Abstract
The MYB proteins constitute one of the largest transcription factor families in plants. Much research has been performed to determine their structures, functions, and evolution, especially in the model plants, Arabidopsis, and rice. However, this transcription factor family has been much less studied in wheat (Triticum aestivum), for which no genome sequence is yet available. Despite this, expressed sequence tags are an important resource that permits opportunities for large scale gene identification. In this study, a total of 218 sequences from wheat were identified and confirmed to be putative MYB proteins, including 1RMYB, R2R3-type MYB, 3RMYB, and 4RMYB types. A total of 36 R2R3-type MYB genes with complete open reading frames were obtained. The putative orthologs were assigned in rice and Arabidopsis based on the phylogenetic tree. Tissue-specific expression pattern analyses confirmed the predicted orthologs, and this meant that gene information could be inferred from the Arabidopsis genes. Moreover, the motifs flanking the MYB domain were analyzed using the MEME web server. The distribution of motifs among wheat MYB proteins was investigated and this facilitated subfamily classification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sessile plants are highly successful terrestrial inhabitants on earth, probably due to their broad adaptation to environmental challenges. As such, they must possess mechanisms for responding to their environment, among which transcriptional regulation plays a crucial step in these elaborate systems [1]. Transcription factors can be categorized into different families according to conservation in their DNA-binding domains [2, 3]. Examples include helix–loop–helix, zinc finger, helix–turn–helix, MADS cassette, and leucine zipper. The MYB transcription factor family is one of the largest families in plants and is involved in many biological processes. The MYB family can be divided into three subfamilies depending on the number of adjacent repeats of the MYB domain: 1RMYB, R2R3-type MYB, and 3RMYB contain one, two, and three repeats, respectively [4, 5]. The 4RMYB sequences which contain four MYB repeats were also reported, but little is known about their functions [6]. 1RMYB genes are quite divergent and these function in the circadian clock [7], cellular morphogenesis [8], and secondary metabolism [9]. 3RMYB genes constitute a much smaller subfamily and they appear to have conserved roles in animals and plants [10]. In contrast, R2R3-type MYB genes constitute the largest subfamily of MYB proteins in plants and these influence various functions, including primary and secondary metabolism, cell shape and morphogenesis, development processes, and responses to biotic and abiotic stresses [6]. Large scale identification of MYB genes in plants has been conducted in Arabidopsis thaliana [3], Populus trichocarpa [11], Vitis vinifera [12], and rice (Oryza sativa) [13], where 126, 192, 108, and 109 R2R3-type MYB genes were identified, respectively. This information is useful for gene cloning and identification of MYB genes in other major crops.
Wheat (Triticum aestivum) is notorious for its large genome size, and genomic and molecular genetic research in wheat has lagged behind other major crop species. However, in plants for which a complete genome is lacking, expressed sequence tags (ESTs) are a suitable alternative that allow for gene discovery, genome annotation, the characterization of single nucleotide polymorphisms, and proteome analyses [14]. By using EST databases, numerous studies have identified gene families in silico. Nagaraj et al. identified 4,710 excreted or secreted (ES) proteins from nearly 500,000 ESTs derived from 39 different species of parasitic nematodes. Subsequently, it was possible to functionally classify these sequences according to gene ontology, establish pathway associations and also identify protein interaction partners [15]. By using a computational pipeline based on ESTs, Xu et al. were able to obtain 142 odorant binding proteins (OBPs) and 177 chemosensory proteins (CSPs) from total 752,841 insect ESTs covering 54 species in eight Orders of insecta. The complete open reading frames (ORFs) were determined by electronic elongation for 88 and 123 of the OBPs and CSPs, respectively [16]. By comparative analysis of the non-specific lipid transfer protein (nsLtp) genes in rice and the ESTs indexed in the Unigene database for wheat, Boutrot et al. [17] identified 156 putative nsLtp genes in wheat.
Despite much progress in the identification and functional analyses of MYB genes in plants, there are few studies in wheat on this gene family. Chen et al. [18] used degenerate primers corresponding to the MYB domain to obtain 23 MYB gene fragments and 6 near-complete ORFs. Based on the maize rough sheath2 (RS2) sequence which encodes a MYB transcription factor, Morimoto et al. [19] cloned a wheat ortholog and showed that it had conserved function with RS2. With the increase in the availability of nucleotide sequence data, large scale identification of gene families by bioinformatic approaches is more promising and necessary.
