Abstract
Chikungunya virus (CHIKV) infection and subsequent high patient morbidity is a global threat. The present study aimed to identify the potent antiviral agent against Chikungunya virus, with minimum in vitro cytotoxicity. CHIKV nsP4 3D structure was determined using the I-TASSER server followed by its refinement and pocket determination. Furthermore, high-throughput molecular docking was employed to identify candidate CHIKV nsP4 inhibitors in a library containing 214 compounds. The top ranked compound was evaluated further with various assays, including cytotoxicity, antiviral activity, time of drug addition, viral entry attachment, and microneutralization assays. High-throughput computational screening indicated silibinin to have the best interaction with CHIKV nsP4 protein, immature and mature glycoproteins with highest negative free binding energy, − 5.24 to − 5.86 kcal/mol, and the lowest inhibitory constant, 50.47 to 143.2 µM. Further in vitro analysis demonstrated silibinin could exhibit statistically significant (p < 0.05) dose-dependent anti-CHIKV activity within 12.5–100-µM concentrations with CC50 as 50.90 µM. In total, 50 µM silibinin interfered with both CHIKV attachment (75%) and entry (82%) to Vero cells. Time of addition assay revealed silibinin interfered with late phase of the CHIKV replication cycle. Microneutralization assay revealed that silibinin could inhibit clearing of 50% Vero cell monolayer caused by CHIKV-induced CPE at a minimum dose of 25 µM. These data indicated silibinin to be a promising candidate drug against CHIKV infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Reemergence of Chikungunya virus (CHIKV) infection and its rapid spread across the globe and high patient morbidity demarked it as a global threat (Dutta et al. 2018; Sengupta et al. 2020). It is a mosquito-borne virus belonging to the genus alphavirus of Togaviridae family. Two-thirds of 11.7-kb CHIKV genome encodes nonstructural proteins (nsP1, nsP2, nsP3, nsP4): 5′ capping and subgenomic RNA synthesis, helicase, phosphatase, and RNA-dependent RNA polymerase (RdRp), respectively. The nsP4 is one of the independent functional modules whose relative abundance determines efficiency of viral RNA replication. Lack of RdRp in humans and other animals makes these polymerases an attractive antiviral target (Lello et al. 2021). The remaining one-third of viral genome codes for structural proteins (C, E3, E2, 6K, E1) involved in viral particle assembly and budding (Voss et al. 2010). Primarily, viral envelope complex is made up of E1 and E2 proteins which dissociate due to acidic environment of endosome, resulting in breakdown of heterodimer and release of E1 homotrimer. E2 glycoprotein is responsible for receptor binding, whereas E1 glycoprotein via its fusion peptide interacts with targeted cellular membrane leading to release of nucleocapsid into host cell cytoplasm. E3 plays an important role by protecting E2-E1 heterodimer from premature fusion with cellular membrane and also mediates correct folding of pE2. Overall, glycoproteins play a critical role in both early and late stages of CHIKV infection.
Currently, no FDA-approved specific drug/vaccine is available for effective cure of CHIKV infection, a high-priority disease declared by National Institute of Health, USA (Akahata et al. 2010). CHIKV infection is primarily controlled by reducing viral transmission to the host by using protective measures against mosquito bites (Cunha and Trinta 2017). Primarily, treatment is based on analgesics, antipyretics, and anti-inflammatory drugs for treatment of symptoms associated with CHIKV infection, viz., non-steroidal anti-inflammatory drugs, which might have side effects. Due to the explosive nature of recent CHIKV epidemics across the globe, development of potent anti-CHIKV drug is urgently needed (Wahid et al. 2017). Among broad-spectrum antiviral compounds, ribavirin, arbidol, harringtonine, silymarin, and suramin showed potent anti-CHIKV effect in vitro (Parashar and Cherian 2014). Plants are sources of natural compounds with antiviral properties, which can protect the human body against pathogens and have minimal toxicity to host cells (Oliveira et al. 2017). Most of them are readily absorbed in the bloodstream and reach concentrations that have potential to exert effects in vivo. Previous studies have suggested antiviral activity of andrographolide, arbidol, baicalin, benzofuran, cephalotoxin, chloroquine, coumarin, fluorouracil, harringtonine, kaempferol, luteolin, pyropropyridin, quercetin, and silymarin by inhibiting RdRp of different viruses (Ahmed-Belkacem et al. 2010, 2014; Alam et al. 2012; Anusuya and Gromiha 2017; Subudhi et al. 2018).
The present study was aimed to identify compound(s) of these groups with potent anti-CHIKV activity and preferably with less cytotoxicity in vitro. High-throughput virtual screening of similar compounds, retrieved from PubChem database, followed by testing of in vitro anti-CHIKV activity of the top-ranked compound was performed for this purpose.
Materials and methods
Retrieval of target sequence of nsP4 and crystal structure of glycoprotein complex
The 3D structure CHIKV nsP4 protein was determined based on NCBI Reference Sequence (RefSeq) with GenBank accession ID: NP_690588 (CHIKV reference sequence for nonstructural polyprotein), using the I-TASSER server (Ann Arbor, MI). The crystal structures of immature and mature envelope glycoprotein complexes (E3-E2-E1) of CHIKV were retrieved from Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) (pdb file: 3N40 and 3N42 respectively).
