Abstract
Myeloblastosis (MYB) proteins constitute one of the largest transcription factor (TF) families in plants and play crucial roles in regulating plant physiological and biochemical processes, including adaptation to diverse abiotic stresses. These TF families contain highly conserved MYB repeats (1R, R2R3, 3R and 4R) at the N-terminus. Roles for MYB TFs have been reported in response to such stresses as dehydration, salt, cold, and drought. The characterization of Masson pine (Pinus massoniana) MYB TFs are reported, including the analysis of MYB TFs expression in seedlings under controlled conditions and two different phosphate (Pi) deficient treatments. By searching for conserved MYB motifs in full transcriptomic RNA sequencing data for P. massoniana, 59 sequences were identified as MYB TFs. Conserved domain structures and comparative functional and phylogenetic relationships of these MYB TFs with those in Arabidopsis were assessed using various bioinformatics tools. Based on microarray analysis, P. massoniana MYB genes exhibited different expression patterns under the two Pi deficiency conditions. Genes encoding MYB TFs that showed increased expression under critical Pi deficiency were identified, and some MYBs were differentially expressed only under conditions of severe Pi starvation. These results are useful for the functional characterization of MYB TFs that may be involved in the response to Pi deficiency and play divergent roles in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MYB (myeloblastosis) transcription factors (TFs) comprise a large and functionally diverse group of proteins found in all eukaryotic organisms (Dubos et al. 2010). As one of the largest TF families in the plant kingdom, MYB TFs play important roles in regulating physiological and biochemical processes (Du et al. 2012; Su et al. 2015; Liao et al. 2016). MYB proteins are characterized by a highly conserved DNA-binding domain (DBD) located at the N-terminus that consists of 1–4 imperfect tandem repeats, (R0, R1, R2, and R3), of approximately 52 amino acids, each of which forms three α-helices. The MYB repeat contains regularly spaced tryptophan residues that form a hydrophobic core and stabilize the 3D structure of the protein. MYB TFs have been divided into four groups based on the number of repeated MYB domains: 4R-MYB, 3R-MYB (R1R2R3-MYB), 2R-MYB (R2R3-MYB), and 1R-MYB or MYB-related, containing four, three, two and single or partial repeats, respectively (Rosinski and Atchley 1998; Dubos et al. 2010). Among these groups, R2R3-MYBs are the most common TFs in the family and appear to be specific to plants and to yeast (Jin and Martin 1999).
The first gene described as containing a MYB domain was the v-myb oncogene, which is derived from the avian myeloblastosis virus (Klempnauer et al. 1982). Among plants, the first cloned MYB gene, C1, was identified in maize (Zea mays). This gene encodes a c-myb-like TF involved in the regulation of anthocyanin biosynthesis (Paz-Ares et al. 1987). Since then, an increasing number of MYB family members have been identified and characterized in a variety of vascular plants. There are currently 22,032 MYB and 15,369 MYB-related sequences available in the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/) (Jin et al. 2017). Specifically, the diverse functions of a variety of plant MYBs have been investigated through both genetic and molecular analyses, confirming that they are widely involved in regulating hormone signaling, primary and secondary metabolism, and cellular and developmental processes as well as plant responses to biotic and abiotic stresses (Jin and Martin 1999; Dubos et al. 2010; Dai et al. 2012).
Recent studies have shown that plant MYB TFs are involved in regulating plant homeostasis under various environmental stresses. For example, AtMYB2 expression is induced by dehydration and salt stress (Abe et al. 2003), and AtMYB15 plays a role in cold stress and improves drought and salt tolerance in Arabidopsis (Agarwal et al. 2006; Ding et al. 2009). In addition, OsMYB2 is involved in rice tolerance to salt, cold and dehydration stresses (Yang et al. 2012), and AmMYB1 from Avicennia marina enhances tolerance to NaCl stress in transgenic tobacco (Ganesan et al. 2012). Moreover, several reports have indicated that MYB family members are associated with tolerance to phosphorus deficiency. AtMYB62 appears to be involved in phosphate (Pi) starvation (Devaiah et al. 2009), and canola BnPHR1, encoding a MYB-like protein, is regulated by exogenous Pi and promotes uptake and homeostasis of Pi for plant growth (Ren et al. 2012). Furthermore, Wang et al. (2013) investigated the ability of Ta-PHR1, a wheat MYB gene that is transcriptionally activated under Pi deficient conditions, to enhance the efficiency of phosphorus use and yield performance under Pi starvation.
