Abstract
Alternaria helianthiinficiens previously has been found as a pathogen of sunflower and cosmos in northern hemisphere. This fungus comprises a monotypic lineage which obviously is a separate group that has not been formally described as a section. Information about morphology, distribution, and pathogenic characters of this species is very limited. During this study, two taxonomic novelties were entered. A new section, A. sect. Helianthiinficientes, was described. Alternaria simmonsii was acknowledged to be a synonym of A. helianthiinficiens. The present work allowed definition of Arctium sp. and Sonchus sp. (both are family Asteraceae) as new hosts for A. helianthiinficiens. Isolates of this plant pathogenic fungus were obtained from several new places in the Southern European part of Russia. Six strains were tested on nine asteraceous plants and supported pathogenicity of all strains and susceptibility of all hosts. All strains were more aggressive for Helianthus annuus and Xanthium sibiricum than for other plants regardless on host from which they were isolated. Moderate aggressiveness was detected for Cirsium arvense and H. tuberosus while expansion of lesions on Arctium tomentosum, Artemisia vulgaris, Sonchus arvensis, Tanacetum vulgare, and Taraxacum officinale was sufficiently slower.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alternaria Nees is a large, morphologically diverse genus. Recently, a number of molecular phylogenetic studies have attempted to better resolve Alternaria phylogeny (Lawrence et al. 2012, 2013, 2014; Woudenberg et al. 2013, 2014; Al Ghafri et al. 2019). Morphological assessment and phylogenetic analysis allow the conclusion that up to date the genus consists of about 360 species (Wijayawardene et al. 2020). In total, currently, 28 sections were described in the genus Alternaria (Lawrence et al. 2016; Al Ghafri et al. 2019). There are a few monotypic lineages that have not been assigned a status of section. The species A. helianthiinficiens E.G. Simmons, Walcz & R.G. Roberts comprises one of such clade which obviously is a separate group, but formally must be still treated as a member of A. sect. Alternaria.
Alternaria helianthiinficiens was found as a pathogen of sunflower and cosmos in a few locations in North America, Europe, and Asia (Simmons 1986; Aćimović and Lačok 1991; Cho and Yu 2000; Luo et al. 2017). To our knowledge, there are only a few reports about solitary strains of this species isolated from sunflower in Russia (Gannibal 2011; Ivebor et al. 2013, 2014). Morphologically, A. helianthiinficiens resembles a number of A. sect. Porri species which are widely distributed on many asteraceous plants. Likely, this fungus could be misidentified with some of them. Thereby, in general information about its morphology, distribution and pathogenic characters are very limited.
The aims of this study were to clarify A. helianthiinficiens taxonomy, geography, and host specialization.
Materials and methods
Alternaria strains
The collection of micromycetes of the Laboratory of Mycology and Phytopathology of the All-Russian Institute of Plant Protection contained six strains isolated from Asteraceae plants in different geographical locations of European Russia and morphologically similar to A. helianthiinficiens, including two strains of A. simmonsii Gannibal. One representative A. helianthiinficiens strain was used in this work. Information on strains is summarized in the Table 1.
DNA isolation, PCR, and sequencing
Mycelium (10–50 mg per strain) was obtained from cultures incubated on V4 agar medium for 7 days. DNA was extracted with Genomic DNA Purification Kit (Thermo Fisher Scientific) according to a manufacturer protocol.
The primers EF1-728f/EF1-986r (Carbone and Kohn 1999), gpd1/gpd2 (Berbee et al. 1999), and CALDF1/CALDR1 (Lawrence et al. 2013) were used to amplify parts of the gene for translation elongation factor 1-α TEF, the gene for glycerol-3-phosphate dehydrogenase GPD, and the gene for calmodulin CALD, respectively. Amplicons were sequenced by Sanger’s method on ABIPrism 3500 (Applied Biosystems–Hitachi, Japan), with the Big Dye Terminator v3.1 Cycle Sequencing Kit (ABI, Foster City, USA), according to the manufacturer’s instructions. All sequences were deposited in the GenBank (Table 1).
