Abstract
Eucalyptus globulus, an exotic species in Portugal, is one of the dominant and intensively managed forest species in the country. A disease syndrome characterised by leaf necrosis, stem girdling and cutting dieback in eucalyptus and associated with pestalotioid fungi has been detected in nurseries and young plantations in the last years. Twenty-seven isolates were recovered from diseased plants. Phylogenetic analysis based on internal transcribed spacers, partial translation elongation factor 1-α gene and partial β-tubulin gene sequence data grouped the isolates in five separate clades. Combining morphological, cultural and molecular data, five new species of Neopestalotiopsis are described, namely, Neopestalotiopsis eucalyptorum, Neopestalotiopsis hispanica, Neopestalotiopsis iberica, Neopestalotiopsis longiappendiculata and Neopestalotiopsis lusitanica.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Eucalyptus species are among the most widely planted hardwood timber trees in the world (FAO 2001). In Portugal, Eucalyptus globulus is the most important commercial species with a land usage of more than 845,000 ha (ICNF 2019), directed mostly at the papermaking industry (Pirralho et al. 2014).
Eucalyptus globulus was introduced into Portugal in the nineteenth century but only in the middle of the twentieth century did the great increase of plantations occur. In the following decades, eucalyptus plantations were relatively free of pests and diseases. However, since the 1970s, several pests appeared, firstly insects and then fungi (Branco et al. 2014). Leaf diseases caused by Mycosphaerella, Teratosphaeria and Quambalaria species and canker diseases caused by Teratosphaeria gauchensis, Quambalaria eucalypti and various species in Botryosphaeriaceae have been reported to cause important losses in eucalyptus plantations in the last years (Barradas et al. 2016; Bragança et al. 2016; Silva et al. 2015) . Grey mould caused by Botrytis cinerea and anthracnose caused by Hendersonia eucaliptina are also important diseases faced by Portuguese eucalypt nursery growers (Ferreira et al. 1994).
Pestalotioid fungi have received considerable attention for their association with a wide range of cultivated plants, as well as for the production of economically important metabolites (Jayawardena et al. 2019; Maharachchikumbura et al. 2011; Metz et al. 2000; Mishra et al. 2014; Tejesvi et al. 2007; Xu et al. 2014). These fungi have ubiquitous distribution and are mostly regarded as secondary pathogens affecting weakened plants, or opportunistic saprobes found on dead material (Almeida et al. 2003; Bakry et al. 2011; Hopkins and McQuilken 2000; Old et al. 2000; Stone et al. 2004; Tejesvi et al. 2007). However, they have been found to cause damage on economically important plants, both in forestry and agriculture (Akinsanmi et al. 2017; Bakry et al. 2011; Chen et al. 2018; Espinoza et al. 2008; Hopkins and McQuilken 2000; Ismail et al. 2013; Lin et al. 2011; Morales-Rodríguez et al. 2019; Qi et al. 2021; Silva et al. 2020).
Since 2012, a disease causing stem girdling and dieback in young eucalyptus plants has been detected, and pestalotioid fungi are consistently associated with diseased plants. The aim of the work reported here was to identify the fungal species associated with this disease.
Material and methods
Sampling and isolation
In Portugal, the disease syndrome was first detected in November 2012 in a nursery where it caused stem girdling and dieback in young plants of Eucalyptus globulus. Since then, and until 2018, tissues of young eucalypts plants from recent plantations and nursery stocks have been collected and transported to the laboratory for observations and disease diagnosis.
Two methods were used to obtain pure cultures. When sporulation was observed on the plant material, as tendrils exuding from fruiting bodies, the spores were carefully removed with a fine needle avoiding any other sporulation from the host. They were transferred to a 2% water agar plate and streaked with a sterile loop. After 24 to 48 h, single germinating conidia were selected under a compound microscope and aseptically transferred to half strength potato dextrose agar (½PDA) plates prepared from 20 g/L Difco potato dextrose agar (Difco Laboratories, Detroit, MI, EUA), plus 10 g/L agar. When no sporulation could be observed, small fragments (2 mm2) were cut from the margins of the lesions and treated as previously described (Diogo et al. 2010). Cultures resulting from single spores were deposited in the fungal collection of INIAV (formerly Micoteca da Estação Agronómica Nacional, MEAN). Representative isolates were also deposited in the CBS public fungal culture collection at the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. Dried specimens were deposited in the LISE herbarium of INIAV. Nomenclatural novelties and descriptions were deposited in MycoBank (www. MycoBank.org; Crous et al. 2004).