Although phosphorus (P) is an indispensable macronutrient that plays crucial roles in plant development and metabolism, the amount of P available for plant uptake is often limited due to high fixation and slow diffusion in soil (Shen et al. 2011). Masson pine (Pinus massoniana Lamb.), a conifer native to southern China’s tropical and subtropical regions, is severely stricken by phosphorous deficiency (Zhang et al. 2010). In a previous transcriptomic profiling analysis, there was increased expression of several MYB genes in P. massoniana seedlings subjected to phosphorous deficiency (Fan et al. 2014). Nevertheless, the identities and roles of MYB family genes in P. massoniana remain largely unknown. In the present study, whole-transcriptome-wide identification of MYB TFs in P. massoniana were performed. Fifty-nine MYB family members were identified by searching for protein motifs and conserved domains. Phylogenetic analysis was conducted by comparing the retrieved P. massoniana MYB sequences with those identified in Arabidopsis, and expression profiles of these 59 MYB genes in response to different Pi deficient conditions were analyzed by microarray. As MYB TFs are involved in diverse biological processes, our findings should provide a solid foundation for future research into the functional roles of P. massoniana MYB (PmMYB) genes.
Materials and methods
Identification of MYB TFs
Putative MYB TF sequences were retrieved from annotated protein sequences based on P. massoniana transcriptomic data (Fan et al. 2014). Arabidopsis MYB genes were obtained from The Arabidopsis Information Resource (TAIR) (Katiyar et al. 2012), and used as query sequences to ensure that no additional related genes were selected. The identified PmMYBs were rechecked and confirmed to avoid repetition through multiple alignment, and the resulting sequences were subjected to open reading frame (ORF) prediction using OrfPredictor (Muthamilarasan et al. 2015). The peptide sequences were employed for the identification of MYB DBDs using the methodology of Plant Transcription Factor Database (Jin et al. 2017). Amino acid motifs present in the deduced PmMYBs were analyzed using the MEME tool (version 4.11.3) (http://meme-suit.org/tools/meme), and a schematic diagram of these motifs was drawn. The following parameter settings were used: distribution of motifs, zero or one per sequence; maximum number of motifs to find, 10; minimum width of motif, 6; maximum width of motif, 250. For other options, the default values were employed. Only motifs with an e-value of < 1e−20 were retained for further analysis. Structural verification of the MYB domains was performed using Simple Modular Architecture Research Tool (SMART) (Letunic et al. 2012).
Multiple sequence alignment, phylogenetic analysis and digital expression profiles
Multiple sequence alignments of the MYB TFs were performed between Arabidopsis and P. massoniana using ClustalW with default parameters. Phylogenetic trees were generated based on the multiple sequence alignment using MEGA 5.2, and the neighbor-joining (NJ) method was adopted, with 1000 bootstrap replicates. Additionally, the biological functions of some PmMYBs were predicted based on the phylogenetic tree according to orthology.
Custom-designed 8 × 60-K DNA microarray chips (Agilent, Beijng, China) were constructed and used to estimate MYB gene expression based on previous transcriptomic data (Fan et al. 2014). Fluorescent dye (Cy3-dCTP) labeling of P. massoniana cDNA was performed as previously described using a cDNA Amplification and Labeling Kit (CapitalBio Corporation, Beijing, China) (Fan et al. 2014). The ratios of fluorescent signal intensities of each DNA element were measured to determine changes in gene expression. Procedures for microarray hybridization, washing, scanning and data analysis were performed at the Biochip National Engineering Research Center of Beijing, CapitalBio Corporation. Analysis was performed using Feature Extraction and Gene Spring GX software with robust multi- array average (RMA) normalization. Expression clusters of the PmMYB genes under different conditions were analyzed using Cluster, and a diagram was drawn using Tree View.