In the present study, a total of 364 potential MYB genes (contigs and singlets) were identified from wheat ESTs by a computational pipeline; among them, 36 MYB genes had complete ORFs. In order to gain insight into their functions, orthologs in rice and Arabidopsis were assigned based on the phylogenetic tree. Tissue-specific expression patterns of six wheat MYB genes and their orthologs in Arabidopsis were investigated. Moreover, the motifs flanking the MYB domain were analyzed by MEME for the whole MYB family from rice and Arabidopsis and the 36 wheat R2R3-type MYB proteins.
Materials and Methods
Plant Growth
The cv. Yangmai12 of T. aestivum was used in this study. Seeds were sterilized for 5 min with commercial bleach (NaOCl, 30% v/v), then rinsed several times with sterile water, before being immersed in sterile water for germination. Germinated seeds were transferred to pots and grown in a chamber operating at 14:10 h day:night photoperiod at 25°C:18°C (day:night) and 60% humidity. At flowering, root, stem, leaf, and flower tissues were collected separately and frozen immediately in liquid nitrogen for storage at −70°C.
Data Collection
Wheat ESTs were downloaded from the National Center for Biotechnology Information (NCBI) dbEST database (May 2010). The 3RMYB and R2R3-type MYB proteins of Arabidopsis and rice were used as original sequences to perform the BLAST searches. A total of 129 Arabidopsis and 90 rice MYB proteins were extracted from the NCBI non-redundant protein database.
Computational Pipeline for MYB Gene Identification
The computational pipeline used for MYB gene identification is shown in Fig. 1. BLAST (downloaded from NCBI) searches were performed against the wheat ESTs using the MYB protein sequences from Arabidopsis and rice. The E value was set to 10 to ensure that no MYB sequences were missed. The tblastn method was employed, which uses a protein query to search the nucleotide database. The resultant ESTs were dealt with python scripts to remove any repeated sequences. Subsequently, CAP3 software [20] was used to assemble the sequences and default parameters were selected. The resulting contigs from CAP3 were subjected to six-frame translation and analyzed by PROSITE program (www.expasy.org) to confirm the presence of the MYB domain. Putative MYB genes were submitted to electronic sequence elongation which collects the overlapping ESTs to make sequence near to full length based on the present contigs or ESTs, and the resultants were examined to identify any full-length ORFs by DNAMAN software version 6 (Lynnon Biosoft).
Construction of Phylogenetic Trees
Phylogenetic trees were constructed with the MEGA 4.0 software [21] using the amino acid sequences of the conserved MYB domains of R2R3-MYB proteins from wheat (36 putative proteins), Arabidopsis (124 proteins), and rice (85 proteins). The neighbor-joining method with p-distance was used for the construction of the trees. Bootstrap analysis was performed with 1,000 replicates, while all other parameters were default.
Motif Identification
The online MEME program (http://meme.sdsc.edu) [22] was used for motif predictions. Multiple EM for Motif Elicitation (MEME) is a widely used tool for searching for novel motif patterns in the DNA or protein sequences. By inputting sequences and setting parameters, it is effectively to find new sequence patterns in biological sequences and analyze their significance. In this study, the input sequences were C-terminal regions flanking the MYB domains of R2R3-type MYB proteins from wheat, Arabidopsis, and rice. The maximum number of motifs to find was set to 8 and all other parameters were default.
RNA Extraction and First-Strand Synthesis
Total RNA was extracted from samples using TRIzol reagent and according to the manufacturer’s instructions (Invitrogen, USA). RNase-free DNase I (Promega, USA) was added to eliminate DNA contamination. The first-strand cDNA was synthesized with 2.5 μg RNA using the Superscript II First-Strand Synthesis kit for reverse transcription (RT)-PCR (Invitrogen, USA).