Morphology
Colony characteristics were registered after 7 days on PDA plates incubated at 25 °C with 12 h of near ultraviolet light per day. Colours were determined using the colour chart of Rayner (Rayner 1970). Growth rates were determined under the same conditions in the dark.
Fructifications and spores were mounted in water. Observations of micromorphological features were made with Leica MZ95 and Leica DMR microscopes, and digital images were recorded with Leica DC300 and Leica DFC320 cameras, respectively. Measurements were made with the measurement module of the Leica IM500 image management system (Leica Micro-systems GmbH, Wetzlar, Germany). Mean, standard deviation (SD) and 95% confidence intervals were calculated. Thirty spores and at least ten other structures were measured.
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted using the DNA, RNA and Protein Purification—NucleoSpin Plant II (Macherey–Nagel-MN) following the manufacturer’s instructions. Fresh mycelium scraped from the surface of PDA cultures was disrupted by vortexing with approximately 200 µL glass beads (450–600 µm diameter) added to the extraction buffer according to Bragança et al. (2007). Three DNA regions, the internal transcribed spacer (ITS), translation elongation factor 1-α (TEF1) and partial β-tubulin gene (TUB2) were amplified with the primer pairs ITS5 and ITS4 (White et al. 1990), EF1-728F (Carbone and Kohn 1999) and EF2 (O’Donnell et al. 1998) and BT2A and BT2B (Glass and Donaldson 1995), respectively. All PCR reactions were performed in a 25 μL reaction containing DNA template (diluted 10 ×), 10 × PCR reaction buffer, 3 mM MgCL2, 0.5 mM of each deoxyribonucleotide triphosphate, 1 U of Taq DNA polymerase (BioTaqTM DNA polymerase—Bioline, London, UK) and 2 μM each primer. PCR reactions were performed in a Biometra TGradient thermo cycler (Biometra, Göttingen, Germany) as described by Silva et al. (2020). The products were resolved by electrophoresis at 5 Volts.cm−1 in agarose gel (1%) containing 0.5 μg/mL ethidium bromide and 1 × TBE running buffer. Products were visualised by VersaDoc Gel Imaging System (BioRad, USA) and purified with the QIAquick PCR Purification Kit (Qiagen, USA) following the manufacturer’s instructions. Sequencing was performed by STABVIDA (Caparica, Portugal) on an ABI PRISM 3730xl DNA analyser (Applied Bio systems) with the same primers used for amplification. Sequences generated in this study were deposited in GenBank (Table 1) and alignments in TreeBase (study: TB2:S28369).
Phylogenetic analysis
Sequences of Neopestalotiopsis species referred to in recent studies (Table 1), including all sequences of type species available, were retrieved from GenBank. The individual loci were aligned separately with MAFFT V.7 (Katoh et al. 2019), checked and trimmed in MEGA v.7 (Kumar et al. 2016) and concatenated with sequence matrix (Vaidya et al. 2011). The combined dataset was analysed by maximum parsimony (MP), maximum likelihood (ML) and Bayesian Inference (BI). Sequences of Pestalotiopsis diversiseta MFLUCC 12–0287 were used as the outgroup.
MP analysis was performed using PAUP* v4.0a (Swofford 2002) implemented in the CIPRES science gateway (Miller et al. 2010) using the heuristic search option with random stepwise addition and tree bisection reconnection (TBR) as the branch swapping algorithm. Maxtrees were set to 1000, and branches of zero length were collapsed, and alignment gaps were treated as missing data. Clade stability and robustness of the most parsimonious trees were assessed using bootstrap analysis with 1000 pseudoreplicates, each with 10 replicates of random stepwise addition of taxa (Felsenstein 1985). Tree length (TL), consistency index (CI), retention index (RI) and homoplasy index (HI) were calculated.
For BI and ML, a partitioned analysis was performed with three partitions: ITS, TEF1 and TUB2. MrModeltest v2.4 (Nylander 2004) was used to select the best-fit nucleotide substitution model for each partition using the Akaike information criterion (AIC) (Akaike 1974) implemented in PAUP* v. 4.0b10. The HKY + G (Hasegawa et al. 1985) model was selected as the most suitable for all partitions. BI was performed with the Markov chain Monte Carlo method (MCMC) with MrBayes 3.2.7a (Ronquist et al. 2012) implemented in the CIPRES portal. Four MCMC chains were run simultaneously, starting from random trees for 1,000,000 generations. Trees were sampled every 100th generation for a total of 10,000 trees. The first 1000 trees were discarded as the burn-in phase of each analysis. Posterior probabilities (Rannala and Yang 1996) were determined from a majority-rule consensus tree generated with the remaining 9000 trees. This analysis was repeated three times starting from different random trees to ensure trees from the same tree space were sampled during each analysis.