Plant materials and stress treatments
The plant materials and stress treatments were similar to those employed in a previous study (Fan et al. 2014). Masson pine seeds were surface-sterilized with 5% NaOCl for 5 min, followed by 70% ethanol for 10 min, rinsed in sterile water and soaked overnight at 30 °C. Seeds germinated after 7 days.
Three treatments involving different phosphorous levels were applied: (1) a control treatment: 5.0 mM KNO3, 4.5 mM Ca(NO3)2·4H2O, 2.0 mM MgSO4·7H2O, 0.5 mM KH2PO4, 46 µM H3BO3, 10 µM MnCl2·4H2O, 0.8 µM ZnSO4·7H2O, 0.56 µM CuSO4·5H2O, 0.4 µM H2MoO4·4H2O, and 25 µM Fe-NaEDTA; (2) P1—same as the control treatment except that 0.06 mM KH2PO4 was provided; and (3) P2—same as the control treatment except that 0.01 mM KH2PO4 was provided. The low concentration of potassium due to a reduction in KH2PO4 concentration was corrected with the addition of KCl. 10 days after emergence (DAE), the nutrient solutions with different P concentrations were added every 2 days. Seedlings were harvested at 58 DAE, immediately frozen in liquid nitrogen and stored at − 80 °C. All treatments were replicated in three pots.
RNA extraction and quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) validation
Total RNA from collected samples was extracted using an Invitrogen Plant RNA Isolation Kit according to the manufacturer’s protocols. First-strand cDNA was synthesized using an RNA LA PCR Kit (TaKaRa, Dalian, China) and the supplied oligo dT-adaptor primer. To verify the digital expression profiles obtained, six MYBs were randomly selected and quantitatively analyzed via qRT-PCR. Primer Premier 5.0 was used to design the primers for each PmMYB gene (Table S1). The samples and standards were examined in triplicate in each plate using a SYBR Select Master Mix Kit (Applied Biosystems, Carlsbad, USA) and a 7500 Fast Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instructions. The relative transcript levels for each gene were calculated as the comparative cycle threshold (Ct), which was normalized to the geometric mean of the Ct value of two internal control genes (18s-rRNA; UBQ). Expression ratios were calculated via the \(2^{{ - {\Delta \Delta }C_{\text{t}} }}\) method with correction for the PCR efficiency of each gene.
Results
Transcriptome-wide identification of MYB TFs in P. massoniana
To identify MYB-related proteins from the previously reported full-transcriptomic data for Masson pine (Fan et al. 2014), BLASTP searches were performed using consensus motifs for the MYB domain as queries (Altschul et al. 1997). From among 70,896 unigenes, 73 MYB and MYB-related candidate protein sequences were filtered; redundant sequences of candidate MYBs were discarded from the dataset according to similarity. Structural and functional verification of MYB domains was also performed using ScanProsite and SMART tools. The sequences that included an apparent MYB-type DBD were confirmed to be MYB TF members. Fifty-nine non-redundant PmMYB transcription factors were ultimately identified, named and analyzed (Table S2). A total of 10 motifs were identified to illustrate the P. massoniana MYB protein structure using the MEME program. The positions of MYB/SANT domains and any conserved motifs are shown in Fig. 1 and Fig. S1. The number of motifs in PmMYBs ranged from 2 to 6, with lengths ranging from 8 to 223 amino acids. Most of the PmMYBs contained motifs 1 and 2, and most of the close members in the phylogenetic tree exhibited similar motif compositions, suggesting that proteins in the same subgroup may have similar functions (Fig. 1). Additionally, 39 typical R2R3-MYB TFs, three 3R-MYB proteins (PmMYB1, 22, 46) and one 4R-related MYB protein (PmMYB50) were found among these confirmed PmMYBs.