Primary and secondary structure analyses of nsP4
Physiochemical properties of CHIKV nsP4 (NP_690588): theoretical isoelectric point (pI), molecular weight (MW), molecular formula (MF), total number of positive (+R) and negative (-R) residues, instability index (II), extinction coefficient (EC), aliphatic index (AI), and grand average hydropathy (GRAVY) were analyzed using Expasy’s ProtParam server to validate the nature of nsP4 protein (Artimo et al. 2012). The secondary structural feature of nsP4 protein was studied using the PSIPRED server and its disulfide connectivity was predicted with the help of the DiANNA tool to understand proper protein stability and folding (Ferrè and Clote 2006; Buchan et al. 2013).
Modeling, structure refinement, and pocket identification
Ab initio modeling approach via the I-TASSER Web server was used to generate a reliable 3D model of nsP4 with proper folding conformations and with their confidence scores (Yang and Zhang 2015). Furthermore, stereo-chemical parameters, overall folding, and error over localized regions of predicted models were evaluated by VARIFY 3D and ERRAT of Structural Analytical and Verification Server (SAVES) (https://servicesn.mbi.ucla.edu/SAVES/). The Ramachandran plot for each predicted model was constructed using the PROCHECK Web server. The best model was then refined using the ModRefiner server. Finally, the MetaPocket server was used to locate functionally important amino acid residues of predicted nsP4 model and crystal structure of both mature and immature envelope glycoprotein complexes of CHIKV (3N40; 3N42).
Virtual library construction
Previous reports suggested plant-derived compounds, viz., andrographolide, arbidol, baicalin, cephalotoxin, chloroquine, coumarin, fluorouracil, harringtonine, kaempferol, luteolin, pyropropyridin, quercetin, silymarin, and ribavirin, to have antiviral activity against HIV, HCV, and influenza viruses ( Ahmed-Belkacem et al. 2010, 2014; Alam et al. 2012; Anusuya and Gromiha 2017; Subudhi et al.2018). Thus, a virtual library of compounds, having structural similarity with the abovementioned chemical groups, was constructed using PubChem similarity search with Tanimoto cutoff of 0.95. Similarity search was further refined on the basis of “Lipinski’s rule of five” and absorption, distribution, metabolism, and excretion (ADME) analysis. The sdf file of the compound was retrieved from PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and converted to mol2 format using OPEN BABEL software (O’Boyle et al. 2011).
Small molecular docking and pharmacophore development
Preliminary virtual screening of compounds with nsP4 was done using the iGEMDOCK v.2 program, following the below mentioned specifications: population size, 200; number of generations, 70; and number of solutions, 3. After analyzing the interaction profiles, compounds were ranked on the basis of their binding energy (electrostatic force of attraction, hydrogen bond, and Van der Waals interaction) (Hsu et al. 2011).
Top ten compounds were further docked using Autodock tools 1.5.6rc3 for refinement of compounds using a higher population size, 2,500,000; higher number of generations, 100; and higher number of solutions, 100 (Trott and Olson 2010). The nsP4 model was energy minimized. The model was placed within a grid box (48Å × 54Å × 58Å) with grid space of 0.925Å. One hundred runs were generated using Autodock 4.2 Lamarckian genetic algorithm to evaluate the binding energy between compounds and nsP4. Finally, the lowest energy pose of each compound, interacting with nsP4, was further visualized using LigPlot (Laskowski and Swindells 2011).
CHIKV glycoprotein is known to exist in two forms: immature glycoprotein contains furin susceptible peptide, which is absent in its mature form, might affect ligand protein interactions. Therefore, the top-ranked autodocked compound was also docked with X-ray crystal structures of immature (3N40) and mature (3N42) CHIKV glycoproteins using the aforesaid program, with grid boxes, dimensions, and positions of which are mentioned in Table S1. The lowest energy pose of the compound interacting with CHIKV glycoproteins was further visualized using LigPlot (Laskowski and Swindells 2011).
Finally, toxicity of the top ranked ligand was predicted using the PROTOX Web server (Drwal et al. 2014).
Compound preparation
Top-ranked compound of above docking analysis and ribavirin (catalog # R9644), a well-known CHIKV inhibitor, was purchased from Sigma-Aldrich Corporation. In total, 50 mM stock solution of the compound was prepared using dimethyl sulfoxide (DMSO) (Sigma-Aldrich, St. Louis, MO), whereas ribavirin was dissolved in double-distilled Millipore water and stored at −20°C for further use. The working solutions of each compound were prepared by twofold serial dilution in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco-BRL, Waltham, MA), followed by syringe filtration (pore size 0.2 μm). DMSO concentration in working solutions was kept less than 0.1%.
Cells and virus
C6/36 (Aedes albopictus mosquito) and Vero (African green monkey kidney) cell lines, purchased from National Centre for Cell Science, Pune, India, were used in this study. Both cell lines were maintained and propagated in DMEM containing 10% fetal bovine serum (FBS) (Gibco-BRL) and penicillin-streptomycin (Gibco-BRL). Cultured C6/36 and Vero cells were incubated at 28°C and 37°C, respectively, in 5% CO2 humidified incubator. After obtaining approval from the Clinical research ethical board of Calcutta School of Tropical Medicine, written consent was received from the patient, prior to participation in the study. In total, 100 µl of patient serum containing CHIKV, recovered from a patient during 2011 Eastern Indian outbreak [ECSA strain (GenBank ID: KJ679577)], was used as CHIKV stock for infecting C6/36 cell line (Dutta et al. 2018). At the time of CHIKV propagation, FBS concentration was reduced to 2%. To increase CHIKV copy number, cells were harvested after the seventh day post-infection, when full cytopathic effect (CPE) was visible.
