Abstract
Stress leads to generation of reactive oxygen species in living cells, which are scavenged by antioxidant enzymes. In the present study, genes encoding for two different antioxidant enzymes, namely cytosolic superoxide dismutase and cytosolic ascorbate peroxidase isolated from salt-tolerant cell lines of Arachis hypogaea, were simultaneously overexpressed in tobacco (Nicotiana tabacum cv. Xanthi) and the effect of their overexpression on alleviation of salinity stress was evaluated. The co-expression of AhCuZnSOD and AhcAPX in tobacco plants helped the resulting transgenic plants in overcoming salt stress. Compared to the wild-type plants, transgenic plants survived longer periods of salinity stress and displayed improved recovery after removal of the stress. The transgenic tobacco plants exhibited better photosynthetic efficiency, higher germination rate, more chlorophyll content, higher levels of total antioxidant enzyme activities, greater water retention capacity, higher proline accumulation, lower level of malondialdehyde content, and reactive oxygen species accumulation under stress conditions in comparison to the WT plants. Our analysis revealed that combined overexpression of the two antioxidant enzymes improved the salinity tolerance in transgenic plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The development of environment-sturdy crops that are able to withstand abiotic stress is the need of the hour to ensure food security for the growing population. Among the various environmental stresses, salinity is the major factor limiting plant growth and productivity worldwide. Salinity interferes with plant’s vital physiological and biochemical functions of plants such as osmosis, nutrient uptake, translocation leading to ionic toxicity, and cellular disturbances (Ediga et al. 2013). Addressing the problem of increased salinity is more crucial as it is predicted that it is going to affect 30% of arable land in the next 25 yr (Wang et al. 2003). This will further shrink the available land for agriculture, eventually leading to further decrease in crop productivity.
When plants are subjected to stress, the reactive oxygen species (ROS) such as singlet oxygen (O2 −), hydrogen peroxide (H2O2), and hydroxyl radicals (OH) are generated. Enhancement of ROS can cause oxidative damage to cellular components such as nucleic acids, lipids, and proteins. The deleterious effects of ROS are surmounted by an array of enzymatic and non-enzymatic mechanisms (Gill and Tuteja 2010). The enzymatic scavenging involves antioxidant enzymes including superoxide dismutase, ascorbate peroxidase, catalase, guaiacol peroxidase, and glutathione reductase. The antioxidative defense is initiated by superoxide dismutase (SOD), which converts superoxide (O2 −) radicals to H2O2. The H2O2 that is also potentially harmful is converted to non-toxic water and monodehydroascorbate by the ascorbate peroxidase (APX) enzyme utilizing ascorbate as the electron donor (Asada 1997).
To improve tolerance against oxidative stress and maintain the development and advancement of plants under these environmental stress conditions, genetic engineering techniques have been utilized. Many groups have conducted studies on developing transgenic plants by overexpression of antioxidant genes like SOD or APX (Badawi et al. 2004; Wang et al. 2010; Xu et al. 2013) and have successfully demonstrated augmented tolerance of transgenic plants to various stresses.
Earlier, we have shown that overexpression of a single antioxidant gene, cytosolic superoxide dismutase (AhCuZnSOD), resulted in better tolerance against salinity and drought as compared with the wild-type (WT) plants (Negi et al. 2015). Another antioxidant gene, cytosolic ascorbate peroxidase (AhcAPX), was also been reported from our laboratory as one of the several genes induced in response to salinity and drought stress (Shrivastava et al. 2015). To assess the effect of antioxidant gene pyramiding and to evaluate whether the combined overexpression would impart better tolerance in transgenic plants against abiotic stress, we transferred both the genes, i.e., AhCuZnSOD and AhcAPX (AhCuZnSOD + AhcAPX), concurrently into the model plant, tobacco and evaluated the tolerance of the transgenic plants against salinity stress. The resulting transgenic tobacco plants exhibited increased germination and seed production under salinity stress compared to control plants, thus signifying the potential for engineering the antioxidant enzyme pathway for achieving better salinity tolerance in plants.
