebellum removal (Sled et al. 1998; Wang et al. 2011). The affine registration with 12 degrees of freedom was then performed by setting the default parameters to linearly register the 3D structural MR images to a template image by using FSL 5.0 (Jenkinson et al. 2012; Smith et al. 2004; Woolrich et al. 2009).
Hippocampus segmentation
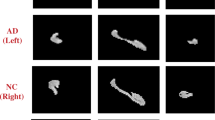
In the existing clinical studies, the hippocampus has been repeatedly proved to be the most closely related to dementia. The decline in cognitive abilities is usually accompanied by some significant hippocampal atrophy. To achieve hippocampus analysis, it is necessary to segment the hippocampus from the provided MR images. Here we develop the 3D deep U-Net with a Dice-like loss function following (Liu et al. 2020) for hippocampus segmentation on structural MR images, by which a binary mask is generated for each hippocampus. Then, the centroids of the two segmentation masks are further computed for the translational alignments of the hippocampus. After segmentation and alignment, a 3D image patch of size 72 × 64 × 64 is cropped to cover each hippocampus mask, which is input into the subsequent CNN model for feature extraction and classification.
Due to the bilateral symmetry of the human brain, the left and right hippocampus are mirror-symmetrical to some extent. To unify the training process, the left hippocampus patch is flipped before fed into the deep CNN model. The asymmetry of the left and right hippocampi is obtained by subtracting the flipped left hippocampus mask and the right one, which is the input to another deep CNN for learning features of hippocampal asymmetry. To alleviate the potential overfitting problem and improve the robustness and generalization performance of the model, we also implement the data augmentation in this study. Specifically, the collected 3D image patches are augmented by shifting the coordinates of the two hippocampi and their margin with 1–2 voxels. Finally, based on the binary masks from the right and flipped left hippocampus as well as their mask margins, we construct and train three-channel CNN models, to respectively extract the features of hippocampal shapes and asymmetries for AD classification as shown in Fig. 1.
Feature learning by multi-channel cascaded convolutional neural networks
After hippocampus segmentation, several convolutional neural networks (CNNs) are investigated to model the hippocampal surface and automatically learn the complex features of the hippocampus. Compared with traditional methods, CNN has stronger feature extraction capabilities and can be automatically optimized through gradient derivation, which greatly reduces human interference. Here we construct multi-channel and cascaded deep CNNs to learn the combined hierarchical features of hippocampal shapes and asymmetries, based on the hippocampus masks and their mask difference for AD classification. The architecture of deep 3D CNNs is shown in Table 2. Since the hippocampal mask is a 3D binary image, the 3D convolutional kernel is used to efficiently encode the spatial features of the hippocampal surface.
The first layer is an input layer that accepts a 3D binary mask image of fixed size (72 × 64 × 64 in the work). Then, the convolutional layer convolves the input image with the kernel filters, followed by a bias term and a non-linear activation function, and a feature map is generated by each filter for the representation. After the convolutional layer, there is a max-pooling layer to down-sample the feature map and generate the compact and efficient features for distinguishing images, which can help to achieve the robustness to some variations. In this way, three convolutional and pooling layers are alternatively stacked to gradually learn the hierarchical representation of 3D hippocampal shapes. Next, a convolutional layer followed by three fully connected layers is added to learn and flatten the features in the high level. In the fully connected layer, the output of each neuron is a linear combination of all inputs from the previous layer, passed through a nonlinearity. Since two hippocampi are symmetrically distributed on both sides of the brain, we build the deep CNNs of the same architecture for both hippocampi, but train them with different masks to learn the features individually.
Besides, some recent studies show that the asymmetry of two hippocampi in the human brain is an important biomarker for AD diagnosis (Gordon et al. 2013; Leung et al. 2011; Shi et al. 2010). Thus, we subtract the right and flipped left hippocampus masks to generate the mask margin for each subject. Then, we build a 3D deep CNN to learn the features of hippocampal asymmetry from the mask differences for AD diagnosis. In our implementation, this deep CNN has the same structure as the deep CNNs for the hippocampal masks. But it is trained with the mask differences instead of the hippocampal masks.