CHIKV RNA was extracted from C6/36 cell supernatant using Trizol reagent according to manufacturer’s protocol (Invitrogen, Waltham, MA). CHIKV titer was determined using Chikungunya Non structural protein 2 (nsp2) genesig Standard Kit according to manufacturer’s protocol (PrimerDesign, UK). In vitro cytotoxicity of the top-ranked compound, its anti-CHIKV activity, and its inhibition towards CHIKV-mediated cytopathicity, virus entry, and attachment were detected on Vero cell lines.
In vitro cytotoxicity assay
In vitro cytotoxicity of top-ranked compound and ribavirin on Vero cells was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay following manufacturer’s instructions (HiMedia, Mumbai, India). Briefly, 104 Vero cells were seeded in a 96-well cell culture plate and treated with twofold serial dilutions of both the compounds (200–1.5μM) in triplicate along with a negative control (DMEM with 0.1% DMSO). The plate was incubated in a humidified incubator at 37°C with 5% CO2 for 48 h. After 48 h post-incubation, cells were treated with 10% MTT solution and were incubated in a humidified incubator at 37°C with 5% CO2 for 4 h. The plate was then observed under a microscope for the presence of formazan crystals, which was dissolved in 100 µl of solubilization solution. Finally, absorbance of each well was detected at 570-nm wavelength with a reference wavelength higher than 650 nm using iMark™ Microplate Absorbance Reader (BioRad, Hercules, CA). Cell viability percentage was determined using:
Cytotoxic concentrations (CC50) of the top-ranked compound and ribavirin were calculated using GraphPad Prism 6.0 software.
Antiviral activity assay
Antiviral activity of the top-ranked compound and ribavirin was evaluated using the qRT-PCR method. Briefly, 105 Vero cells were seeded in a 24-well cell culture plate and were cultured in a humidified incubator at 37°C with 5% CO2 for 24 h (Cruz et al. 2013). Cells were infected with CHIKV culture supernatant with a multiplicity of infection (MOI) of 1 and incubated at 37°C with 5% CO2 for 2 h. After viral incubation, supernatant containing unabsorbed virus was removed and cell monolayer was washed with 1× phosphate buffer saline (PBS) to remove unabsorbed virus partials. Subsequently, top-ranked compound/ribavirin (100–3.12 μM) were twofold serially diluted in complete DMEM and added to respective wells and incubated in a humidified incubator at 37°C with 5% CO2 for 48 h. Cells were harvested after 48 h post infection (hpi), followed by RNA extraction using Trizol reagent according to manufacturer’s protocol (Invitrogen). Absolute quantification of CHIKV was done using Chikungunya nsp2 genesig Standard Kit according to manufacturer’s protocol (PrimerDesign, Chandler’s Ford, UK), in Applied Biosystems 7500 Fast Real-Time PCR (ABI, San Francisco, CA), and each sample was analyzed in triplicate.
Viral attachment assay
To understand whether top-ranked compound could inhibit CHIKV attachment to host cell, viral attachment assay was performed (Moghaddam et al. 2014). Briefly, Vero cells were seeded at a concentration of 105 cells per well in a 24-well cell culture plate for 24 h. Twofold serially diluted top-ranked compound (100–3.12 μM) along with CHIKV-containing cell supernatant (MOI of 1) was added to respective wells and was incubated at 4°C for 2 h. After 2 hpi, virus-containing media were removed and cells were washed with 1× PBS. The wells were replenished with new complete DMEM and incubated under humidified condition at 37°C with 5% CO2 for 48 h. After 48 hpi, absolute quantification of CHIKV RNA within Vero cells was done using Chikungunya nsp2 genesig Standard Kit according to manufacturer’s protocol (PrimerDesign). Cellular attachment was determined by percentage viral load reduction between compound treated and untreated groups. Results were obtained from at least three independent experiments.
Viral entry assay
To evaluate efficiency of top-ranked compound to prevent subsequent CHIKV infection, viral entry assay was carried out (Moghaddam et al. 2014). Briefly, Vero cells were seeded at concentration of 105 cells per well in a 24-well cell culture plate for 24 h. Twofold serially diluted top-ranked compound (100–3.12 μM) along with CHIKV-containing cell supernatant (MOI:1) was added to respective wells and was incubated under humidified condition at 37°C with 5% CO2 for 2 h. After 2 hpi, virus-containing media were removed and cells were washed with ice-cold 1× PBS. Wells were replenished with new complete DMEM and incubated under humidified condition at 37°C with 5% CO2 for 48 h. After 48 hpi, absolute quantification of CHIKV RNA within Vero cells was done using Chikungunya nsp2 Genesig Standard Kit according to manufacturer’s protocol (PrimerDesign). Cellular entry was determined by percentage viral load reduction between compound treated and untreated groups. Results were obtained from at least three independent experiments.