Semi-Quantitative RT-PCR
Semi-quantitative RT-PCR was conducted using samples derived from root, stem, leaf, and flower tissues. The β-actin gene, which is expressed constitutively in wheat, was used as internal control to normalize the data. The primers used for validation and expression analyses were designed using the Primer Premier 5.0 software [23]. All primers used in this study are listed in Table 1.
Results
Identification of MYB Genes in Wheat
The identification of MYB genes in wheat was performed according to the computational pipeline detailed in Fig. 1. A total of 364 fragments, consisting of 158 contigs and 206 singlets, were identified using the MYB protein sequences from Arabidopsis and rice against the wheat ESTs. Subsequently, all candidate MYB protein sequences from wheat were surveyed using the PROSITE program to confirm that they contained the MYB domain. Finally, 125 contigs and 93 singlets were confirmed as putative MYB genes, including 1RMYB, R2R3-type MYB, 3RMYB, and 4RMYB types. Due to the large number and importance of R2R3-type MYB proteins in plants, further analyses focussed on this important MYB subfamily. Electronic elongation and complete ORF finding led to the identification of 36 R2R3-type MYB genes with complete ORFs.
Functional Annotation of R2R3-Type MYB Proteins in Wheat
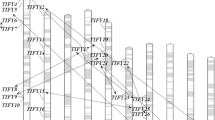
In Arabidopsis, R2R3-type MYB proteins cluster into 25 groups based on the pathways in which they participate [6]. The 36 wheat MYB genes with complete ORFs were translated into proteins, and then phylogenetic trees were constructed with MYB proteins from Arabidopsis, rice, and wheat. All MYB proteins from wheat fell broadly into the same functional groups seen in Arabidopsis. A further phylogenetic tree containing only the MYB proteins identified from wheat was constructed. As shown in Fig. 2, seven groups were identified and their functions included responses to abiotic and biotic stresses, light and other environmental signals, other stress responses, influences on carbon allocation, and acting as repressors of transcription.
Orthologs of Wheat MYB Proteins in Rice and Arabidopsis
Orthologs are pairs of homologous genes in different species that diverged through speciation events. As gene duplication occurs in a single species, orthologs are not just one to one between species. Orthologs exist extensively between species and they are presumed to perform similar biological functions. In order to gain insight into the functions of the MYB proteins identified in wheat, putative orthologs were assigned based on the phylogenetic tree constructed using the MYB proteins from wheat, rice, and Arabidopsis (Table 2). More than one ortholog was found for certain wheat MYB proteins. Such analyses can provide some indications for the putative roles of the putative MYB proteins in wheat.
Shared Expression Patterns Between Orthologs MYB Genes in Wheat and Arabidopsis
To examine whether wheat MYB transcript levels varied between different plant tissues, the tissue-specific expression patterns of phylogenetically related MYB genes of wheat and Arabidopsis were investigated (Fig. 3). Six pairs of genes were chosen at random for these expression studies in root, stem, leaf, and flower tissues. The expression profiles of wheat MYB genes were analyzed by RT-PCR, while for their orthologs in Arabidopsis, these profiles were obtained from microarray data using the GENEVASTIGATOR software [24]. Tissue expression patterns of wheat TaMYB5, TaMYB18, and TaMYB32 correlated well with those of their Arabidopsis orthologs AtMYB15, AtMYB86, and AtMYB91, respectively. TaMYB5 showed relatively high expression in leaf tissue, and transcripts of this gene were also detected in root, flower, and stem tissue. This was also the case for the AtMYB15 ortholog from the Arabidopsis microarray data. For TaMYB18 and TaMYB32, most of organs showed highly similar expression patterns with their respective putative orthologs in Arabidopsis. Both TaMYB11 and TaMYB12 genes exhibited an expression pattern that partially correlated with their Arabidopsis ortholog. The TaMYB11 gene was expressed in root and flower tissues, while its ortholog AtMYB19 had a high level of transcripts in root (transcript levels were about twice those seen in stem, leaf and flower tissues). The TaMYB16 and its ortholog AtMYB26 seemed to have opposing expression patterns. TaMYB16 had a root-specific pattern of expression, whereas AtMYB26 was expressed at high levels in leaves and flowers, less so in stem, and at lowest levels in the roots. Nevertheless, most of these pairs of genes have relative consistent tissue-specific expression patterns, and this suggested that similar biological functions may be conserved among the pairs.