Materials and Methods
Construction of plant transformation vector
The cytosolic superoxide dismutase isolated from Arachis hypogaea (DQ 499511) was cloned into plant expression vector pGREEN 0029 as described in a previous report (Negi et al. 2015). The cytosolic ascorbate peroxidase complementary DNA (cDNA) isolated from A. hypogaea (EF165068) cloned in PRT101 was available in the lab. A PCR-amplified product of 1.7 kb carrying CaMV 35S-AhAPX gene with ClaI and SalI overhangs was subcloned in PRT101 vector. The fragment (CaMV 35S-AhAPX gene) cloned in PRT101 vector was released by digestion with ClaI and SalI and cloned at the ClaI and SalI site of the construct pGREEN0029-SOD. The construct was transformed and multiplied in E. coli, and the double construct was introduced into Agrobacterium tumefaciens (GV3101) by freeze-thaw method (Hofgen and Willmitzer 1988). The cloning of genes was confirmed by PCR amplification with gene specific primers and restriction digestion with PstI and ClaI-SalI.
Plant material and tobacco transformation
Young leaves of tobacco (Nicotiana tabacum cv. Xanthium) were used as explants for tobacco transformation. The transformants were selected on MSB1N.1 medium-containing kanamycin (100 mg/l) and cefotaxime (400 mg/l). Single shoots regenerating from leaf discs were excised and rooted on basal MS medium containing kanamycin and cefotaxime. For hardening, the rooted plantlets were transferred to pots containing autoclaved agropeat in culture room for 1 wk and finally transferred to glasshouse (Horsch et al. 1985).
PCR and Southern blot analysis
Genomic DNA was extracted from 2-wk-old tobacco plants by CTAB method (Rogers and Bendich 1994). The presence of the transgenes was confirmed by PCR analysis using gene specific primers (for SOD: FP 5′-AAATGGTGAAGGCTGTGGC-3′) and RP 5′-CCAAACAACGGAAAGGGGT-3′; (for APX: FP 5′-AAGAACGTCAAGCCATG-3′ and RP 5′-AACTTCTTGCATTCATCACT-3′) were used. The PCR products were visualized using ethidium bromide (EtBr) staining on a 0.8% (w/v) agarose gel. The stable integration and copy number of the transgene in the PCR positive lines was confirmed by Southern blot analysis (Sambrook et al. 1989). Genomic DNA (20 μg) from transgenic and WT plants was digested with BamHI overnight at 37°C, and the digested DNA was loaded on a 0.8% (w/v) agarose gel and subjected to electrophoresis at 50 V. After denaturation and neutralization, the digested DNA was transferred from the gel to a nylon membrane (Amersham Pharmacia, Piscataway, NJ). Hybridization was performed using standard procedures (Sambrook et al. 1989) and probed using 32P-dCTP radiolabeled AhCuZnSOD. The same blot was stripped and probed with radiolabeled AhcAPX.
RT-PCR analysis
Leaves from mature transgenic and WT plants were used for total RNA isolation using RNeasy extraction kit (Qiagen, Valencia, CA), following which RT-PCR was carried out according to manufacturer’s instructions (AccuScript, Stratagene, Foster City, CA). First strand cDNA was generated using 1 μg of total RNA and reverse transcriptase. The gene specific primers used for amplification of AhCuZnSOD and AhcAPX cDNA were the same as described above. The primers for tobacco actin gene used as an internal control were FP 5′-TGGACTCTGGTGATGGTGTC-3′ and RP 5′-CCTCCAATCCAAACACTGTA-3′.
Assessment of salinity stress tolerance in transgenic seeds
The seeds from transgenic and WT plants were germinated on half MS medium supplemented with 100, 200, and 300 mM NaCl for inducing salinity stress. The plain MS medium was used for germination of seeds that served as the experimental control. Germination rate was recorded after 15 d of germination.
Leaf disc senescence assay and estimation of chlorophyll content
Leaf discs of transgenic and WT tobacco plants were floated on 200, 400, and 600 mM NaCl solution for 4 d at 25 ± 2°C under 16 h photoperiod. Extraction of chlorophyll was carried out in 80% (v/v) acetone, and the chlorophyll content was measured spectrophotometrically (Shrivastava et al. 2015).