Time of drug addition assay
Optimal time of CHIKV inhibition by top-ranked compound was determined by performing time of drug addition assay (Kaur et al. 2013). Briefly, Vero cells were seeded at a concentration of 105 cells per well in a 24-well cell culture plate for 24 h. For pretreatment assay, Vero cells were treated with top-ranked compound for 2 h before infection with CHIKV (MOI: 1). For co-treatment assay, Vero cells were treated with top-ranked compound and CHIKV (MOI: 1) at the same time. For post-treatment assay, Vero cells were infected with CHIKV and treated with top-ranked compound after 2 and 4 h of infection, respectively. For virus control, CHIKV along with complete DMEM with 0.1% DMSO was used. Plate was then incubated under humidified condition at 37°C with 5% CO2 for 48 h. After 48 hpi, absolute quantification of CHIKV RNA within Vero cells was done using Chikungunya nsp2 Genesig Standard Kit according to manufacturer’s protocol (PrimerDesign).
Microneutralization assay
To evaluate protection conferred by the top-ranked compound against CHIKV induced CPE, microneutralization assay was carried out based on a previously described method (Sui et al. 2004; Cruz et al. 2013). Briefly, 104 Vero cells per well were seeded in a 96-well cell culture plate and were incubated in a humidified incubator at 37°C with 5% CO2 for 24 h. After 24 h incubation, twofold serially diluted top-ranked compound (100 μM, 50 μM, 25 μM, 12.5 μM, 6.25 μM, 3.12 μM, and 1.5 μM) was added to respective wells along with CHIKV culture supernatant at MOI of 1 and incubated in a humidified incubator at 37°C with 5% CO2 for 2 h. The supernatant containing unbounded virus was removed and was replaced with new complete DMEM containing same amount of top-ranked compound, followed by incubation under a humidified condition at 37°C with 5% CO2 for 48 h. After 48 hpi, cells were fixed with 10% formaldehyde and stained using 0.1% crystal violet solution to visualize CHIKV-induced CPE. Optical density at 570 nm was measured using an EPOCH Microplate spectral photometer. Concentration that achieved 50% of the maximal effect (end point of microneutralization: EC50) was calculated using GraphPad Prism Version 6.0. The assay was performed in quadruplicate. Selectivity index (SI = CC50/EC50) value was calculated.
Results
No crystal structure of CHIKV nsP4 protein is available in RCSB PBD. Therefore, in this study, CHIKV nsP4 3D structure was predicted based on CHIKV reference sequence NP_690588, available in GenBank database. To validate the nature of CHIKV nsP4 protein, primary structural information predicted using the Expasy’s ProtParam server that revealed the protein with molecular formula C2212H3523N603O657S24 had the following properties: molecular weight (MW), 49,846.39 Dalton; isoelectric point (PI), 6.04; instability index (II), 45.11; average antigenicity (AI), 98.85; and negative grand average of hydropathicity (GRAVY), −0.050 (Fig. 1). Secondary structure of nsP4, predicted using the PSIPRED Web server, showed that it contained 16 helices, 30 coils, and 12 beta sheets. Since disulfide bonds influence protein stability and folding, disulfide bonds were predicted using the Diaana server, which showed CHIKV nsp4 to contain five disulfide bonds between cysteines at position 8-174, 57-264, 77-370, 83-356, and 310-333.
To obtain a good-quality model of nsP4, its three-dimensional structure was predicted based on an ab initio concept. I-TASSER generated three models of NP_690588 along with scoring values (viz., C-score) based on the five best matched templates (Table 1 and 2). Furthermore, stereo-chemical properties of three models were verified using PROCHEK Web server and Ramachandran plots were obtained based on φ and ψ angles of amino acids. Comparing stereo-chemical parameters and based on Ramachandran plot, Model-1 was proved to be of good quality and with proper folded conformation (Fig. 1). MetaPocket 2.0 Web server identified 214 ligand interacting amino acid residues, among which 25 residues were located in the catalytic domain (215-330 amino acid) of nsP4.
To identify potent anti-CHIKV compound, PubChem similarity search was performed to generate a virtual library of 214 compounds from different chemical groups (andrographolide, 46; arbidol, 7; baicalin, 4; cephalotoxin, 17; chloroquine, 39; coumarin, 30; fluorouracil, 7; harringtonine, 3; kaempferol, 8; luteolin, 4; pyropropyridin, 2; quercetin, 13; silymarin, 33; and ribavirin, 1), based on “Lipinski’s rule of five” and ADME analysis (Table S2). Protein-compound interaction profiles, generated after iGEMDOCK, were ranked based on binding energy, ranging from 0 to −25.3 kcal/mol. None of the compounds showed electrostatic force of attraction. Ribavirin was found to be top ranked with binding energy: −25.3 kcal/mol. Top ten compounds with the highest negative free binding energy, ranging from −25.3 to −19.8 kcal/mol, were selected for further analysis. For further refinement of iGEMDOCK data, top ten compounds were again docked with nsP4 using Autodock Vina software and their interaction profiles were viewed using LIGPLOT software (Table 3). Ligand with PubChem ID CID_16211710 (silibinin) showed the highest negative binding energy, −5.36 kcal/mol, and the lowest inhibition constant (Ki), 117.06 µM, while ribavirin (CID_37542), a well-known antiviral drug, showed binding energy −3.66 kcal/mol and Ki 2.08 mM (Fig. 2). Silibinin was found to form a hydrogen bond with Phe283 and hydrophobic interactions with residues: Ser280, Met282, Thr285, and Leu286, located within nsP4 catalytic domain, whereas none of the nsP4 catalytic domain residues interacted with ribavirin. Predicted toxicity of silibinin, using the ProTox Web server, was 2000mg/kg in a rodent model.