Tissue expression profiles of selected wheat MYB genes and their Arabidopsis orthologs. The transcripts level of wheat MYB genes were measured by RT-PCR and that of Arabidopsis orthologs were obtained from microarray data by GENEVESTIGATOR. The Gene Atlas tool of the microarray database GENEVESTIGATOR was used to search the expression levels of the MYB genes AtMYB9 (At5g16770), AtMYB15 (At3g23250), AtMYB19 (At5g52260), AtMYB26 (At3g13890), AtMYB86 (At5g26660), and AtMYB91 (At2g37630) in different plant tissues. For chip type, “ATH1:22k array” was selected. The bar indicates standard error
Motif Discovery of R2R3-Type MYB Proteins From Wheat, Rice, and Arabidopsis
R2R3-type MYB proteins are found in both monocotyledons and dicotyledons of higher plants, and their functions are various. Except for the conserved MYB domain, there are other motifs that flank the MYB domain, which may determine their specific functional roles. In order to provide further insight on the relationship between structure and function of MYB proteins, a motif discovery program was undertaken using the online MEME server. In total, eight motifs were obtained and their patterns are shown in Fig. 4. Interestingly, motifs 1 and 2 were found closely adjacent to the MYB domain, but they never both appeared in the same sequence. According to MYB protein structure analysis, this particular location should play a role in activation. The presence of motif 1 in a variety of sequences (the set of sequences submitted to MEME server as described in “Materials and methods” section), may suggest its role in activation, while the role of the less abundant motif 2 requires further study. Another interesting discovery was that motif 8 was distributed only in rice and wheat MYB proteins, and no MYB proteins contained this motif in Arabidopsis, which perhaps indicates that these sequences are specific for monocotyledons.
Motif patterns flanking the R2R3-type MYB proteins. The wheat, rice and Arabidopsis R2R3-type MYB proteins were combined into one set of sequences and then submitted to MEME server. The number of motif to find was set to 8. E value is the statistical significance of the motif. The E value is an estimate of the expected number of motifs with the given log-likelihood ratio (or higher), and with the same width and site count, that one would find in a similarly sized set of random sequences. The motifs are displayed as “sequence LOGOS,” containing stacks of letters at each position in the motif. The total height of the stack is the “information content” of that position in the motif in bits. The height of the individual letters in a stack is the probability of the letter at that position multiplied by the total information content of the stack
In Arabidopsis, a total of 47 insertions among 36 members of the R2R3-type MYB gene family were introduced using a reverse genetic approach, but none of insertions gave rise to visible morphological phenotypes in soil-culture conditions [25]. The distributions of the eight motifs among the identified wheat MYB proteins were located in their protein sequences (Fig. 5). Those that share the same motif pattern probably serve overlapping functions, which is especially useful for predicting protein function.
Discussion
The complete genome sequences of Arabidopsis and rice not only facilitate bioinformatic and molecular studies of these model plants, but also aid the exploration of other important crops, such as wheat. The MYB transcription factors constitute one of the largest transcription factor families in Arabidopsis, and over half of these proteins have been studied in detail [6]. MYB proteins are involved in a variety of important physiological processes in plants. In the present study, using information on the MYB proteins in Arabidopsis and rice, a computational pipeline was designed to identify MYB genes in wheat. Using the ORF finding and PROSITE programs, a total of 36 wheat R2R3-type MYB genes with complete ORFs were identified.
The R2R3-type MYB proteins are specific to plants and they participate in many plant-specific processes. These proteins have been classified into 25 subgroups according to the functions with which they are associated [6]. Phylogenetic analyses were performed with wheat and Arabidopsis MYB proteins to assign the putative wheat MYB proteins to appropriate subgroups. This is especially important for functional annotation of the wheat proteins, as sufficient other data to this end is lacking.
In order to analyze further the putative functions of the wheat MYB proteins, orthologs from Arabidopsis and rice were assigned. Although a variety of methods have been developed for identifying orthologs [26–29], the precision and their validity need to be investigated, and perhaps the most useful and simple method for this purpose is phylogenetic analysis. Therefore, phylogenetic analyses with MYB proteins from wheat, Arabidopsis, and rice were performed to assign orthologs based on evolutionary distances.