The ML analysis was performed in the CIPRES portal (Miller et al. 2010) using RAxML-HPC2 on XSEDE (8.2.12) (Stamatakis 2014). The optimal ML tree search was conducted with 1000 rapid bootstrap inferences. The final tree was selected among suboptimal trees from each run by comparing likelihood scores under the GTR + GAMMA substitution model. The resulting trees were visualised with MEGA v. 70.026 (Kumar et al. 2016) and edited in Microsoft PowerPoint.
Results
Symptoms, fungal isolation and identification
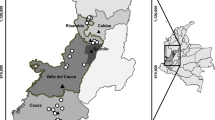
Most eucalypt plants with symptoms consistent with pestalotioid fungi that were analysed were very young plants of E. globulus (Fig. 1). Symptoms were typically found in one of three situations: (1) in the nursery, sometimes associated with intense mortality in early stages of plant production; (2) in forest yards, where young eucalypts awaited plantation; or (3) shortly after plantation, usually within a few months. In forest yards and plantations, mortality was variable, ranging from a few plants to 30–40%. In most cases, sporulation was observed on the lesions especially on the nursery plants. In total, 27 isolates were identified from their morphology as belonging to the genus Neopestalotiopsis. From these, only 23 were included in the phylogenetic analysis due to the failure of amplification of the β-tubulin gene.
Phylogenetic analysis
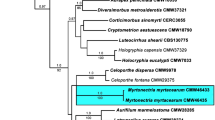
The concatenated dataset of 100 ingroup taxa (representing 56 species) and 1 outgroup taxon consisted of 1512 characters including alignment gaps (527 for ITS, 550 for TEF1 and 435 for TUB2). Of these characters, 1037 were constant, and 242 variable characters were parsimony uninformative. MP analysis of the remaining 233 parsimony informative characters resulted in 1000 equally most parsimonious trees of 940 steps (CI = 0.661, RI = 0.742, HI = 0.339). Topology of the trees generated by MP and BI (Treebase study TB2: S28369) did not differ significantly from the tree generated by ML (Fig. 2, final ML optimization likelihood = − 7379.899056).
Consensus phylogram of 1000 trees resulting from a RAxML analysis of the (ITS + TUB2 + TEF1) alignment of the analysed Neopestalotiopsis sequences. RAxML bootstrap support values (MLB), maximum parsimony bootstrap supports (MPB) and Bayesian posterior probabilities are given at the nodes (MLB/MPB/BPP). Strains sequenced in this study are in bold and T denotes ex-type strains. Scale bar corresponds to 0.02 substitutions per sites. The tree is rooted with Pestalotiopsis diversiseta MFLUCC 12–0287
Of the 22 isolates from E. globulus and one from E. nitens, isolates MEAN 1328 and MEAN 1329 grouped with N. zimbabwana, in a clade containing N. hadrolaeliae and N. honoluluana (Fig. 2). Isolates MEAN 1325, MEAN 1327 and MEAN 1228 grouped close to N. rosae and N. javensis in an unresolved clade. The remaining eighteen isolates grouped in five well-resolved clades and are here described as new species.
Taxonomy
Neopestalotiopsis eucalyptorum E. Diogo, M. H. Bragança & A. J. L. Phillips sp. nov., Figure 3
MycoBank: MB839457.
Holotype: LISE 96318, (dried culture), ex-type culture MEAN 1308 = CBS 147684.
Etymology: Named after Eucalyptus, the host genus from which it was isolated.