Molecular docking of CHIKV nsP4 protein with silibinin (CID_16211710) and ribavirin (CID_37542). (a) Docking solution of CID_16211710 in the catalytic domain of nsP4. (b) 2D representation of CID_16211710 and nsP4 interaction as analyzed using LIGPLOT. (c) Docking solution of CID_37542 in the catalytic domain of nsP4. (d) 2D representation of CID_37542 and nsP4 interaction as analyzed using LIGPLOT.
MTT assay was used to determine in vitro cytotoxicity of silibinin and ribavirin (Sigma-Aldrich). The CC50 of silibinin and ribavirin on Vero cells were 50.90 µM and 272.1 µM, respectively (Fig. 3). Hence, antiviral activity of silibinin was performed at concentrations less than CC50. The highest 0.1%DMSO was used as solvent of silibinin with no cytotoxic effect. Silibinin exhibited statistically significant (p<0.05) dose-dependent anti-CHIKV activity within 6.25–100-µM concentrations (30.98 to 82.03% of inhibition); similar statistically significant (p<0.05) dose-dependent inhibition was observed in the case of ribavirin (6.25–100µM concentrations) with 28.10 to 64.47% inhibition (Fig. 4a). Therapeutic index of silibinin was calculated as follows:
(a) Dose-dependent inhibition of CHIKV infectivity using silibinin (Sil) and ribavirin (Riv) (antiviral activity assay). Vero cells were infected with CHIKV for 2 h. Silibinin/ribavirin (100–3.12 μM) were added and incubated for 48 h. Each sample was analyzed in triplicates. Antiviral activity was evaluated using qRT-PCR method. (b) Effect of different concentrations of silibinin on CHIKV attachment to Vero cells (viral attachment assay). Vero cells were incubated with silibinin (100–3.12 μM) along with CHIKV and incubated at 4°C for 2 h. After 48 hpi, absolute quantification of CHIKV RNA was done. Results were obtained from three independent experiments. (c) Effect of different concentration of silibinin on CHIKV entry to Vero cells (viral entry assay). Vero cells were incubated with silibinin (100–3.12 μM) along with CHIKV and incubated at 37°C for 2 h. After 48 hpi, absolute quantification of CHIKV RNA was done. Results were obtained from three independent experiments. (d) Inhibition pattern of CHIKV infection by addition of silibinin at different time points (time of drug addition assay). Pretreatment assay: Vero cells were treated with silibinin for 2 h before infection with CHIKV. Co-treatment assay: Vero cells were treated with silibinin and CHIKV at the same time. Post-treatment assay: Vero cells were infected with CHIKV and then treated with silibinin after 2 and 4 h of infection, respectively. All were incubated at 37°C for 48 h. After 48 hpi, absolute quantification of CHIKV RNA within Vero cells was done.
Therapeutic index of silibinin is within the same range of drugs, viz., amphetamine, dextroamphetamine, and digoxin, currently used in clinical practice (Saganuwan 2020).
Since virus envelope protein plays an important role in fusion process, therefore docking analysis was conducted using different grid co-ordinates to understand whether silibinin could interact with both mature and immature CHIKV glycol-proteins, responsible for receptor binding and membrane fusion during early stage of infection. Silibinin exhibited both hydrogen bonding and hydrophobic interactions with different amino acids of immature (grid-1: binding energy −5.86 kcal/mol, Ki 50.47 µM; grid-2: binding energy −5.42 kcal/mol, Ki 105.8 µM) and mature (grid-1: binding energy −5.82 kcal/mol, Ki 54.10 µM; grid-2: binding energy −5.24 kcal/mol, Ki 143.2 µM) glycoprotein as described in Fig. 5. Furthermore, results obtained from docking study were validated using in vitro binding and attachment assay. Results from the in vitro study clearly depicted silibinin at concentration 50 µM could inhibit maximum 75% CHIKV attachment to Vero cells, whereas the same concentration could prevent 82% viral entry within host cells (Fig. 4b,c. Time of addition assay was conducted to find out the maximum inhibition activity of silibinin at early or late phase of CHIKV infection. Results from this assay revealed that silibinin exerted its antiviral effect against CHIKV infection within 0–2 hpi, with maximum anti-CHIKV activity at 2 hpi (Fig. 4d). Endpoint of microneutralization (EC50) assay was defined as the dilution at which minimum 50% of testing cells are not protected from infection (Sui et al. 2004). This assay revealed that silibinin could inhibit clearing of 50% cell monolayer caused by CHIKV-induced CPE at minimum dose of 25 µM (Fig 6a and b). Thus, SI value of silibinin was 50.90 µM/25 µM = 2.036.