Orthologs are invaluable for the annotation of protein function, and this information provides a foundation for further functional analysis. Phylogenetic tree analysis is a precise method that is especially useful for difficult cases [30]. Due to the difficulties in validating the ortholog predictions, tissue expression pattern analyses were also performed comparing orthologs in wheat and Arabidopsis. Six pairs of orthologs were selected and tested for their tissue-specific expression profiles. Although some showed a little different or contrast expression patterns in the same organs, the information could help to validate the prediction of orthologs, and is also useful for conferring gene functions from Arabidopsis to wheat genes. In addition to this, the results also raise questions in the accuracy of the orthologs-predicting methods.
Due to the modular nature of proteins, they are usually constructed by one or a few building blocks, namely the functional unit motif(s). Motifs are essential elements in determining the function of proteins. Related sequences that share the same motif probably perform similar functions. This could explain the functional redundancy seen among R2R3-type MYB proteins. One motif pattern determines the special role for the biochemical reaction of the protein [31]. Many basic processes of life are conserved across species boundaries, and it is not surprising that protein motifs can be well conserved. Most genes identified from other species function in Arabidopsis, and this suggests that at least some proteins are conserved between species. In the present study, wheat, rice, and Arabidopsis MYB proteins were submitted to the MEME server. Eight motifs were formulated and the potential roles of some of these were proposed, although experimentation is needed to confirm the assigned biological functions of these novel sequence signatures. The motif patterns could be adopted for the classification of MYB gene subfamilies.
The reliability of the computational pipeline for the identification of wheat MYB genes in this study was confirmed by sequencing and by using wheat MYB genes that have been cloned previously. Ten putative MYB sequences were selected at random for laboratory confirmation, although two of these were not obtained by RT-PCR, which is probably due to the high GC content in the wheat genome, or were false positives from the computational methods.
In summary, by a computational pipeline based on EST database and a series of bioinformatics analyses, the putative MYB genes in wheat were identified. This will be useful for the further study of the functions of these genes in wheat, the signal pathways in which they participate, and the evolution of this gene family. A very recent research conducted by Zhang et al. [32] focus on identification of MYB genes from full-length wheat cDNA libraries. In their study, 60 full-length MYB genes (containing 1 3RMYB, 22 R2R3-type MYBs, and 37 1RMYBs) were isolated and their expression profiles under abiotic stress were measured by RT-PCR methods. As relative small number of sequences comparing to EST database located in NCBI was used and a number of genes exhibited characteristics of low copy numbers and inducible, the results would miss a handful of genes. Thus, only with the complete of wheat genome sequencing project, can we have a sound and comprehensive overview on the largest gene family in this “giant genome.”
References
Singh, K., Foley, R. C., & Onate-Sanchez, L. (2002). Transcription factors in plant defense and stress responses. Current Opinion in Plant Biology, 5, 430–436.
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science, 290, 2105–2110.
Stracke, R., Werber, M., & Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology, 4, 447–456.
Jin, H., & Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Molecular Biology, 41, 577–585.
Rosinski, J. A., & Atchley, W. R. (1998). Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. Journal of Molecular Evolution, 46, 74–83.
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., & Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends in Plant Science, 15, 573–581.
Schaffer, R., Landgraf, J., Accerbi, M., Simon, V., Larson, M., & Wisman, E. (2001). Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell, 13, 113–123.
Simon, M., Lee, M. M., Lin, Y., Gish, L., & Schiefelbein, J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Developmental Biology, 311, 566–578.
Matsui, K., Umemura, Y., & Ohme-Takagi, M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant Journal, 55, 954–967.
Ito, M. (2005). Conservation and diversification of three-repeat Myb transcription factors in plants. Journal of Plant Research, 118, 61–69.
Wilkins, O., Nahal, H., Foong, J., Provart, N. J., & Campbell, M. M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiology, 149, 981–993.
Matus, J. T., Aquea, F., & Arce-Johnson, P. (2008). Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biology, 8, 83.
Yanhui, C., Xiaoyuan, Y., Kun, H., Meihua, L., Jigang, L., Zhaofeng, G., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology, 60, 107–124.