Associated with leaf necrosis and stem basal cankers of Eucalyptus globulus young plants. Sexual state not observed. Asexual state: Conidiomata in culture on PDA pycnidial, globose, black, up to 470 µm diam., covered with white mycelium, aggregated or discrete and scattered, semi-immersed, exuding black globose, glistening, conidial masses. Conidiophores indistinct and reduced to conidiogenous cells. Conidiogenous cells discrete, ampulliform, mostly cylindrical to subcylindrical, smooth, thin-walled, simple, 7.1–20 × 2.4–5.8 µm, apex 1.7–2.6 µm diameter. Conidia fusoid to ellipsoid, straight or slightly curved, 4 septate, (22.6) 27.5–29.2 (33.2) × (6.4) 7.6–8.1(9.5) µm, \(\overline{x }\) ± SD = 28.3 ± 2.4 × 7.9 ± 0.7 µm; basal cell obconic to subcylindrical with a truncate base, hyaline to pale brown, smooth, and thin-walled, 3.2–7.3 µm long, three median cells doliiform to subcylindrical, (15.1) 17.6–18.6 (22.1) µm long, \(\overline{x }\) ± SD = 18.1 ± 1.5 µm, smooth-walled, versicoloured, septa darker than the rest of cell, second cell from the base pale brown, 5.1–8.6 µm long, third cell honey brown 4.6–6.8 µm long, fourth cell honey brown to brown, 4.6–7.7 µm long, apical cell 3.9–7.2 µm long, hyaline, subcylindrical to conical, smooth, thin-walled, with 2–4 apical appendages (mostly 3) arising from the apical crest, unbranched, filiform, flexuous, (12.7)16.2–18.8(27.7) µm long \(\overline{x }\) ± SD = 17.5 ± 3.7 µm; basal appendage single, filiform, unbranched, centric, 3.4–8.1 µm long.
Culture characteristics: Colony on PDA attaining 76 mm diameter after 7 days at 25 °C with 12 h of near UV light per day, with undulate edge, pale rosy buff with fluffy white aerial mycelium, conidiomata scattered, isolated. Reverse pale buff.
Habitat: On leaves and stems of Eucalyptus globulus.
Distribution: Fundão, Guarda, Portugal.
Specimens examined: PORTUGAL, Fundão, on Eucalyptus globulus, March 2016, leg. C. Valente, holotype LISE 96318, culture ex-type MEAN 1308 = CBS 147684; Guarda, Rochoso, on Eucalyptus globulus, September 2013, leg. E. Diogo, culture MEAN 1309 = CBS 147685; Pegões, on Eucalyptus globulus, April 2013, leg. C. Valente, cultures MEAN 1322, MEAN 1323; Tondela, on Eucalyptus globulus, June 2014, leg. C. Valente, culture MEAN 1324; Paredes de Coura, on Eucalyptus globulus, March 2016, leg. C. Valente, culture MEAN 1326.
Notes: Neopestalotiopsis eucalyptorum is phylogenetically closely related to, but distinct from N. foedans and can be distinguished by its larger conidia (N. eucalyptorum (22.6)27.5–29.2(33.2) × (6.4)7.6–8.1(9.5) and N. foedans 19–24 × 5.7–6.9 (\(\overline{x }\)= 20.7 × 6.4). Neopestalotiopsis foedans has a wide host range (Maharachchikumbura et al. 2012) but to the best or our knowledge was never isolated from Eucalyptus species. Blast searches in GenBank restricted to type species showed that N. eucalyptorum differs from N. foedans in ITS (NR_111785; identities 463/467 (99%), no gaps); TUB2 (JX399022; identities 414/417 (99%), no gaps) and TEF1 (JX399053; identities 467/482 (97%), 5 gaps (1%)).
Neopestalotiopsis hispanica E. Diogo, M. H. Bragança & A.J.L.Phillips sp. nov., Fig. 4
MycoBank MB839458.
Holotype: LISE 96319 (dried culture), ex-type culture MEAN 1310 = CBS 147686.
Etymology: Named after Hispania, the old roman name for the Iberian Peninsula.
Associated with leaves and stem necrosis of Eucalyptus globulus young plants and nursery plants for planting. Sexual state unknown. Asexual state: Conidiomata in culture on PDA pycnidial, globose, black, up to 690 µm, aggregated or discrete and scattered, semi-immersed, exuding black globose, glistening, conidial masses. Conidiophores indistinct and reduced to conidiogenous cells. Conidiogenous cells discrete, proliferating 1–2 times percurrently, collarete inconspicuous, cylindrical to subcylindrical, smooth, thin-walled, simple, 7–14.7 × 2.4–4.1 µm, apex 1.5–2.1 µm diameter. Conidia fusoid to ellipsoid, straight, 4 septate, (21.5)24.4–25.3(27.1) × (5.9)7.2–7.8(8.7) µm, \(\overline{x }\) ± SD = 24.9 ± 1.3 × 7.5 ± 0.8 µm; basal cell obconic to subcylindrical with a truncate base, hyaline, smooth, and thin-walled, 3.3–5.5 µm long, three median cells doliiform, (13.5)15.8–16.5(17.8) µm long, \(\overline{x }\) ± SD = 16.2 ± 1.0, wall smooth, versicoloured, septa darker than the cell, second cell from the base pale brown, 4.8–6.8 µm long, third cell honey brown 4.2–6.1 µm long, fourth cell brown to pale brown, 3.7–6.5 µm long, apical cell 2.8–6.2 µm long, hyaline, conical, smooth and thin-walled with 3–4 apical appendages arising from the apical crest, unbranched, filiform, flexuous, (30.7)19.5–22.6(30.7) µm long \(\overline{x }\) ± SD = 21.0 ± 4.4 µm; basal appendage single, filiform, unbranched, centric, 5.1–15.5 µm long.