In this work, each deep CNN consists of 4 convolutional layers, 3 max-pooling layers, and 3 fully connected layers, as illustrated in Table 2. The sizes of the first three convolutional filters are 3 × 3 × 3, the last convolution filter is 1 × 1 × 1, and the numbers of kernels are set to 15, 25, 40 and 50 for 4 convolution layers, respectively. Max-pooling is applied for each 2 × 2 × 2 region, and Tanh is adopted as the activation function in these layers because of its good performance for CNNs. During the pre-training period, each CNN is optimized individually to output the class probabilistic score by a softmax layer. After the convolutional and max-pooling layers, all the 3D feature maps are flattened to 1D vector followed by two fully connected layers and a top-most softmax output layer. The Adadelta gradient descent is used to do the back-propagation and tune the trainable parameters. The dropout strategy is also applied to reduce the overfitting problem.
The final ensemble classification
In the above sections, the three-channel CNNs are trained with the left and right hippocampus masks, and their mask difference to capture the features of 3D hippocampal shapes and asymmetry. To make use of these features, we first build a cascaded 2D CNN to combine the features learned by deep 3D CNNs from two hippocampi as shown in Fig. 2. The output of 3D CNN is a feature vector of length 60. The cascaded 2D CNN can learn the high-level correlation features between two hippocampi. The cascaded 2D CNN consists of two convolutional layers and two fully connected layers, followed by a softmax classifier for AD classification.
Second, a fully connected layer is further appended in the high level to combine the features learned by three-channel CNNs and the cascaded 2D CNN to improve the AD classification. There are 60 features generated by each deep CNN and 40 features learned by the cascaded 2D CNN. To train the proposed multi-channel cascaded convolutional neural networks, the three 3D CNNs are individually trained. Meanwhile, the 2D CNN and high-level fully connected layer followed by a softmax layer are finely tuned to make the final classification. Our proposed method is an end-to-end deep learning network, which can gradually capture the combined and hierarchical features of hippocampal shapes and asymmetry for the classification of disease status. The hippocampal features and disease classifier are jointly learned with the proposed deep learning framework.
Experimental results
In this section, we first introduce the preparation of datasets and the implementation of the proposed method. Then, the extensive experiments on the classifications of different groups of subjects are presented. Finally, we compare the proposed method with other competing methods, followed by the discussions on the salient regions for disease diagnosis.
Datasets and implementation
The proposed classification algorithm is evaluated on the T1-weighted MR brain images from the ADNI dataset. The size of MR images after initial preprocessing is 256 × 256 × 256 voxels. After hippocampus segmentation, a 3D image patch of size 72 × 64 × 64, centered on the centroids of hippocampus mask, is cropped to cover each hippocampus mask for feature extraction.
The proposed method is implemented with the Keras framework, accelerated by GPU of NVIDIA GeForce GTX 1080Ti and powered by Ubuntu16.04-×64. To train the 3D CNNs, the initial weights are uniformed for the whole network following default rules in Keras. Adadelta gradient descent optimizer is adopted with a learning rate of 0.7. The batch size is set to 32. The L2 regularization and dropout are also applied to address the overfitting problem.
To better evaluate the classification performance, a 5-fold cross-validation strategy is applied for the proposed method to reduce the effects of random factors. Specifically, we randomly and evenly divided the dataset under study into 5 groups. For each fold, we use one group as testing, another group as validation and the rest for traning the model. In the experiments, we use five classification performance measures for evaluation, which are classification accuracy (ACC), sensitivity (SEN), specificity (SPE), receiver operating characteristic (ROC) curve and the area under curve (AUC). ACC, SEN and SPE respectively reflect the proportion of all samples, positive samples (AD/MCI/pMCI subjects) and negative samples (NC/sMCI subjects) being correctly classified. The ROC curve is a comprehensive indicator that reflects the sensitivity and specificity of continuous variables. AUC is a summary of the ROC curve to measure the ability of a classifier to distinguish two classes.
Evaluations on multi-channel cascaded CNNs
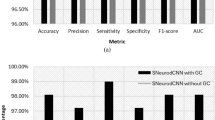
The first conducted experiment is to evaluate the performance of multi-channel cascaded CNNs for AD classification. Two 3D CNNs are trained with the left and right hippocampus masks to extract the features of hippocampal shapes. One 3D CNN is used to learn the asymmetrical features from the mask difference between the left and right hippocampus. The proposed method with multi-channel cascaded CNNs can learn the combined hierarchical features of hippocampal shapes and asymmetry. Thus, we compare the proposed cascaded CNNs for extraction of the combined features with the 3D CNNs for extraction of shape and asymmetry features, which are denoted as “Proposed method”, “Shape” and “Asymmetry”, respectively. Table 3 demonstrates the comparison of the classification performances using the hippocampal shapes, asymmetry and the proposed combination method for classifications of AD vs. NC, MCI vs. NC and pMCI vs. sMCI. Accordingly, the ROC curves for the classifications of AD vs. NC, MCI vs. NC and pMCI vs. sMCI are displayed in Fig. 3a, b and c, respectively. It can be observed that the proposed method by multi-channel cascaded CNNs performs better than the individual 3D CNNs for learning the combined shape and asymmetry features.