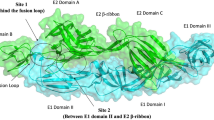
Molecular docking of immature (3N40) and mature (3N42) CHIKV glycoprotein with silibinin (CID_16211710) using Autodock 4.2. (a) Docking solution of CID_16211710 with 3N40 considering grid-1. (b) Interaction analyzed between CID_16211710 and 3N40 within grid-1, using LIGPLOT showing hydrogen bond interactions of CID_16211710 with E1 residues: Tyr233, His230, Ser238, Thr53, Tyr51, and hydrophobic interactions with E1 residues: Val231, Gln235, Pro237, Lys52, and E3 His191 with binding energy: − 5.86 kcal/mol and Ki: 50.47 µM. (c) Docking solution of CID_16211710 with 3N40 considering grid-2. (d) Interaction analyzed between CID_16211710 and 3N40 within grid-2, using LIGPLOT, showing hydrogen bond interaction of CID_16211710 with E1 residues Gly274, Glu366; and hydrophobic interactions with El residues: Val273, Ala272, Thrl0, Phe257; E2 residues: Glu388, Gly365, Glu367 with binding energy − 5.42 kcal/mol and Ki: 105.8 µM. (e) Docking solution of CID_16211710 with 3N42 considering grid-1. (f) Interaction analyzed between CID_16211710 and 3N42 within grid-I, using LIGPLOT, showing hydrogen bond interaction of the compound with El residues: Val231, Tyr51; E2 residue: Ile37 and hydrophobic interaction with El residues: His230, Pro232, Tyr233; E2 residue: Arg36, Glu35, Lys52, Pro240, Leu24l with binding energy − 5.82 kcal/mol and Ki: 54.10 µM. (g) Docking solution of CID_16211710 with 3N42 considering grid-2. (h) Interaction analyzed between CID_16211710 and 3N42 within grid-2, using LIGPLOT, showing hydrogen bond interaction with El residues: Asp292, Ala342, Val28, Gln138; and hydrophobic interaction with El residues: Lys325, Gly326, Glu345, Lys327, Thr288, Ala329, Glu343, Met27 with binding energy − 5.24 kcal/mol and Ki: 143.2 µM. (3N40-El region: colored yellow; E2 region colored gray; E3 region colored red).
(a) Photograph representing micro-neutralization assay of silibinin against CHIKV. Vero cells were infected with CHIKV in the presence of silibinin (100 μM, 50 μM, 25 μM, 12.5 μM, 6.25 μM, 3.12 μM, and 1.5 μM) and incubated for 2 h. The supernatant was removed and was replaced with new complete DMEM containing silibinin, and incubated for 48 h and fixed with 10% formaldehyde and stained using 0.1% crystal violet solution to visualize CHIKV-induced CPE. The assay was performed in quadruplicates. (b) Graph representing effect of micro-neutralization assay.
Discussion
Though explosive CHIKV epidemics have been reported worldwide, treatments of infected patients have been carried out using broad-spectrum antivirals only, due to the lack of effective anti-Chikungunya drugs (Akahata et al. 2010). Ribavirin (200 mg twice for 7 d) was known to reduce CHIKV symptoms, though this dose level was considered to be genotoxic and cytotoxic (Ravichandran and Manian 2008). Thus, identification of compound(s) with potent anti-CHIKV activity is urgently needed. In the present study, 214 bioactive compounds were in silico screened against CHIKV nsP4 protein to identify the lead compound with the least binding energy—which was further validated by in vitro studies.
Since X-ray crystallographic/NMR structure of CHIKV nsP4 protein was unavailable, ab initio modeling was performed to determine 3D structure of CHIKV nsP4 protein, followed by its stereo-chemical quality checking and its docking with 214 compounds. High-throughput screening identified silibinin to have the best nsP4 interaction profile with the highest negative free binding energy and lowest Ki—thus making it an attractive candidate for anti-CHIKV activity. Previous in silico analysis of chikungunya nsP4 protein by Ghildiyal et al. identified two pockets in its catalytic core (Ghildiyal et al. 2019). The top ten compounds identified in the current study interacted with one and six amino acids of pocket 1 and pocket 2, respectively. Silibinin formed hydrogen bond with amino acid at position 283 (Pocket 2) and demonstrated hydrophobic interactions with those at 280, 282, 285, and 286 positions within the same pocket. Similarly, silibinin exhibited hydrophobic interaction with Pro95 and Ser94 allosteric sites within the palm domain of RdRp and hydrogen bonded with Lys141 at the core cleft of nsP4 (Kumar et al. 2012). Silibinin, a major component of silymarin, has been previously reported to inhibit Orthohepadnavirus (e.g., HBV), Hepacivirus (e.g., HCV), and Lentivirus (e.g., HIV) replication (McClure et al. 2012; Umetsu et al. 2018). In vitro analysis of the present study identified anti-CHIKV activity of silibinin. Silibinin showed the lowest cytotoxicity against Vero cells at CC50: 50.90 µM. It significantly (p<0.05) reduced more than 80% of CHIKV replication between concentrations of 12.5 and 100 µM—as determined by the qRT-PCR method. In total, 50 µM silibinin was below the toxicity range of silibinin as predicted by the ProTox Web server for rodents. A docking study revealed more favorable binding of silibinin with different amino acid residues of CHIKV glycoprotein responsible for viral-host interactions than CHIKV nsP4 RdRp protein (Fig. 5). Silibinin (50 µM) showed to inhibit both CHIKV entry and attachment to Vero cells, indicating that silibinin might have an ability to bind with the viral envelope. This has been validated by the docking study, suggesting interaction of silibinin with His230 of CHIKV E1, the amino acid previously known to play an important role in membrane fusion, thereby restricting its interaction with targeted cellular membrane and inhibition in release of nucleocapsid into host cell cytoplasm. The results from time of addition assay suggested that silibinin might interfere with late phase (0–2 hpi) of the CHIKV replication cycle (Fig. 4). EC50 of silibinin indicated selectivity index of this compound was better than that of ribavirin, a common anti-viral drug used to treat viral hemorrhagic fever, as mentioned by previous studies (Kitaoka et al. 1986). Previous reports suggested flavonoids, viz., baicalein, quercetagetin, and hesperetin to inhibit early stage of CHIKV replication, while fisetin, naringenin, and silymarin to inhibit early post entry stages of CHIKV replication in vitro. Wada Y et al. have previously demonstrated inhibition of CHIKV infection by a benzimidazole-related Compound-A that targets functional domain of nsP4 (Wada et al. 2017). The anti-inflammatory effect of silibinin by reducing production of inflammatory cytokines and induced apoptosis in rheumatoid arthritis pathogenesis-related cells might assist in attenuating CHIKV-associated clinical symptoms (Tong et al. 2018).