Stress treatment
The seeds of T2 generation transgenic tobacco plants were used to investigate the salt stress. For salt tolerance analysis, 1-mo-old transgenic and WT tobacco plants were watered every alternate day with 200 mM NaCl for 15 d and were further grown for 7 d without the supplementation of NaCl. Physiological and biochemical analyses were done with these stress-treated plants. Further, we additionally completed the appraisal for resistance of the transgenic plants in the presence of salt for the duration of their life cycle. For this reason, the plants were grown in a glasshouse until maturity both with and without salt stress. The parameters that mark the extent of wellbeing of plants such as plant height, fresh weight of plant, days to flower, and pod weight were considered for the evaluation of salinity stress tolerance.
Ascertaining of relative water content (RWC), photosynthetic efficiency, proline, and malondialdehyde (MDA) contents
RWC was measured in the leaves of plants according to the procedure described by Nayyar and Gupta (2006). The leaves of transgenic and WT were detached, and fresh weight (FW) was recorded. The leaves were hydrated for 12 h in water, and the turgid weights (TW) were recorded. They were then dried in an oven at 70°C for 24 h, and dry weight (DW) was recorded. The RWC was calculated using the equation: RWC = (FW − DW) × 100/(TW − DW). Photosynthetic activity of tobacco plants was measured as photochemical yield (Fv/Fm), using a chlorophyll fluorescence meter according to the procedure described by Yusuf et al. (2010). Proline content of plants was determined as described by Lei et al. (2007). MDA content in the leaves of transgenic and WT tobacco plants were measured according to the procedure of Heath and Packer (1968).
Antioxidant enzyme assays
The specific activities of the antioxidant enzymes were measured in transgenic and WT tobacco plants under stress and non-stress conditions. The superoxide dismutase (SOD) activity was determined using the protocol described by Gupta et al. (1993). The reaction was performed in 3 ml of reaction mixture having 50 mM phosphate buffer (pH 7.8), 63 μM NBT, 13 μM methionine, 1.3 μM riboflavin, and 100 μl of the enzyme extract. The ascorbate peroxidase (APX) activity was assayed as described by Chen and Asada (1989). The specific activity of catalase (CAT) was measured following the procedure of Aebi (1984), and the glutathione reductase (GR) activity was assayed according to the protocol described by Smith et al. (1989).
Histochemical staining
For visualization of O2 − and H2O2 production under salinity stress, detached leaves were infiltrated with 8–10 ml diaminobenzidine (DAB; 1 mg mL−1, pH 3.8, Sigma) and nitroblue tetrazolium (NBT) solutions, respectively, according to the methods described by Kumar et al. (2013). NBT reacts with O2 − forming a dark blue insoluble formazan compound whereas DAB gives a reddish-brown coloration denoting the presence of H2O2.
Statistical analysis
The effect of the overexpression of AhCuZnSOD + AhcAPX on the antioxidative metabolism in transgenic lines and WT under salinity and normal conditions was tested by a two-way analysis of variance (ANOVA) and subsequent post hoc multiple comparisons by Duncan’s test. All the experiments were performed in triplicate.
Results
Identification of transgenic lines
Tobacco was transformed with the gene construct having AhCuZnSOD + AhcAPX cDNA under the control of a constitutive cauliflower mosaic virus 35S (CaMV 35S) promoter (Fig. 1 a ). The integration of transgene in the PCR positive lines was confirmed by Southern blot analysis. Four Southern positive transgenic lines, Dc1, Dc2, Dc3, and Dc4, were identified by probing the blots for AhCuZnSOD and AhcAPX; no signal was obtained in WT plants. The positions of the bands were similar in the transgenic plants when probed with AhCuZnSOD (Fig. 1 b ) and AhcAPX (Fig. 1 c ) genes. This indicated that both the genes were inserted together. A single copy transgene insertion was seen in the lines Dc1 and Dc3, and a double copy insertion was found in the lines Dc2 and Dc4. The expression of transgene AhCuZnSOD + AhcAPX was confirmed by RT-PCR analysis in the transgenic lines where all lines showed expression. The lines Dc1 and Dc3 showed high expression of the transcript. As expected, no expression was found in the WT plants (Fig. 1 f ). All the transformed lines showed higher SOD and APX activity as compared to the WT plants (Fig. 1 d, e ). The transgenic lines having single copy of the transgenes, namely Dc1 and Dc3, were used for further analysis.