Structural analysis of MYBs in P. massoniana
As shown in Fig. 1, motif 2 was found in all 59 MYB proteins. SMART analysis revealed four motifs (motif 1, 2, 7 and 10) conserved in the MYB DBD; motif 3 was an HMG 17 domain; motif 5 an S 15 domain; motif 8 a coiled coil region; motif 9 a TFIIA domain; and motifs 4, 5, and 6 domains of unknown function.
Multiple amino acid sequence alignments using ClustalX2 showed that the 59 P. massoniana MYBs could be separated into four groups based on sequence similarity (Fig. S2). Further sequence alignment revealed that the 45 proteins of group 1 share the R2 and R3 MYB repeats and contain characteristic amino acids, including a series of highly conserved tryptophan (Trp, W) residues (Fig. 2a) in each DBD repeat. The five proteins of group 2 share a MYB-like domain containing a SHAQKYF amino acid signature motif (Fig. 2b). Sequence alignment of group 3 proteins indicated that the four MYBs contain a MYB-CC domain (Fig. 2c) located at the C-terminus (Ruan et al. 2017). The other five members of group 4 harbor non-conserved domains, except for motif 2.
Highly conserved amino acid residues are present among the R2R3 (a), 1R (b) and MYB-CC (c) domains of the PmMYBs. The regularly spaced Trp residues present in each repeat are labeled with asterisks. The shading of the alignment indicates identical residues in black, conserved residues in dark gray, and similar residues in light gray
Phylogenetic analysis and functional prediction of the PmMYBs
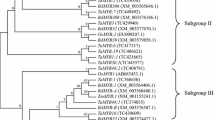
To evaluate the evolutionary relationships and functions of the PmMYBs, comparative phylogenetic analysis was conducted with the identified P. massoniana MYB TFs and 127 Arabidopsis MYB TFs using the neighbor-joining method. The MYBs were divided into 19 groups, (designated Group 1–Group 19), according to the alignment based on the sequence similarity and topology of the encoded proteins (Fig. 3). The 59 PmMYBs were dispersed among 15 MYB groups. Groups 2, 6, 11 and 16 comprise only Arabidopsis MYBs. Furthermore, the number of homologs from the two species was highly asymmetrical. For example, one PmMYB and 12 AtMYBs were included in Group 1, and 15 PmMYBs and eight AtMYBs were included in Group 8. The results of phylogenetic comparative analysis revealed considerable conservation and diversification of the MYB gene family in these two plant species.
Homologous genes are widely distributed in different species and are generally assumed to perform equivalent or similar biological functions (Cai et al. 2012). The functions of most Arabidopsis MYB TFs have been well characterized experimentally, and phylogenetic analysis has identified several functional groups (Peng et al. 2016). Accordingly, PmMYBs were clustered into different Arabidopsis functional groups, including meristem formation, cell wall biogenesis, stress response, anthocyanin biosynthesis, cell differentiation and stomata development (Fig. 3). The results were consistent with the Gene Ontology annotation.
Expression profiles of identified PmMYB genes under different Pi-deficient conditions
To analyze PmMYB gene expression profiles under different Pi-deficient conditions, P. massoniana transcriptome microarray analyses were performed. Genes were considered differentially expressed if they were up-regulated by > twofold or down-regulated by < 0.5-fold (P < 0.05); the data are presented with clustering using fold-change values and displayed in a heat map. The results reveal altered expression numbers and levels in association with the degree of Pi deficiency. Fourteen PmMYB genes were differentially expressed under both P1 and P2 stress, and 10 others were differentially expressed exclusively under the P2 treatment (Fig. 4 and Table S3), suggesting differential sensitivity of stress-induced genes to the stress severity. The expression pattern of PmMYBs showed that some that clustered in the same group or subgroup of the phylogenetic tree (Fig. 3) and exhibited similar expression changes. For example, the expression level was increased for most PmMYBs in Group 4 and a subgroup (including PmMYB51, -52, -53) of Group 8, and functional prediction revealed that these genes play an important role in stress response. However, some PmMYBs within Group 7 showed completely different expression patterns, even though they were predicted to have similar functions.