Comparison with other methods
Next, we further compare our method with other methods proposed in the literature. The hippocampal volumes are firstly computed based on hippocampus masks of every subject. Besides, the shape analysis of the bilateral hippocampi is also performed using the SPHARM-MAT (a toolbox in Matlab) on the hippocampal mask, by which morphological shape features of the hippocampus are extracted for AD classification. In our implementation, 507 SPHARM coefficients are obtained for each hippocampus mask to train a Multi-Layer Perceptron (MLP) model, which is made up of two fully connected layers for feature reduction and one softmax layer for classification. For a fair comparison, we implement these methods using the same training and test data. The same image preprocessing processes are also applied for all the compared methods. Table 4 shows the comparison of the classification performances with the methods mentioned above. Furthermore, the ROC curves for classifications of different groups are illustrated in Fig. 4a, b and c, respectively. It turns out that our proposed method has an obvious advantage over the hippocampal volumes and shape analysis methods, which are also based on the binary hippocampus segmentation masks. Compared with our previous works in (Cui and Liu 2018), which built a DenseNet to extract the features of MR images in the local hippocampus, the proposed method has lower classification performances. This is because the proposed method focuses on hippocampal surface and extracts features only from the binary hippocampus masks, which have much less visual and appearance information than the MR images. Nevertheless, the proposed multi-channel cascaded CNNs achieves better classification performances (about 3.5–7.5% improvements of AUC) than the traditional SPHARM-PDM method, which also used the binary hippocampal mask for shape analysis. The results demonstrate that the proposed deep learning method works well on modeling the 3D surface of the hippocampal structure for disease diagnosis.
Discussion
In clinical application, in addition to the performance of disease diagnosis, how to identify the relevant biomarkers is also important for medical interpretation. Although the proposed cascaded convolutional neural networks can jointly extract features from both hippocampus masks and asymmetry to achieve the optimal classification performance of AD diagnosis, the learned features cannot provide the important information for medical interpretation. To better identify the most significant biomarkers related to brain diseases, a visualization technique proposed in (Simonyan et al. 2013) is applied to provide convincing clinical evidence to better understand the abnormalities of the hippocampus surface caused by the disease. Specifically, a class saliency map is generated by calculating the gradient of the normalized score in the correct classification corresponding to the input image. The map with high saliency indicates that the brain area is more relevant to diseases for a given image. In the experiments, we generate the saliency maps of the hippocampus masks for all test images, and then calculate the average of them for illustration as shown in Fig. 5. The saliency maps can characterize the contribution of different sub-regions to the classification task, and gauge the importance of corresponding hippocampal surface locations in the prediction of disease for interpretation. Figure 5 (a), (b) and (c) show the saliency maps of the left hippocampus, right hippocampus and their asymmetry for the classifications of AD vs. NC, MCI vs. NC, and pMCI vs. sMCI, respectively. The warmer color corresponds to the larger salience in the hippocampal surface area for disease diagnosis, while blue and purple colors correspond to the smaller salience. It is observed that the larger saliences are often present in the global shape of the left and right hippocampi and their asymmetry, especially in their medial parts for classification of AD vs. NC than those of MCI vs. NC and pMCI vs. sMCI.
Conclusion
In this paper, we develop a novel deep learning classification method based on multi-channel cascaded convolutional neural networks to learn the combined hierarchical features of hippocampal shapes and asymmetry from binary hippocampal masks for AD and MCI diagnosis. The multi-channel 3D CNNs are constructed and trained with the 3D hippocampus binary masks and their mask difference to learn the features of hippocampal shapes and asymmetry. Then, a 2D CNN is cascaded on the 3D CNNs to learn the high-level features between two hippocampus masks. The final classification is made by the combination of all features from 3D CNNs and cascaded 2D CNN. The proposed method can make full use of the asymmetry and shape features to improve the disease classification. Experimental results and comparison on structural MR images from the ADNI dataset demonstrate that the proposed method has promising performances for disease classification. Besides, the saliency maps are also investigated to gauge the importance of hippocampal surface locations in disease diagnosis for interpretation.