Conclusion
In conclusion, based on both in silico and in vitro analyses, it can be postulated that silibinin might act as a new candidate molecule against CHIKV infection and also could help reduce CHIKV-associated clinical symptoms. Further in vitro studies should be needed to understand mode of CHIKV inhibition by silibinin. However, in vivo studies should be conducted to understand the effect of silibinin on CHIKV-mediated disease symptoms.
References
Ahmed-Belkacem A, Ahnou N, Barbotte L, Wychowski C, Pallier C, Brillet R, Pohl RT, Pawlotsky JM (2010) Silibinin and related compounds are direct inhibitors of hepatitis C virus RNA-dependent RNA polymerase. Gastroenterology 138(3):1112–1122. https://doi.org/10.1053/j.gastro.2009.11.053
Ahmed-Belkacem A, Guichou JF, Brillet R, Ahnou N, Hernandez E, Pallier C, Pawlotsky JM (2014) Inhibition of RNA binding to hepatitis C virus RNA-dependent RNA polymerase: a new mechanism for antiviral intervention. Nucleic Acids Res. 42(14):9399–409. https://doi.org/10.1093/nar/gku632
Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, Lewis MG, Higgs S, Rossmann MG, Rao S, Nabel GJ (2010) A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat Med. 16(3):334–8. https://doi.org/10.1038/nm.2105
Alam I, Lee JH, Cho KJ, Han KR, Yang JM, Chung MS, Kim KH (2012) Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase in complex with 2-thiouridine or ribavirin. Virology 426(2):143–151. https://doi.org/10.1016/j.virol.2012.01.016
Anusuya S, Gromiha MM (2017) Quercetin derivatives as non-nucleoside inhibitors for dengue polymerase: molecular docking, molecular dynamics simulation, and binding free energy calculation. J Biomol Struct Dyn 35(13):2895–2909. https://doi.org/10.1080/07391102.2016.1234416
Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H (2012) ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40(Web Server issue):W597–W603. https://doi.org/10.1093/nar/gks400
Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41(Web Server issue):W349–W357. https://doi.org/10.1093/nar/gkt381
Cruz DJ, Bonotto RM, Gomes RG, da Silva CT, Taniguchi JB, No JH, Lombardot B, Schwartz O, Hansen MA, Freitas-Junior LH (2013) Identification of novel compounds inhibiting chikungunya virus-induced cell death by high throughput screening of a kinase inhibitor library. PLoSNegl Trop Dis 7(10):e2471. https://doi.org/10.1371/journal.pntd.0002471
Cunha RVD, Trinta KS (2017) Chikungunya virus: clinical aspects and treatment - a review. Mem Inst Oswaldo Cruz 112(8):523–531. https://doi.org/10.1590/0074-02760170044
Drwal MN, Banerjee P, Dunkel M, Wettig MR, Preissner R (2014) ProTox: a web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 42(Web Server issue):W53–W58. https://doi.org/10.1093/nar/gku401
Dutta SK, Bhattacharya T, Tripathi A (2018) Chikungunya virus: genomic microevolution in Eastern India and its in-silico epitope prediction. 3 Biotech 8(7):318. https://doi.org/10.1007/s13205-018-1339-3
Ferrè F, Clote P (2006) DiANNA 1.1: an extension of the DiANNA web server for ternary cysteine classification. Nucleic Acids Res 34(1):W182–W185. https://doi.org/10.1093/nar/gkl189
Ghildiyal R, Gupta S, Gabrani R et al (2019) In silico study of chikungunya polymerase, a potential target for inhibitors. Virusdisease 30(3):394–402. https://doi.org/10.1007/s13337-019-00547-0
Hsu KC, Chen YF, Lin SR, Yang JM (2011) iGEMDOCK: a graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinformatics 12 Suppl 1(Suppl 1):S33. https://doi.org/10.1186/1471-2105-12-S1-S33
Kaur P, Thiruchelvan M, Lee RC, Chen H, Chen KC, Ng ML, Chu JJ (2013) Inhibition of chikungunya virus replication by harringtonine, a novel antiviral that suppresses viral protein expression. Antimicrob Agents Chemother. 57(1):155–67. https://doi.org/10.1128/AAC.01467-12
Kitaoka S, Konno T, De Clercq E (1986) Comparative efficacy of broad-spectrum antiviral agents as inhibitors of rotavirus replication in vitro. Antiviral Res 6(1):57–65. https://doi.org/10.1016/0166-3542(86)90039-2
Kumar P, Kapopara RG, Patni M, Pandya H, Jasrai Y, Patel S (2012) Exploring the polymerase activity of chikungunya viral non structural protein 4 (nsP4) using molecular modeling, e- pharmacophore and docking studies. Int J Pharm Life Sci 3:1752–1765
Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51(10):2778–2786. https://doi.org/10.1021/ci200227u
Lello LS, Bartholomeeusen K, Wang S et al (2021) nsP4 is a major determinant of alphavirus replicase activity and template selectivity. J Virol 95(20):e0035521. https://doi.org/10.1128/JVI.00355-21
McClure J, Lovelace ES, Elahi S, Maurice NJ, Wagoner J, Dragavon J, Mittler JE, Kraft Z, Stamatatos L, Horton H, De Rosa SC, Coombs RW, Polyak SJ (2012) Silibinin inhibits HIV-1 infection by reducing cellular activation and proliferation. PLoS One 7(7):e41832. https://doi.org/10.1371/journal.pone.0041832. Epub 2012 Jul 25. Erratum in: PLoS One. 2012;7(10). doi: https://doi.org/10.1371/annotation/78ba072a-6b7a-430a-8fcd-ef020e4fc458. Stamatatos, Leonidis [corrected to Stamatatos, Leonidas].