Molecular analysis of transgenic tobacco plants overexpressing AhCuZnSOD and AhcAPX gene. (a) Schematic diagram of the T-DNA structure of the plant expression vector. (b) Southern blot analysis from independent tobacco transformants (Dc1, Dc2, Dc3, and Dc4) digested with BamHI. The blot was hybridized with a probe specific for AhCuZnSOD gene. WT showed no detectable hybridization. (c) DNA blot analysis from independent tobacco transformants (Dc1, Dc2, Dc3, and Dc4) digested with BamHI. The blot was hybridized with a probe specific for AhcAPX gene, (d) SOD enzyme activity, (e) APX enzyme activity. (f) RT-PCR analysis confirming the expression of AhCuZnSOD and AhcAPX gene. Actin gene was used as an internal control. Values are given as means ± SD of three independent experiments.
Overexpression of AhCuZnSOD and AhcAPX genes together in tobacco plants increases salinity tolerance
To evaluate the salinity stress tolerance of T1 generation transgenic seedlings, the seeds were grown on half MS medium supplemented with different concentrations of NaCl and their growth was monitored for 15 d. All transgenic as well as WT seedlings indicated comparable growth when NaCl was absent. The pictures representing the germination of highest overexpressing line Dc1 and WT under different salt treatments are shown in Fig. 2 a . Under 100 mM NaCl stress, the germination observed was ~10% more in AhCuZnSOD + AhcAPX expressing line compared to that in the WT plant. In the presence of 200 mM NaCl, the germination of transgenic seeds was ~26% more than those of WT seeds. However, in the presence of 300 mM NaCl, ~45% of the transgenic seeds were able to germinate and grow well, but the WT seeds were poorly germinated (Fig. 2 b ).
Seed germination and growth of seedlings under stress condition: (a) germination of seeds from WT and T1 transgenic lines grown on MS medium supplemented with 100, 200, and 300 mM NaCl is shown; seed sown on plain MS medium served as the control; and (b) germination was scored after 15 d. Wild-type (WT) tobacco control, independently transformed T1 transgenic lines (Dc1 and Dc3) of tobacco. Data points with different lowercase letters indicate significant difference (p ≤ 0.05) between transgenic lines and wild type exposed to same treatment as determined by two-way ANOVA and Duncan’s multiple range test.
To investigate the relative tolerance of transgenic and WT plants to salinity stress, leaf discs of transgenic and WT plants were floated on different concentration (200, 400, and 600 mM) of NaCl for 4 d. The transgenic lines (Dc1, Dc3) showed considerably higher chlorophyll retention and delayed senescence compared to WT plants under similar conditions (Fig. 3 a ). The chlorophyll content in the transgenic lines was higher than that in the WT by ~1.7–1.8-fold in the 200 mM, ~2.1-fold in the 400 mM, and ~2.3-fold in the 600 mM NaCl treatment as shown in Fig. 3 b .
Leaf disc senescence assay showing (a) bleaching and loss of chlorophyll under 200, 400, and 600 mM NaCl; and (b) the data were scored after 4 d of treatment for WT (WT), transgenic (Dc1, Dc3). For control, experiment leaf discs were floated on distilled water. Data points with different lowercase letters indicate significant difference (p ≤ 0.05) between transgenic lines and wild type exposed to same treatment as determined by two-way ANOVA and Duncan’s multiple range test.
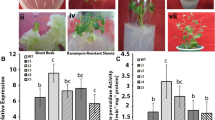
The development and phenotype of T2 transgenic and WT plants were compared by exposing the 1-mo-old plants to 200 mM NaCl, for up to 15 d. All the plants (WT and transgenic lines) grew normally under control conditions (without salinity). Following 15 d of salinity stress, the WT plants showed significant damage and severe yellowing (Fig. 4 ). On rehydration, the revival was observed to be faster in the transgenic lines compared to the WT plants.