References
Barnes, J., Scahill, R. I., Schott, J. M., et al. (2005). Does Alzheimer's disease affect hippocampal asymmetry? Evidence from a cross-sectional and longitudinal volumetric MRI study. Dementia and Geriatric Cognitive Disorders, 19(5–6), 338–344.
Beg, M. F., Raamana, P. R., Barbieri, S., & Wang, L. (2013). Comparison of four shape features for detecting hippocampal shape changes in early Alzheimer's. Statistical Methods in Medical Research, 22(4), 439–462.
Chupin, M., Gérardin, E., Cuingnet, R., Boutet, C., Lemieux, L., Lehéricy, S., ... Colliot, O. (2010). Fully automatic hippocampus segmentation and classification in Alzheimer's disease and mild cognitive impairment applied on data from ADNI. Hippocampus, 19(6), 579–587.
Cui, R., & Liu, M. (2018). Hippocampus analysis by combination of 3D DenseNet and shapes for Alzheimer’s disease diagnosis. IEEE Journal of Biomedical & Health Informatics, 23(5), 2099–2107.
Emilie, G., Gaël, C., Marie, C., Rémi, C., Béatrice, D., Ho-Sung, K., et al. (2009). Multidimensional classification of hippocampal shape features discriminates Alzheimer's disease and mild cognitive impairment from normal aging. Neuroimage, 47(4), 1476–1486.
Gordon, E., Barnes, J., Bartlett, J., Rohrer, J., Cardoso, M., Ourselin, S., & Leung, K. (2013). Alzheimer's disease can be accurately differentiated from semantic dementia using automated measurement of hippocampal asymmetry. Alzheimers & Dementia the Journal of the Alzheimers Association, 9(4), P34–P35.
Herrup, K. (2011). Commentary on “Recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease.” Addressing the challenge of Alzheimer's disease in the 21st century. Alzheimers Dement, 7(3), 335–337.
Ho, A. J., Raji, C. A., Priya, S., Andrew, D. G., Madsen, S. K., Hibar, D. P., ... Toga, A. W. (2011). Hippocampal volume is related to body mass index in Alzheimer's disease. Neuroreport, 22(1), 10–14.
Hou, G., Yang, X., Yuan T. (2013). Hippocampal asymmetry: Differences in structures and functions. Neurochemical Research, 38(3), 453–460.
Jacka, C. R., Knopman, D. S., Mckhann, G. M., Sperling, R. A., Carrillo, M. C., Thies, B., & Phelps, C. H. (2011). Introduction to the recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement, 7(3), 257–262.
Jenkinson, M., Beckmann, C. F., Behrens, T. E., Woolrich, M. W., & Smith, S. M. (2012). FSL. Fsl. Neuroimage, 62(2), 782–790.
Jyrki, L. T. N., Robin, W., Juha, K., Valtteri, J., Lennart, T., Roger, L., ... Daniel, R. (2011). Fast and robust extraction of hippocampus from MR images for diagnostics of Alzheimer's disease. Neuroimage, 56(1), 185–196.
Leung, K., Mahoney, C., Barnes, J., Ourselin, S., & Fox, N. (2011). Automated quantification of hippocampal asymmetry in MRI in semantic dementia and Alzheimer's disease. Alzheimers Dement, 7(4), S16–S16.
Lindberg, O., Walterfang, M., Looi, J. C., Malykhin, N., Ostberg, P., Zandbelt, B., ..., Orndahl, E. (2012). Hippocampal shape analysis in Alzheimer's disease and frontotemporal lobar degeneration subtypes. Journal of Alzheimers Disease Jad, 30(2), 355.
Liu, F., Suk, H. I., Wee, C. Y., Chen, H., & Shen, D. (2013). High-order graph matching based feature selection for Alzheimer’s disease identification. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 311–318). Berlin, Heidelberg: Springer.
Liu, F., Wee, C.-Y., Chen, H., & Shen, D. (2013b). Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer\"s disease and mild cognitive impairment identification. Neuroimage, 84, 466–475.
Liu, S., Liu, S., Cai, W., Che, H., Pujol, S., Kikinis, R., & Feng, D. (2015). Multimodal neuroimaging feature learning for multiclass diagnosis of Alzheimer’s disease. IEEE Transactions on Biomedical Engineering, 62(4), 1132–1140.