Phylogenetic analysis
Sequences were assembled, edited, and aligned using Vector NTI advance 10 (Thermo Fisher Scientific) and MEGA X 10.1 software. Sequences of representative strains and type species were obtained from GenBank (Table 1). To implement phylogenetic analysis, two different datasets were made due to sequences of the same gene and species were represented in the GenBank by different strains. The first set was based on combined gpd and TEF sequences, whereas the second set included cald sequences. Alternaria solani strains were used as an outgroup in both sets. Phylogenetic analysis consisted of maximum likelihood (ML) and maximum parsimony (MP) was performed with MEGA X 10.1 (Kumar et al. 2018). Bootstrap support values with 1000 replications were calculated. Bayesian analyses and Bayesian probability calculation were carried out by Mr. Bayes v. 3.2.1. in Armadillo v. 1.1 (Lord et al. 2012).
Alignment and phylogenetic tree were deposited in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S28125).
Morphology characterization
For examination of micromorphological structures, the strains were grown on PCA (potato carrot agar) (Simmons 2007) and V-4 (Mikhailova et al. 2002) that is analogue of V-8 media using for description of large-spored and other Alternaria species (Simmons 2007). Strains were incubated for 14 days at 23 ± 1 °С without expose under light (for cultural study) or under an alternating light/dark cycle consisting of 12 h of cool-white fluorescent daylight. Species identification was performed with the Alternaria identification manual (Simmons 2007).
Pathogenicity test
Young wild plants (Arctium tomentosum, Artemisia vulgaris, Cirsium arvense, Sonchus arvensis, Tanacetum vulgare, Taraxacum officinale) with no visible symptoms were taken from the experimental field of the All-Russian Institute of Plant Protection (St. Petersburg, Russia). Helianthus annuus, H. tuberosus, and Xanthium sibiricum were grown in a greenhouse in pots. Plants with 4–6 pairs of true leaves were used. Leaf disks 10 mm in diameter or similar pieces of leaves were used for inoculation.
Strains were incubated in 250-mL flasks with 50-mL liquid soybean media (per 1 L: KH2PO4 2 g, (NH4)2SO4 1 g, MgSO4 1 g, glucose 20 g, soybean flour 10 g; pH 6) for 3 days at room temperature with permanent shaking. Mycelium was filtered with a fabric, briefly dried, grinded with a pestle, and diluted with sterile water to get a suspension 50 mg/mL. A drop (10 mL) of suspension was placed on a reverse leaf disk (piece) side (4 disks per test, 3 replicates). Inoculated leaf disks were incubated on wet filter paper in Petri dishes at 24 °C under an alternating light/dark 12/12 h cycle. After 2 days, leaf disks were turned over. Diameter of necrosis was measured 2, 3, 4, 5, 9 dpi. All tests were repeated twice. The fungus was isolated from the blights and subjected to microscopic analysis to support identity of the pathogen and to abide Koch’s postulates.
Results
Molecular phylogeny
The adjusted and aligned sequences in the phylogenetic analysis had the following lengths: TEF, 236 bp; gpd, 580 bp; and cald, 784 bp. The number of parsimony-informative sites per genome locus was 38 (15.7%), 40 (6.9%), and 129 (16.5%), respectively.
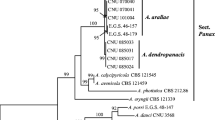
All six analyzed strains formed a compact clade with high bootstrap support containing all reference A. helianthiinficiens strains (Figs. 1 and 2). The topology of trees build by different methods was identical. Also, it was concordant with that reconstructed previously (Woudenberg et al. 2013).
Morphology
Strain MF P283031 was characterized by the best sporulation. It formed moderate sporulation on V-4 and it was only one strain with abundant sporulation on PCA after 5–7 days of incubation. Therefore, it was used for morphological description of A. helianthiinficiens.