Effect of AhCuZnSOD + AhcAPX expression on relative water content, photosynthetic efficiency, proline content, and membrane damage of transgenic plants
RWC of leaves from plants exposed to salt stress was measured to check their capacity to hold water. It was observed that RWC in the transgenic lines showed higher (p ≤ 0.05) water holding capacity during salt stress compared to WT (Fig. 5 a ). Photosynthetic efficiency (Fv/Fm) of transgenic and WT plants were estimated by measuring the chlorophyll fluorescence using Handy-PEA. It has been widely used to study the extent of damage caused by photoinhibition. It was observed that under well-watered conditions, the transgenic plants had slightly enhanced photosynthetic efficiency when compared to the WT plants. Under salinity stress conditions, the Fv/Fm ratio of WT plants reduced (~1.3-fold) in comparison to the transgenic plants (Fig. 5 b ). Under salinity conditions, higher proline accumulation (~2.0-fold) was observed in the transgenic lines compared to the WT plants (Fig. 5 c ). After salinity treatment, the accumulation of MDA in the transgenic lines was less (~2.0–1.7-fold) compared to the WT plants (Fig. 4 d ).
Physiological and biochemical analysis of wild type (WT) and transgenic plants (Dc1, Dc3) grown under salt stress of 200 mM NaCl; (a) RWC, (b) photosynthetic efficiency (Fv/Fm), (c) proline content, and (d) MDA content. Data points with different lowercase letters indicate significant difference (p ≤ 0.05) between transgenic lines and wild type exposed to same treatment as determined by two-way ANOVA and Duncan’s multiple range test.
Antioxidant defenses
As the objective of the present study was to evaluate the degree of detoxification of reactive oxygen species aided through the over expression of antioxidant enzymes, we compared the activity of superoxide dismutase, ascorbate peroxidase, catalase, and GR in the transgenic and WT plants exposed to salinity stress. As speculated, the total enzyme activities of superoxide dismutase, ascorbate peroxidase, catalase, and GR were observed to be higher in the transgenic plants in comparison to WT plants, even under non-stress conditions. However, under stress conditions, the transgenic lines had increased activity of SOD (~3.0-fold) and APX (~3.1-fold) as compared to the WT plants (Fig. 6a, b ). The activity of CAT and GR also increased (~2.4–2.6-fold) in transgenic plants as compared to the WT plants (Fig. 6c, d ).
Specific activities of antioxidant enzymes between wild-type (WT) tobacco and transgenic plants (Dc1, Dc3) grown under salt stress (salt) compared to no salt stress (water) (a) SOD activity, (b) APX activity, (c) CAT activity, and (d) GR activity. Data represent mean of three replicates (n = 3), with error bars representing standard error of the mean. Data points with different lowercase letters indicate significant difference (p ≤ 0.05) between transgenic lines and wild type exposed to same treatment as determined by two-way ANOVA and Duncan’s multiple range test.
Histochemical staining
We examined the status of ROS in the transgenic and WT plants through DAB and NBT staining and conducted histochemical assays under 200 mM NaCl stress. Under non-stress conditions, staining revealed similar levels of ROS in the transgenic and WT plants. Under stress condition, the WT plants were able to make more purple stain by NBT as compared to transgenic plants, indicating higher accumulation of superoxide radical (Fig. 7 a ). Similarly, there was more brown coloration by DAB under stress conditions, indicating higher hydrogen peroxide accumulation in the WT as compared to the transgenic lines (Fig. 7 b ).
Growth and yield of transgenic plants expressing AhCuZnSOD + AhcAPX under salinity stress
Under salinity stress, the seedlings from transgenic plants expressing AhCuZnSOD + AhcAPX exhibited better rate of germination than WT. Furthermore, we tested the functioning of these plants under prolonged period of stress. For this purpose, plants were grown under 200 mM salinity stress, and it was given to the plants until maturity. Under 200 mM NaCl, the transgenic plants grew, flowered, and set seeds. On the other hand, the growth of WT plants was affected and they completely senesced before reaching maturity. Comparison of a few developmental parameters, such as plant height, foliar biomass, number of days to flower, and pod weight of transgenic and WT plants grown under salinity stress and non-stress conditions is shown in Table 1 .