To verify the observed levels of expression, six MYB genes (Table S1) were randomly selected and quantitatively analyzed by qRT-PCR. As the results were consistent (Fig. 5), the data obtained for expression analysis are reliable.
Expression profiles of six selected MYB TFs determined via microarray and qRT-PCR analysis. The P1 and P2 conditions are described in “Materials and methods” section
Discussion
MYB proteins regulate multiple biological processes and constitute the most abundant transcription factor (TF) family in plants. MYB proteins can be divided into different classes depending on the number of adjacent repeats, including 1R-MYB or MYB-related R2R3-MYB, 3R-MYB and 4R-MYB groups (Dubos et al. 2010). The 1R-MYB or MYB-related group is a heterogeneous class comprising proteins with a single or a partial MYB repeat. These proteins are involved in the control of cellular morphogenesis (Simon et al. 2007), secondary metabolism (Dubos et al. 2008), organ morphogenesis (Kerstetter et al. 2001), chloroplast development (Waters et al. 2009) and the response to Pi starvation (Rubio et al. 2001). In this study, four putative PHRs (PmMYB4, PmMYB19, PmMYB21 and PmMYB28), which are conserved MYB TFs involved in Pi starvation signaling, were identified in the P. massoniana transcriptome. Most plant MYB genes encode proteins of the R2R3-MYB class, which are thought to have evolved from a 3R-MYB gene ancestor (Rosinski and Atchley 1998), although the number of R2R3-MYB proteins varies among plant species. For instance, there are 126 R2R3-MYB proteins in Arabidopsis (Dubos et al. 2010), 109 in rice (Katiyar et al. 2012), 192 in Populus (Wilkins et al. 2009), and 108 in grape (Matus et al. 2008). In our study, 39 putative R2R3-MYB proteins were identified from P. massoniana transcriptomic data, and three typical 3R-MYB proteins and one 4R-related MYB protein. Genes encoding 3R-MYB proteins, which regulate progression through cell cycle transitions, are considered an ancient and evolutionarily conserved family (Peng et al. 2016). Conversely, little is known about the role of 4R-MYB proteins in plants.
In the present study, 10 motifs were predicted by MEME, four of which are MYB/SANT domains according to the SMART analysis. However, these motifs did not exhibit the basic features of whole MYB DBD domains, indicating that there are fewer well-conserved DBD domain sequences among the diverse PmMYB proteins. Most of these PmMYB proteins share the R2 and R3 MYB repeats and contain characteristic amino acids, including a series of highly conserved W residues (Fig. 2a). These conserved W residues are known to play key roles in sequence-specific DNA binding and to serve as landmarks of plant MYB proteins (Ogata et al. 1992; Wang et al. 2015), including a series of highly conserved W residues (Fig. 2a) in each DBD repeat. The proteins of group 2 share a MYB domain similar to the second MYB repeat in tomato LeMYB, which acts as a transcriptional activator in yeast and plants (Rose et al. 1999; Wang et al. 2015), and contains a SHAQKYF amino acid signature motif in the MYB-like repeat (Fig. 2b). Group 3 proteins exhibit a MYB-CC domain (Fig. 2c) located at the C-terminal end of the protein (Ruan et al. 2017).