On PCA, conidiophores are solitary 20–80 × 4–6 μm. Conidia are mainly solitary but sometimes in chains of two due to apical or lateral secondary conidiophores (Fig. 3). Mature conidia have narrow- to broad-ovoid or ellipsoid body, reaching a size of ca. 35–65(75) ×16–20 μm with 7–9 transverse septa and 1–3 longitudinal septa in 4–7 transverse segments. Conidia are not or slightly constricted by most of transverse septa. Many conidia have narrow or wide conical apical cell. Some conidia produce short or long filiform beak (up to 300 × 2–4 μm or longer). Solitary conidia have apical secondary conidiophore 5–30 × 3–6 μm. Rarely, conidia produce one or two short lateral secondary conidiophores.
On V-4, conidiophores are solitary or in small groups, simple or slightly branched 50–90(200) × 5–6 μm with 1–2(3) conidiogenous loci. Conidia are solitary or sometimes in chains of 2(3). Basal conidia in a chain can form one apical and 1–4 lateral secondary conidiophores 5–30 × 4–6 μm long; each of them bear 1–2(3) conidiogenous loci. Due to formation of secondary conidiophores with several conidiogenous loci, 2–3-week-old cultures contain “bushes” of 10–30 conidia. Conidia body on V-4 is slightly wider than that produced on PCA, mainly 30–60 × 14–22(30) μm ovoid to broad-ovoid. Some conidia have broad-ovoid or saccular body 50–80(105) × 18–23(29) μm and filiform apical beak 20–150 × 2–4 μm. Conidial body has 4–7(9) transverse and 4–11 longitudinal or oblique septa, i.e., 1–3 in several of the broadest transverse segments. Conidia are slightly or moderately constricted by most of transepta.
Other strains on PCA and V-4 formed weak or moderate sporulation. In comparison with MF-P283-031, conidia were in general slightly smaller (40–105 × 17–26(28) μm on V-4) and less percentage of conidia looks mature. Only comparatively short beaks 20–100(140) μm long were observed in some conidia even after 2 weeks of growth.
Taxonomy
Section Helianthiinficientes Gannibal, sect. nov.
MycoBank: MB839327.
Type species: Alternaria helianthiinficiens E.G. Simmons, Walcz & R.G. Roberts.
Diagnosis: Primary conidiophores are simple or branched, with one or a few conidiogenous loci. Conidia are solitary or in short chains. Body of mature conidia is moderately large, narrow- to broad-ovoid or ellipsoid, constricted near septa. Conidia have several transverse and longitudinal septa. Some conidia are non-beaked when other form apical secondary conidiophore or short to very long filiform beak. Conidia can produce one or a few lateral secondary conidiophores. Sexual morph is not known.
Notes: Molecular phylogenetic analyses performed by a number of authors (Woudenberg et al. 2013; Al Ghafri et al. 2019) based on three to six (SSU+LSU+ITS+GPDH+TEF+RPB2) genomic loci unambiguously demonstrated that A. helianthiinficiens formed an independent, well-separated lineage within the genus Alternaria and could not be assigned to any of the known sections. Its nearest neighbors were another monotypic lineage represented by Alternaria brassicae and sect. Sonchi.
Ex-type A. simmonsii strain MF P024011 (VKM F-4110) and similar strain MF P024021 obtained from the same place and date and all our other strains have TEF, gpd, and cald sequences identical to that of ex-type A. helianthiinficiens strain CBS 208.86. Strain MF P024011 and MF P024021 had insufficient morphological difference with A. helianthiinficiens strains. Thus we postulate that Alternaria simmonsii Gannibal (Gannibal 2010) [MB#518504] is a synonym of Alternaria helianthiinficiens E.G. Simmons, Walcz & R.G. Roberts (Simmons 1986) [MB#534400].