Moghaddam E, Teoh BT, Sam SS, Lani R, Hassandarvish P, Chik Z, Yueh A, Abubakar S, Zandi K (2014) Baicalin, a metabolite of baicalein with antiviral activity against dengue virus. Sci Rep 26(4):5452. https://doi.org/10.1038/srep05452.PMID:24965553;PMCID:PMC4071309
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: an open chemical toolbox. J Cheminform 7(3):33. https://doi.org/10.1186/1758-2946-3-33
Oliveira AF, Teixeira RR, Oliveira AS, Souza AP, Silva ML, Paula SO (2017) Potential antivirals: natural products targeting replication enzymes of dengue and chikungunya viruses. Molecules 22(3):505. https://doi.org/10.3390/molecules22030505
Parashar D, Cherian S (2014) Antiviral perspectives for chikungunya virus. Biomed Res Int. 2014:631642. https://doi.org/10.1155/2014/631642
Ravichandran R, Manian M (2008) Ribavirin therapy for Chikungunya arthritis. J Infect Dev Ctries 2(2):140–142
Saganuwan SA (2020) Comparative therapeutic index, lethal time and safety margin of various toxicants and snake antivenoms using newly derived and old formulas. BMC Res Notes 13(1):292. https://doi.org/10.1186/s13104-020-05134-x
Sengupta S, Mukherjee S, Haldar SK, Bhattacharya N, Tripathi A (2020) Re-emergence of Chikungunya virus infection in Eastern India. Braz J Microbiol. 51(1):177–182. https://doi.org/10.1007/s42770-019-00212-0
Subudhi BB, Chattopadhyay S, Mishra P, Kumar A (2018) Current strategies for inhibition of chikungunya infection. Viruses 10(5):235. https://doi.org/10.3390/v10050235
Sui J, Li W, Murakami A, Tamin A, Matthews LJ, Wong SK, Moore MJ, Tallarico AS, Olurinde M, Choe H, Anderson LJ, Bellini WJ, Farzan M, Marasco WA (2004) Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci U S A 101(8):2536–2541. https://doi.org/10.1073/pnas.0307140101
Tong WW, Zhang C, Hong T, Liu DH, Wang C, Li J, He XK, Xu WD (2018) Silibinin alleviates inflammation and induces apoptosis in human rheumatoid arthritis fibroblast-like synoviocytes and has a therapeutic effect on arthritis in rats. Sci Rep 8(1):3241. https://doi.org/10.1038/s41598-018-21674-6
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334
Umetsu T, Inoue J, Kogure T, Kakazu E, Ninomiya M, Iwata T, Takai S, Nakamura T, Sano A, Shimosegawa T (2018) Inhibitory effect of silibinin on hepatitis B virus entry. BiochemBiophys Rep 31(14):20–25. https://doi.org/10.1016/j.bbrep.2018.03.003
Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA (2010) Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468(7324):709–712. https://doi.org/10.1038/nature09555
Wada Y, Orba Y, Sasaki M et al (2017) Discovery of a novel antiviral agent targeting the nonstructural protein 4 (nsP4) of chikungunya virus. Virology 505:102–112. https://doi.org/10.1016/j.virol.2017.02.014
Wahid B, Ali A, Rafique S, Idrees M (2017) Global expansion of chikungunya virus: mapping the 64-year history. Int J Infect Dis. 58:69–76. https://doi.org/10.1016/j.ijid.2017.03.006
Yang J, Zhang Y (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43(W1):W174–W181. https://doi.org/10.1093/nar/gkv342
Funding
This study was partly funded by Department of Biotechnology, Government of West Bengal, India [No.132-BT (Estt.) /RD-1/10].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
11626_2022_666_MOESM1_ESM.doc
Supplementary file1 Different grid coordinates x, y, z; spacing; grid box size and location of immature and mature glycoprotein structure considered for autodock analysis. (DOC 29 KB)
11626_2022_666_MOESM2_ESM.doc
Supplementary file2 Binding energy of CHIKV nsP4 protein with selected 214 compounds considered for primary screening by iGEMDOCK v.2. (DOC 257 KB)
Rights and permissions
About this article
Cite this article
Dutta, S.K., Sengupta, S. & Tripathi, A. In silico and in vitro evaluation of silibinin: a promising anti-Chikungunya agent. In Vitro Cell.Dev.Biol.-Animal 58, 255–267 (2022). https://doi.org/10.1007/s11626-022-00666-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-022-00666-x