Considering all MYB-related proteins, it is clear that a wide diversity of functions exists. As the biological functions of proteins are correlated with the primary structures within subgroups (Dubos et al. 2010), it is useful to predict gene function by identifying orthologs between plants based on evolutionary relationships. Arabidopsis MYB TFs have been well characterized experimentally and in recent years have been employed to predict the functional annotations of MYB TFs in other plants, such as barley (Tombuloglu et al. 2013), sugar beet (Stracke et al. 2014), pear (Feng et al. 2015) and Jatropha curcas L. (Peng et al. 2016). To deduce the biological functions of P. massoniana MYBs, comparative phylogenetic analysis was conducted with Arabidopsis MYB TFs. Referring to the functional categories for Arabidopsis (Dubos et al. 2010), all PmMYB proteins clustered into Arabidopsis functional groups (Fig. 2). A small number of three MYB repeat-containing plant proteins, known as MYB3R or R1R2R3-MYB, are related to the regulation of mitosis (Ito 2005). Kobayashi et al. (2015) found that AtMYB3R TFs are important for repressing the expression of specific genes during the cell cycle and establishing a post-mitotic quiescent state during organ size determination. Three of the confirmed P. massoniana 3R-MYB proteins clustered into the same subgroup with AtMYB3R TFs, which suggests a similar function in regulating growth in P. massoniana. Gene duplication is considered to be the primary driving force underlying new gene functions (Liao et al. 2016), and indeed, the expansion of the MYB family in plants suggests that MYB genes may play a variety of roles in plant-specific processes. The R2R3-MYB family is large, and its functions in plant development and environmental responses are diverse (Jin and Martin 1999). In the present study, many P. massoniana R2R3-MYBs clustered into Group 4, Group 7 and Group 8, members of which have been shown to be involved in stress responses in Arabidopsis, and this facilitated the identification of PmMYB genes that may play a role in the response to stress conditions.
Previous studies in plants have revealed that numerous MYB family members are widely involved in responding to diverse abiotic stresses and are differentially expressed under drought, salt and other stress conditions. In Arabidopsis, AtMYB2, AtMYB74 and AtMYB102 are up-regulated by drought stress (Urao et al. 1993; Abe et al. 2003; Denekamp and Smeekens 2003). In addition, AtMYB96 contributes to drought resistance by regulating lateral root meristem activation or transcriptional activation of cuticular wax biosynthesis (Seo and Park 2009; Seo et al. 2011). Kim et al. (2013) reported that AtMYB73 is a negative regulator of salt-sensitive induction in response to salt stress; AtMYB73 is strongly induced by salt stress but not by other stresses. Lotkowska et al. (2015) recently reported that AtMYB112, a formerly unknown regulator, promotes anthocyanin accumulation under salt stress and conditions of high-light stress. In addition, AtMYB62 expression has been reported to be induced in response to Pi starvation (Devaiah et al. 2009), and AtMYB2 regulates the plant response to Pi deficiency by regulating expression of the miR399 gene (Baek et al. 2013). All of these findings confirm that MYB genes are involved in plant responses to abiotic stress. A gene expression pattern similar to a closely grouping paralog suggests functional similarity (Tombuloglu et al. 2013). Based on microarray analyses, we observed 24 P. massoniana MYB genes to be differentially expressed under Pi deficiency stress, including 17 up-regulated genes and seven down-regulated genes, and the similarity in expression of these PmMYB genes was found to be related to their phylogeny (Figs. 2, 3). These results suggest that these PmMYB genes are transcriptionally regulated by Pi starvation signaling. In addition, examination of the expression patterns of PmMYB TFs showed some MYBs to be differentially expressed only under severe Pi starvation conditions, suggesting differential sensitivity of stress-induced genes to stress severity. Interestingly, four putative PHRs (PmMYB4, -19, -21 and -28), which are conserved MYB TFs, are involved in Pi starvation signaling (Mehra et al. 2017; Pandey et al. 2017), showed no significant differences in expression under either the P1 or P2 conditions. Rubio et al. (2001) reported that PHR1, which acts downstream in the Pi starvation signaling pathway, is expressed under Pi-sufficient conditions and only weakly responsive to Pi starvation.
These results provide insight into the structural and functional framework of P. massoniana MYB proteins and may be used as a reference for future functional investigations.