Pathogenicity
Any or all analyzed A. helianthiinficiens strains demonstrated pathogenic properties in artificial inoculation of each tested plant species. All analyzed strains were highly aggressive to Helianthus annuus and Xanthium sibiricum (both belong to tribe Heliantheae), and caused 100% necrosis of leaf discs on 4 dpi (Fig. 4). The inoculation of Helianthus tuberosus leaf discs (also tribe Heliantheae) by the analyzed strains led to development of necrosis from 4.4 ± 1.1 to 8.7 ± 0.5 mm (on average 5.6 ± 1.3 mm). The exception was A. helianthiinficiens strain MF P024021 that was not pathogenic for Helianthus tuberosus. The analyzed A. helianthiinficiens strains also turned out to be highly aggressive when infected the leaf discs of Cirsium arvense (tribe Cynareae) and caused necrosis from 5.5 ± 2.2 to 10.0 mm (on average 8.2 ± 0.7 mm) on 4 dpi.
The analyzed A. helianthiinficiens strains were, on average, weakly aggressive to other tested asteraceous plants. The size of necrosis did not exceed 1.7 mm on 4 dpi. On 9 dpi, all analyzed strains caused significant necrosis 6.5 ± 2.3–10 mm on leaf discs of Taraxacum officinale (tribe Lactuceae). Only three A. helianthiinficiens strains induced necrosis from 5.5 ± 2.2 to 10 mm on leaf discs of Sonchus arvensis (also tribe Lactuceae) on 9 dpi. Five A. helianthiinficiens strains of six were pathogenic to Artemisia vulgaris and four strains were pathogenic to Tanacetum vulgare (both belong to tribe Anthemideae) and caused necrosis 6.3 ± 1.7 and 2.7 ± 1.2 mm on 9 dpi, respectively. Four strains were weakly pathogenic to Arctium tomentosum (tribe Cynareae); the size of necrosis varied between 1.8 ± 1.2 and 6.5 ± 0.8 mm on 9 dpi.
Strains MF P283031 and MF P437021 isolated from Arctium tomentosum and Helianthus annuus, respectively, were the most aggressive in majority of the tests. No interrelation between host origin of strains and their pathogenicity to tested plants was revealed.
Discussion
Accurate identification of A. helianthiinficiens as well as many other Alternaria species is a bit troublesome. Careful adherence to standard protocols of strain cultivation is essential. However, even if the conditions are met, conidia size and shape can vary between strains and between different passages of the same strain.
Alternaria helianthiinficiens morphologically is very similar to many species of Alternaria sect. Porri. Strains of at least 21 Alternaria sect. Porri species were obtained from asteraceous plants (Woudenberg et al. 2014). There are two large-spored Alternaria species excepting A. helianthinficiens detected on sunflower—Alternaria carthami S. Chowdhury (A. heliophytonis E.G.Simmons) and A. protenta E.G.Simmons (Simmons 1986, 1997, 2007). Taxonomy of both species was supported by molecular phylogenetic approach (Woudenberg et al. 2014). The first species was found on Helianthus and Carthamus plants (both Asteraceae) while the second species was isolated worldwide from at least five species of four families. Neither A. carthami nor A. protenta were found on the territory of Russia.
Another large-spored species, A. zinnia M.B. Ellis, was repeatedly reported as sunflower-associated fungus (Neergaard 1945; McDonald and Martens 1963; Rao 1971; Carson 1987; Gulya et al. 1991; Prathuangwong et al. 1991). Most likely, all those cases were result of misidentification since those reports have been done primarily before A. helianthiinficiens, A. carthami, and A. protenta were described. The concept of the species A. zinnia was previously rather wide and resulted in combination of several morphologically similar species under this name (Simmons 1986, 1997).
Isolates of Alternaria helianthiinficiens were obtained from several locations in the South of European Part of Russia (Table 1). Recently, a few times, this fungus was isolated from sunflower leaves from other regions of European Russia (Lipetsk, Samara, and Kursk regions), as well as in Siberia (Altai Krai) (Gomzhina, Orina, unpubl.). Previously, Alternaria helianthiinficiens was also isolated from sunflower seeds grown in Altai Krai (Gannibal 2011) and Saratov region (Ivebor et al. 2014). Also A. helianthiinficiens was found on sunflower in North Dakota, USA (Simmons 1986), former Yugoslavia (Aćimović and Lačok 1991), South Korea (Cho and Yu 2000), and on cosmos in China (Luo et al. 2017). Obviously, this fungus has worldwide distribution.