Nagaraj, S. H., Gasser, R. B., & Ranganathan, S. (2007). A hitchhiker’s guide to expressed sequence tag (EST) analysis. Brief Bioinformatics, 8, 6–21.
Nagaraj, S. H., Gasser, R. B., & Ranganathan, S. (2008). Needles in the EST haystack: large-scale identification and analysis of excretory-secretory (ES) proteins in parasitic nematodes using expressed sequence tags (ESTs). PLoS Neglected Tropical Diseases, 2, e301.
Xu, Y. L., He, P., Zhang, L., Fang, S. Q., Dong, S. L., Zhang, Y. J., et al. (2009). Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genomics, 10, 632.
Boutrot, F., Chantret, N., & Gautier, M. F. (2008). Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics, 9, 86.
Chen, R. M., Ni, Z. F., Nie, X. L., Qin, Y. X., Dong, G. Q., & Sun, Q. X. (2005). Isolation and characterization of genes encoding Myb transcription factor in wheat (Triticum aestivem L.). Plant science, 169, 1146–1154.
Morimoto, R., Nishioka, E., Murai, K., & Takumi, S. (2009). Functional conservation of wheat orthologs of maize rough sheath1 and rough sheath2 genes. Plant Molecular Biology, 69, 273–285.
Huang, X., & Madan, A. (1999). CAP3: A DNA sequence assembly program. Genome Research, 9, 868–877.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599.
Bailey, T. L., Williams, N., Misleh, C., & Li, W. W. (2006). MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Research, 34, W369–W373.
Singh, V. K., Mangalam, A. K., & Dwivedi, S. (1998). S. Naik, Primer premier: Program for design of degenerate primers from a protein sequence, 24, 318–319.
Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., & Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database, analysis toolbox. Plant Physiology, 136, 2621–2632.
Meissner, R. C., Jin, H., Cominelli, E., Denekamp, M., Fuertes, A., Greco, R., et al. (1999). Function search in a large transcription factor gene family in Arabidopsis: Assessing the potential of reverse genetics to identify insertional mutations in R2R3 MYB genes. Plant Cell, 11, 1827–1840.
Remm, M., Storm, C. E., & Sonnhammer, E. L. (2001). Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. Journal of Molecular Biology, 314, 1041–1052.
Li, L., Stoeckert, C. J., Jr., & Roos, D. S. (2003). OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Research, 13, 2178–2189.
Deluca, T. F., Wu, I. H., Pu, J., Monaghan, T., Peshkin, L., Singh, S., et al. (2006). Roundup: A multi-genome repository of orthologs and evolutionary distances. Bioinformatics, 22, 2044–2046.
Jensen, L. J., Julien, P., Kuhn, M., von Mering, C., Muller, J., Doerks, T., et al. (2008). eggNOG: Automated construction and annotation of orthologous groups of genes. Nucleic Acids Research, 36, D250–D254.
Altenhoff, A. M., & Dessimoz, C. (2009). Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Computational Biology, 5, e1000262.
Fang, J., Haasl, R. J., Dong, Y., & Lushington, G. H. (2005). Discover protein sequence signatures from protein–protein interaction data. BMC Bioinformatics, 6, 277.
Zhang, L., Zhao, G., Jia, J., Liu, X., & Kong, X. (2011). Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. Journal of Experimental Botany. doi:10.1093/jxb/err264.
Acknowledgment
This work was supported by Key Basic Scientific Study and Development Plan (973 plan; grant no. 2012CB114003), Natural Science Foundation (grant no. 31171830), and Jiangsu Provincial Priority Academic Program Development of Higher Education Institutions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
12033_2011_9486_MOESM4_ESM.tif
AF4. Phylogenetic tree constructed with MYB domains from wheat, rice and Arabidopsis R2R3-type MYB proteins. (TIFF 1667 kb)
Rights and permissions
About this article
Cite this article
Cai, H., Tian, S. & Dong, H. Large Scale In Silico Identification of MYB Family Genes from Wheat Expressed Sequence Tags. Mol Biotechnol 52, 184–192 (2012). https://doi.org/10.1007/s12033-011-9486-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9486-3