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281:37636–37645
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Baek D, Kim MC, Chun HJ, Kang S, Park HC, Shin G, Park J, Shen M, Hong H, Kim WY, Kim DH, Lee SY, Bressan RA, Bohnert HJ, Yun DJ (2013) Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol 161(1):362–373
Cai H, Tian S, Dong H (2012) Large scale in silico identification of MYB family genes from wheat expressed sequence tags. Mol Biotechnol 52(2):184–192
Dai X, Wang Y, Yang A, Zhang WH (2012) OsMYB2P-1, and R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol 159:169–183
Denekamp M, Smeekens SC (2003) Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol 132:1415–1423
Devaiah BN, Madhuvanthi R, Athikkattuvalasu R, Karthikeyan S, Raghothama KG (2009) Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant 2:43–58
Ding Z, Li S, An X, Liu X, Qin H, Wang D (2009) Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics 36:17–29
Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, Tang YX (2012) Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol 12:106
Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55(6):940–953
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15(10):573–581
Fan F, Cui B, Zhang T, Qiao G, Ding G, Wen X (2014) The temporal transcriptomic response of Pinus massoniana seedlings to phosphorus deficiency. PLoS ONE 9(8):e105068
Feng S, Xu Y, Yang L, Sun S, Wang D, Chen X (2015) Genome-wide identification and characterization of R2R3-MYB transcription factors in pear. Sci Hortic 197:176–182
Ganesan G, Sankararamasubramanian HM, Harikrishnan M, Ganpudi A, Parida A (2012) A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. J Exp Bot 63:4549–4561
Ito M (2005) Conservation and diversification of three-repeat Myb transcription factors in plants. J Plant Res 118:61–69
Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41:577–585
Jin JP, Tian F, Yang DC, Meng YQ, Kong L, Luo JC, Gao G (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res D45(11):D1040–D1045
Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom 13(1):544
Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411(6838):706–709
Kim JH, Nguyen NH, Jeong CY, Nguyen NT, Hong SW, Lee H (2013) Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J Plant Physiol 170(6):1461–1465
Klempnauer K, Gonda T, Bishop J (1982) Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell 31:453–463
Kobayashi K, Suzuki T, Iwata E, Nakamichi N, Suzuki T, Chen P, Ohtani M, Ishida T, Hosoya H, Müller S, Leviczky T, Pettkó-Szandtner A, Darula Z, Iwamoto A, Nomoto M, Tada Y, Higashiyama T, Demura T, Doonan JH, Hauser MT, Sugimoto K, Umeda M, Magyar Z, Bögre L, Ito M (2015) Transcription repression by MYB3R proteins regulates plant organ growth. EMBO J 34:1992–2007
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40(1):302–305
Liao W, Yang Y, Li Y, Wang G, Peng M (2016) Genome-wide identification of cassava R2R3 MYB family genes related to abscission zone separation after environmental stress-induced abscission. Sci Rep 6:32006
Lotkowska ME, Tohge T, Fernie AR, Xue GP, Balazadeh S, Mueller-Roeber B (2015) The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol 169:1862–1880
Matus JT, Aquea F, Arce-Johnson P (2008) Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol 8:83
Mehra P, Pandey BK, Giri J (2017) Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnol J. https://doi.org/10.1111/pbi.12699
Muthamilarasan M, Bonthala VS, Khandelwal R, Jaishankar J, Shweta S, Nawaz K, Prasad M (2015) Global analysis of WRKY transcription factor superfamily in Setaria identifies potential candidates involved in abiotic stress signaling. Front Plant Sci 6:910
Ogata K, Hojo H, Aimoto S, Nakai T, Nakamura H, Sarai A, Ishii S, Nishimura Y (1992) Solution structure of a DNA-binding unit of MYB: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc Natl Acad Sci USA 89:6428–6432
Pandey BK, Mehra P, Verma L, Bhadouria J, Giri J (2017) OsHAD1, a haloacid dehalogenase-like APase enhances phosphate accumulation. Plant Physiol. https://doi.org/10.1104/pp.17.00571
Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J 6:3553–3558
Peng X, Liu H, Wang D, Shen S (2016) Genome-wide identification of the Jatrophacurcas MYB family and functional analysis of the abiotic stress responsive gene JcMYB2. BMC Genom 17:251
Ren F, Guo QQ, Chang LL, Chen L, Zhao CZ, Zhong H, Li XB (2012) Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation. PLoS ONE 7(8):e44005
Rose A, Meier I, Wienand U (1999) The tomato I-box binding factor LeMYBI is a member of a novel class of MYB-like proteins. Plant J 20:641–652
Rosinski JA, Atchley WR (1998) Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol 46:74–83
Ruan W, Guo M, Wu P, Yi K (2017) Phosphate starvation induced OsPHR4 mediates Pi-signaling and homeostasis in rice. Plant Mol Biol 93(3):327–340
Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15(6):2122–2133
Seo PJ, Park CM (2009) Auxin homeostasis during lateral root development under drought condition. Plant Signal Behav 4:1002–1004
Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23:1138–1152
Shen J, Yan L, Zhang J, Li H, Bai Z, Chen X, Zhang W, Zhang F (2011) Phosphorus dynamics: from soil to plant. Plant Physiol 156(3):997–1005
Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J (2007) Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol 311(2):566–578
Stracke R, Holtgräwe D, Schneider J, Pucker B, Sörensen TR, Weisshaar B (2014) Genome-wide identification and characterization of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol 14(1):249
Su L, Wang Y, Liu D, Li X, Zhai Y, Sun X, Li X, Liu Y, Li J, Wang Q (2015) The soybean gene, GmMYNJ2, encodes a R2R3-type transcription factor involved in drought stress tolerance in Arabidopsis thaliana. Acta Physiol Plant 37:138
Tombuloglu H, Kekec G, Sakcali MS, Unver T (2013) Transcriptome-wide identification of R2R3-MYB transcription factors in barley with their boron responsive expression analysis. Mol Genet Genomics 288:141–155
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homologs is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529–1539
Wang J, Sun J, Miao J, Guo J, Shi Z, He M, Chen Y, Zhao X, Li B, Han F, Tong Y, Li Z (2013) A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signaling and increases grain yield in wheat. Ann Bot 111:1139–1153
Wang F, Suo Y, Wei H, Li M, Xie C, Wang L, Chen X, Zhang Z (2015) Identification and characterization of 40 isolated Rehmannia glutinosa MYB family genes and their expression profiles in response to shading and continuous cropping. Int J Mol Sci 16:15009–15030
Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21(4):1109–1128
Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM (2009) Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149(2):981–993
Yang A, Dai XY, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bor 63:2541–2556
Zhang Y, Zhou Z, Ma X, Jin G (2010) Foraging ability and growth performance of four subtropical tree species in response to heterogeneous nutrient environments. J For Res 15:91–98
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Project funding: This study was funded by the Natural Science Foundation of Guizhou Province, P. R. China (20161051), the National Natural Science Foundation of China (31660185) and the Doctoral Fund Project of Guizhou University, P. R. China (201464).
The online version is available at http://www.springerlink.com
Corresponding editor: Yu Lei.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11676_2019_911_MOESM5_ESM.tif
Supplementary Fig. S2 Conserved amino acid residues are present among the 59 proteins of the P. massoniana MYBs. The amino acid sequences are aligned, and gaps (dots) have been introduced to maximize the alignment (TIFF 3687 kb)
Rights and permissions
About this article
Cite this article
Fan, F., Wang, Q., Wen, X. et al. Transcriptome-wide identification and expression profiling of Pinus massoniana MYB transcription factors responding to phosphorus deficiency. J. For. Res. 31, 909–919 (2020). https://doi.org/10.1007/s11676-019-00911-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-019-00911-2