Alternaria helianthiinficiens is a more aggressive pathogen for sunflower than another wider distributed Alternaria blight pathogen, Alternariaster helianthi (Alternaria helianthi), but it has a longer incubation period in the laboratory tests (2–3 days instead of 1–2 days for Alternariaster helianthi) (Cho and Yu 2000). During winter time, A. helianthiinficiens can survive as mycelium on plant residues and in seeds (Aćimović and Lačok 1991).
The present work adds Arctium sp. and Sonchus sp. to previously known hosts of A. helianthiinficiens—Helianthus annuus and Cosmos bipinnatus. However, it demonstrates different aggressiveness of pathogen to different hosts when sunflower can be affected in higher degree. It is interesting that A. helianthiinficiens strain MF P024021 isolated from Sonchus was nonpathogenic to its host plant but simultaneously was highly aggressive to Helianthus annuus. Similarly, Arctium-borne strains MF P283031 and MF P283041 very weakly infected their host plant, but induced a significant necrosis on sunflower leaf discs. These observations, as well as the analysis of the pathogenicity of strains to other tested plant species, suggest that A. helianthiinficiens may appear on some other plant species of the Asteraceae family. The ability to infect weeds made this fungus potentially common sunflower pathogen and complicates the control of disease caused by A. helianthiinficiens in the field.
Data availability
All new sequences are deposited in GenBank (https://www.ncbi.nlm.nih.gov) as specified in Table 1. Other data generated or analyzed during this study are included in this published article and its supplementary information file.
References
Aćimović M, Lačok N (1991) Alternaria helianthiinficiens Simmons, Walcz and R. Roberts sp. Nov.: the causal agent of brown-red spot, a new sunflower disease. Helia 14:129–145
Al Ghafri AA, Maharachchikumbura SS, Hyde KD, Al-Saady NA, Al-Sadi AM (2019) A new section and a new species of Alternaria from Oman. Phytotaxa 405:279–289. https://doi.org/10.11646/phytotaxa.405.6.1
Berbee ML, Pirseyedi M, Hubbard S (1999) Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 91:964–977. https://doi.org/10.1080/00275514.1999.12061106
Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556. https://doi.org/10.1080/00275514.1999.12061051
Carson ML (1987) Effect of two foliar pathogen on seed yield of sunflower. Plant Dis 71:549–551. https://doi.org/10.1094/pd-71-0549
Cho HS, Yu SH (2000) Three Alternaria species pathogenic to sunflower. Plant Pathol J 16(6):331–334
Gannibal PB (2010) Taxonomic studies of Alternaria from Russia: new species on Asteraceae. Mycotaxon. 114:109–114. https://doi.org/10.5248/114.109
Gannibal PB (2011) Species composition, systematics and geography of agents of sunflower alternarioses in Russia. Plant Protection News 1:13–19 (In Russian)
Gannibal PB, Orina AS, Mironenko NV, Levitin MM (2014) Differentiation of the closely related species, Alternaria solani and A. tomatophila, by molecular and morphological features and aggressiveness. Eur J Plant Pathol 139(3):609–623. https://doi.org/10.1007/s10658-014-0417-6
Gulya TJ, Woods DM, Bell R, Mankl MK (1991) Diseases of sunflower in California. Plant Dis 75:572–574. https://doi.org/10.1094/PD-75-0572
Ivebor MV, Antonova TS, Saukova SL (2013) On agents of sunflower Alternaria. Oil crops. Sci Techn Bull VNIIMK (1):153–154 (In Russian)
Ivebor MV, Saukova SL, Antonova TS, Araslanova NM (2014) Fungi of Alternaria Nees genus in sunflower seeds. Oil crops. Sci Techn Bull VNIIMK (1):157–158 (In Russian)
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549 https://doi.org/10.1093/molbev/msy096