Discussion
In recent years, salinity has degraded agricultural land, reducing the crop production due to shrinkage of arable land area (Ashraf 2009). It is well known that salt stress triggers oxidative stress in plants, which in turn leads to excessive production of ROS. The ROS, when present in low levels, helps induce signals for defense mechanisms but during stress condition, the ROS production increases and they act as toxic molecules. To combat the harmful affects of ROS, plants are fortified with several antioxidant enzymes. Among an array of antioxidative enzymes, SOD and APX play crucial roles in detoxifying ROS, as has been revealed by earlier reports (Prashanth et al. 2008; Negi et al. 2015; Shrivastava et al. 2015). Therefore, we anticipated that transgenic plants that express both AhCuZnSOD and AhcAPX gene together might prompt synergistic effects resulting in increased stress tolerance. Earlier investigations on antioxidant gene pyramiding also support this hypothesis (Kwon et al. 2002; Tang et al. 2006; Faize et al. 2011). Here, we evaluated the efficacy of gene pyramiding in providing protection against salt stress throughout the life cycle. This was confirmed as the transgenic plants having both these transgene showed enhanced salt tolerance. Our findings are in agreement with those of Lee et al. (2007), where it was shown that the overexpression of SOD and APX genes in transgenic fescue enhanced tolerance to abiotic stress. Although, these workers used SWAP2 promoter to express these genes in chloroplasts, we could achieve salt stress tolerance by using a constitutive promoter to express these genes. The higher SOD and APX enzyme activities led to an efficient ROS scavenging mechanism, under salinity stress, in transgenic plants.
Under salt stress, transgenic plants expressing AhCuZnSOD + AhAPX exhibited higher chlorophyll, suggestive of better photosynthetic efficiency, than the WT plants. Under non-stress conditions, the WT and transgenic plants showed similar morphology. This result is in agreement with the prior report where Tang et al. (2006) have shown that the overexpression of SOD and APX in potato chloroplast enhanced the tolerance to heat stress.
Higher RWC, better photosynthetic efficiency, higher proline content, lower MDA content, increased antioxidant enzymes activity and lower O2 −, and H2O2 accumulation, confirmed the relatively higher tolerance to salt stress of transgenic plants compared to the WT plants. Maintenance of high water status, expressed as RWC, is a reliable indicator of increased stress tolerance (Nayyar and Gupta 2006). Our results show that RWC in transgenic plants was maintained at much higher levels; it decreased drastically in WT plants grown under stress conditions. Fv/Fm value reflects the maximum quantum efficiency of photosystem II and is widely used for stress detection in plants. The higher efficiency with which PSII channels light into photochemical process, increases the salt tolerance of plants, resulting in higher yields (Yusuf et al. 2010). In this investigation, transgenic plants were able to maintain better photosynthetic efficiency as compared to WT plants. For osmotic adjustments, accumulation of proline in plants offers protection against physiological disturbances induced by ROS and is known to be included in ROS scavenging (Hayat et al. 2010). In the present study, transgenic plants overexpressing AhCuZnSOD + AhcAPX showed better photosynthetic activity and accumulated higher proline compared to the WT plants during salt stress.
MDA is the final product of lipid peroxidation of biomembranes, and it reflects the extent of stress induced and membrane injury (Sharma et al. 2012). Salinity stress causes membrane-lipid peroxidation, which causes increased accumulation of MDA. The transgenic lines developed in the present study had remarkably lower levels of MDA, than in the WT plants, which indicated the involvement of antioxidant activity in reduction of MDA and provided stress tolerance to the plants. This result suggests lesser damage by lipid peroxidation in the transgenic plants, which was also attributed to a more efficient ROS scavenging mechanism. This was further supported by SOD and APX enzyme activities that were significantly higher after the exposure to stress treatment. ROS levels are indicative of the extent of injury caused to the vital components of the plant cells (Miller et al. 2010), so we checked the level of ROS (O2 − and H2O2) in these plants after salt stress, to assess the extent of antioxidant defense induced by the overexpression of two important antioxidant genes. Under salinity stress, the transgenic plants showed lower levels of H2O2 and O2 − compared to the wild-type plants, as was evident by NBT and DAB staining. Other groups have also reported similar results (Faize et al. 2011; Lee et al. 2013).