Liu, M., Li, F., Yan, H., Wang, K., Ma, Y., Shen, L., ... Initiative, A. s. D. N. (2020). A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer’s disease. NeuroImage, 208, 116459.
Maruszak, A., & Thuret, S. (2014). Why looking at the whole hippocampus is not enough-a critical role for anteroposterior axis, subfield and activation analyses to enhance predictive value of hippocampal changes for Alzheimer's disease diagnosis. Frontiers in Cellular Neuroscience, 8, 95.
Minati, L., Edginton, T., Bruzzone, M. G., & Giaccone, G. (2009). Current concepts in Alzheimer's disease: A multidisciplinary review. American Journal of Alzheimers Disease & Other Dementias, 24(2), 95–121.
Prince, M. J., Wimo, A., Guerchet, M. M., Ali, G. C., Wu, Y-T., & Prina, M. (2015). World Alzheimer Report 2015 - The Global Impact of Dementia: An analysis of prevalence, incidence, cost and trends. Alzheimer's Disease International. http://www.alz.co.uk/research/world-report-2015.
Shen, K. K., Fripp, J., Mériaudeau, F., Chételat, G., Salvado, O., & Bourgeat, P. (2012). Detecting global and local hippocampal shape changes in Alzheimer's disease using statistical shape models. Neuroimage, 59(3), 2155–2166.
Shi, F., Liu, B., Zhou, Y., Yu, C., & Jiang, T. (2010). Hippocampal volume and asymmetry in mild cognitive impairment and Alzheimer's disease: Meta-analyses of MRI studies. Hippocampus, 19(11), 1055–1064.
Simonyan, K., Vedaldi, A., & Zisserman, A. (2013). Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv preprint arXiv:1312.6034.
Sled, J. G., Zijdenbos, A. P., & Evans, A. C. (1998). A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Transactions on Medical Imaging, 17(1), 87–97.
Smith, S. M., Jenkinson, M., Woolrich, M. W., Beckmann, C. F., Behrens, T. E., Johansen-Berg, H., et al. (2004). Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage, 23, S208–S219.
Suk, H. I., Lee, S. W., & Shen, D. (2015). Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Structure & Function, 220(2), 841–859.
Wang, Y., Nie, J., Yap, P. T., Shi, F., Guo, L., & Shen, D. (2011). Robust deformable-surface-based skull-stripping for large-scale studies. Med Image Comput Comput Assist Interv, 14(3), 635–642.
Woolrich, M. W., Jbabdi, S., Patenaude, B., Chappell, M., Makni, S., Behrens, T., Beckmann, C., Jenkinson, M., & Smith, S. M. (2009). Bayesian analysis of neuroimaging data in FSL. Neuroimage, 45(1), S173–S186.
Yue, L., Wang, T., Wang, J., Li, G., Wang, J., Li, X., Li, W., Hu, M., & Xiao, S. (2018). Asymmetry of Hippocampus and amygdala defect in subjective cognitive decline among the community dwelling Chinese. Frontiers in Psychiatry, 9, 226.
Zhang, D., Wang, Y., Zhou, L., Yuan, H., Shen, D., & Initiative, A. s. D. N. (2011). Multimodal classification of Alzheimer's disease and mild cognitive impairment. NeuroImage, 55(3), 856–867.
Funding
This study was supported in part by the National Natural Science Foundation of China (NSFC) under grants (6181101049, 61981340415, 61773263), Natural Science Foundation of Shanghai (20ZR1426300), Shanghai Jiao Tong University Scientific and Technological Innovation Funds (2019QYB02), and an ECNU-SJTU joint grant from the Basic Research Project of Shanghai Science and Technology Commission (No.19JC1410102).
Data collection and sharing were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). We just use the data from the ADNI dataset for this study. The ADNI investigators did not participate in the analysis or writing of this study. A complete list of ADNI investigators can be found online at http://adni.loni.usc.edu/about/governance/principal-investigators/.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest. All authors participated in experiment design, data acquisition and analysis, and wrote the manuscript. All authors approved the final version of the manuscript.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, A., Li, F., Elahifasaee, F. et al. Hippocampal shape and asymmetry analysis by cascaded convolutional neural networks for Alzheimer’s disease diagnosis. Brain Imaging and Behavior 15, 2330–2339 (2021). https://doi.org/10.1007/s11682-020-00427-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11682-020-00427-y