Lawrence DP, Park MS, Pryor BM (2012) Nimbya and Embellisia revisited, with nov. comb for Alternaria celosiae and A. perpunctulata. Mycol Prog 11:799–815. https://doi.org/10.1007/s11557-011-0793-7
Lawrence DP, Gannibal PB, Peever TL, Pryor BM (2013) The sections of Alternaria: formalizing species-group concepts. Mycologia. 105(3):530–546. https://doi.org/10.3852/12-249
Lawrence DP, Gannibal PB, Dugan FM, Pryor BM (2014) Characterization of Alternaria isolates from the infectoria species-group and a new taxon from Arrhenatherum. Pseudoalternaria arrhenatheria sp nov Mycol Prog 13:257–276. https://doi.org/10.1007/s11557-013-0910-x
Lawrence DP, Rotondo F, Gannibal PB (2016) Biodiversity and taxonomy of the pleomorphic genus Alternaria. Mycol Prog 15:3. https://doi.org/10.1007/s11557-015-1144-x
Lord E, Leclercq M, Boc A, Diallo AB, Makarenkov V (2012) Armadillo 1.1: an original workflow platform for designing and conducting phylogenetic analysis and simulations. PLoS One 7:e29903. https://doi.org/10.1371/journal.pone.0029903
Luo H, Jia G, Fan X, Liu H, Pei D, Deng J, Zhou Y (2017) Isolation and identification of a new pathogen causing leaf blight on Cosmos bipinnatus. Plant Prot 43(6):182–186 (In Chinese)
McDonald WC, Martens JW (1963) Leaf and stem spot of sunflowers caused by Alternaria zinnia. Phytopathol. 53(1):93–96
Mikhailova LA, Gogoleva SG, Gultyaeva EI (2002) Interaction of strains of Bipolaris sorokiniana and wheat samples. Mikol Fitopatol 36(2):63–66 (In Russian)
Neergaard P (1945) Danish species of Alternaria and Stemphylium. Oxford University Press, London, 560 pp
Prathuangwong S, Kao SW, Sommartya T, Sinchaisri P (1991) Role of four Alternaria spp. causing leaf and stem blight of sunflower in Thailand and their chemical controls. Kasetsart J Natur Sci 25:112–124
Rao VG (1971) An account of fungus the genus Alternaria Nees from India. Mycopathol Mycol Appl 43(1–3):361–374. https://doi.org/10.1007/BF02051759
Simmons EG (1986) Alternaria themes and variations (17-21). Mycotaxon. 25:203–216
Simmons EG (1997) Alternaria themes and variations (151-223). Mycotaxon. 65:1–91
Simmons EG (2007) Alternaria. An Identification Manual, CBS, Utrecht, 775 pp
Wijayawardene NN, Hyde KD, Al-Ani LKT et al (2020) Outline of fungi and fungus-like taxa. Mycosphere 11(1):1060–1456. https://doi.org/10.5943/mycosphere/11/1/8
Woudenberg JHC, Groenewald JZ, Binder M, Crous P (2013) Alternaria redefined. Stud Mycol 75:171–212. https://doi.org/10.3114/sim0015
Woudenberg JHC, Truter M, Groenewald JZ, Crous PW (2014) Large-spored Alternaria pathogens in section Porri disentangled. Stud Mycol 79:1–47. https://doi.org/10.1016/j.simyco.2014.07.003
Acknowledgements
The authors are grateful to Maria M. Gomzhina for providing data about a few recent findings of Alternaria helianthiinficiens.
Funding
This work was supported by the Russian Science Foundation (project no. 19-76-30005).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Philipp B. Gannibal contributed in sample collection, fungi isolation, morphology study, and taxonomic conclusions. Phylogenetic analysis was carried out by Aleksandra S. Orina. Elena L. Gasich performed all experiments to study pathogenicity. The manuscript was written by Philipp B. Gannibal and all authors provided critical feedback and helped shape the research, analysis, and manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Gerhard Rambold
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 30 kb)
Rights and permissions
About this article
Cite this article
Gannibal, P.B., Orina, A.S. & Gasich, E.L. A new section for Alternaria helianthiinficiens found on sunflower and new asteraceous hosts in Russia. Mycol Progress 21, 34 (2022). https://doi.org/10.1007/s11557-022-01780-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-022-01780-6