The present investigation showed that the WT plants were sensitive to salt stress as they showed reduced growth and low biomass accumulation. In contrast, the AhCuZnSOD + AhcAPX overexpressing plants demonstrated better growth, increased biomass, and improved yields, thus, exhibiting the traits of ‘salt tolerance’. This was attributed to overexpression of antioxidant enzymes AhCuZnSOD + AhcAPX that could lead to ROS detoxification.
In conclusion, our results illustrate that pyramiding of antioxidant genes (AhCuZnSOD and AhcAPX) resulted in scavenging of ROS in plants exposed to salinity stress. The increased activities of antioxidant enzymes and decreased MDA content in transgenic plants indicated high salinity tolerance. Other findings, such as improved photosynthetic activity, increased water retention, proline accumulation, antioxidant capacity, and a reduction in lipid peroxidation further confirmed the increased stress tolerance of transgenic plants. Thus, we recommend that this strategy could be useful for improvement of crop plants, and further investigations with crop plants are in progress in our laboratory.
References
Aebi H (1984) Catalase in vitro. Methods Enzymol 105:121–126
Asada K (1997) Role of ascorbate peroxidase and monodehydrascorbate reductase in H2O2 scavenging in plants. In: Scandalios JG (ed), Oxidative stress and Molecular biology of antioxidant defenses. Cold Spring Harbor Laboratory press, pp 715-735
Ashraf M (2009) Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol Adv 27:84–93
Badawi GH, Kawano N, Yamauchi Y, Shimada E, Sasaki R, Kubo A, Tanaka K (2004) Overexpression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. Physiol Plant 121:231–238
Chen GX, Asada K (1989) Ascorbate peroxidase in tea leaves: occurrence of two isozymes and the differences in their enzymatic and molecular properties. Plant Cell Physiol 30:987–998
Ediga A, Hemalatha S, Meriga B (2013) Effect of salinity stress on antioxidant defense system of two finger millet cultivars (Eleusine coracana (L.) Gaertn) differing in their sensitivity. Adv Biol Res 7(5):180–187
Faize M, Burgos L, Faize L, Piqueras A, Nicolas E, Barba-Espin G, Clemente-Moreno MJ, Alcobendas R, Artlip T, Hernandez JA (2011) Involvement of cytosolic ascorbate peroxidase and Cu/Zn-superoxide dismutase for improved tolerance against drought stress. J Exp Bot 62(8):2599–2613
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48(12):909–930
Gupta AS, Webb RP, Holaday AS, Allen RD (1993) Overexpression of superoxide dismutase protects plants from oxidative stress. Plant Physiol 103:1067–1073
Hayat S, Hasan SA, Yusuf M, Hayat Q, Ahmad A (2010) Effect of 28- homobrassinolide on photosynthesis, fluorescence and antioxidant system in the presence or absence of salinity and temperature in Vigna radiate. Environ Exp Bot 69:105–112
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125:189–198
Hofgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16:9877
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Kumar D, Yusuf MA, Singh P, Sardar M, Sarin NB (2013) Modulation of antioxidant machinery in α-tocopherol-enriched transgenic Brassica juncea plants tolerant to abiotic stress conditions. Protoplasma 250(5):1079–80
Kwon SY, Jeong YJ, Lee HS, Kim JS, Cho KY, Allen RD (2002) Enhanced tolerances of transgenic tobacco plants expressing both superoxide dismutase and ascorbate peroxidase in chloroplasts against methyl viologen-mediated oxidative stress. Plant Cell Environ 25:873–882
Lee SH, Ahsan N, Lee KW, Kim DH, Kwak SS, Kwon SY, Kim TH, Lee BH (2007) Simultaneous overexpression of both Cu/Zn superoxide dismutase and ascorbate peroxidase in transgenic tall fescue plants confers increased tolerance to a wide range of abiotic stresses. J Plant Physiol 164:1626–1638
Lee YP, Ahmad R, Lee HS, Kwak SS, Shafqat MN, Kwon SY (2013) Improved tolerance of Cu/Zn superoxide dismutase and ascorbate peroxidase expressing transgenic tobacco seeds and seedlings against multiple abiotic stresses. Int J Agric Biol 624:1814–9596
Lei Y, Yin C, Ren J, Li C (2007) Effect of osmotic stress and sodium nitroprusside pretreatment on proline metabolism of wheat seedlings. Biol Plant 51:386–390
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ 33:453–457
Nayyar H, Gupta D (2006) Differential sensitivity of C3 and C4 plants to water deficit stress: association with oxidative stress and antioxidants. Environ Exp Bot 58:106–113
Negi NP, Shrivastava DC, Sharma V, Sarin NB (2015) Overexpression of AhCuZnSOD alleviates salinity and drought stress in tobacco. Plant Cell Rep 15:1770–1774
Prashanth SR, Sadhasivam V, Parida A (2008) Overexpression of cytosolic copper⁄zinc superoxide dismutase from a mangrove plant Avicennia marina in indica rice var Pusa Basmati-1 confers abiotic stress tolerance. Transgenic Res 17:281–291
Rogers SO, Bendich AJ (1994) Extraction of total cellular DNA from plants, algae and fungi. Plant Mol Biol Manual D1:1–8
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, New York
Sharma P, Jha AB, Dubey RS, Pessarakli M (2012) Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Bot 12:1–26
Shrivastava D, Kisku AV, Deswal R, Sarin NB (2015) Stress inducible cytosolic ascorbate peroxidase (Ahcapx) from Arachis hypogaea cell lines confers salinity and drought stress tolerance in transgenic tobacco. Nuclease 58(1):3–13
Smith IK, Vierheller TL, Thorne CA (1989) Properties and functions of glutathione reductase in plants. Physiol Plant 77:449–456
Tang L, Kwon SY, Kim SH, Kim JS, Choi JS (2006) Enhanced tolerance of transgenic potato plants expressing both superoxide dismutase and ascorbate peroxidase in chloroplasts against oxidative stress and high temperature. Plant Cell Rep 25:1380–1386
Wang HS, Miyazaki K, Kawai M, Deyholos DW, Glabraith, Bohnert HJ (2003) Temporal progression of gene expression responses to salt shock in maize roots. Plant Mol Biol 52:873–891
Wang YC, Qu GZ, Li HY, Wu YJ, Wang C, Liu GF, Yang CP (2010) Enhanced salt tolerance of transgenic poplar plants expressing a manganese superoxide dismutase from Tamarix and rossowii. Mol Biol Rep 37(2):1119–1124
Xu J, Duan X, Yang J, Beeching JR, Zhang P (2013) Coupled expression of Cu/Zn- superoxide dismutase and catalase in cassava improves tolerance against cold and drought stresses. Plant Signal Behav 8:24–25
Yusuf MA, Kumar D, Rajwanshi R, Strasser RJ, Tsimilli-Michael M, Govindjee, Sarin NB (2010) Overexpression of gamma-tocopherol methyl transferase gene in transgenic Brassica juncea plants alleviates abiotic stress: physiological and chlorophyll a fluorescence measurements. Biochim Biophys Acta 1797:1428–1438
Acknowledgments
NPN acknowledges the financial support from the Department of Science and Technology. Partial funds from Department of Science and Technology (D.S.T.-PURSE, DST Women Scientist-A) UPOE, UGC-RNW and Jawaharlal Nehru University are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: Ewen Mullins
Rights and permissions
About this article
Cite this article
Negi, N.P., Shrivastava, D., Shekhar, S. et al. Simultaneous overexpression of CuZnSOD and cAPX from Arachis hypogaea leads to salinity stress tolerance in tobacco. In Vitro Cell.Dev.Biol.-Plant 52, 484–491 (2016). https://doi.org/10.1007/s11627-016-9764-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11627-016-9764-7