Culture characteristics: Colony on PDA attaining 74 mm diameter after 7 days at 25 °C, with smooth to slightly undulate edge, whitish with sparse aerial mycelium. Conidiomata scattered, isolated with concentric rings of more abundant conidiomata. Reverse pale buff to ochreous.
Habitat: On leaves and stems of Eucalyptus globulus.
Distribution: Fundão, Portugal and an unknown location in Spain.
Specimens examined: Portugal, Fundão, on one year old plants, March 2016, leg. C. Valente, holotype LISE 96319, culture ex-type MEAN 1310 = CBS 147686; idem MEAN 1311; Spain, unknown location, on imported plants for plantation, May 2018, leg. C. Valente, culture MEAN 1312 = CBS 147687.
Notes: N. hispanica formed a sister clade with N. longiappendiculata sp. nov. These two species are easily distinguished because N. longiappendiculata has very long apical appendages (Table 2). Alignments of the sequences of these two species showed that ITS are identical, in TUB2 differs in 2 b.p. and in TEF1 differs in 15 b.p. with 5 gaps.
Neopestalotiopsis iberica E. Diogo, M. H. Bragança & A. J. L. Phillips sp. nov., Figure 5
MycoBank: MB839459.
Holotype: LISE 96320 (dried culture), ex-type culture MEAN 1313 = CBS 147688.
Etymology: Named after the region where it was found, the Iberian Peninsula.
Associated with leaves and stem necrosis of Eucalyptus globulus seedlings. Sexual state unknown. Asexual state: Conidiomata in culture on PDA picnidial, globose, black, up to 540 µm, aggregated or scattered, semi-immersed, exuding black, globose, glistening, conidial masses. Conidiophores indistinct and reduced to the conidiogenous cell. Conidiogenous cells discrete, ampulliform to lageniform, smooth, thin-walled, simple, 6.9–10.9 × 1.9–5.6 µm, apex 1.7–2.5 µm diameter. Conidia ellipsoid to obovoid, straight, 4 septate, (21.4) 22.9–24.1 (29.4) × (7.2) 8.2–8.7(9.8) µm, \(\overline{x }\) ± SD = 23.5 ± 1.6 × 8.4 ± 0.7 µm; basal cell obconic to subcylindrical with a truncate base, hyaline to pale brown, smooth, and thin-walled, 3.4–5.6 µm long, three median cells doliiform, (13)14.8–15.5(17.7) µm long, \(\overline{x }\) ± SD = 15.1 ± 0.9, smooth-walled, versicoloured, third septa from the base darker than the cell, second cell from the base pale brown, 3.9–6.7 µm long, third cell honey brown 3.7–6.2 µm long, fourth cell honey brown to brown, 4.5–6.6 µm long, apical cell 3.4–6.2 µm long, hyaline, subcylindrical to conical, smooth and thin-walled; with 3, rarely 2 apical appendages arising from the apical crest, unbranched, filiform, flexuous, (13)18.2–20.3(24.6) µm long \(\overline{x }\) ± SD = 19.3 ± 3 µm; basal appendage single, filiform, unbranched, centric, 3.1–8.8 µm long.
Culture characteristics: Colony on PDA attaining 78 mm diameter after 7 days at 25 °C, with smooth to slightly undulate edge, pale rosy-buff with fluffy white aerial mycelium; conidiomata scattered, isolated with concentric rings of more abundant conidiomata. Reverse pale ochreous.
Habitat: On leaves and stems of Eucalyptus globulus.
Distribution: Pegões, Portugal and an unknown location in Spain.
Specimens examined: PORTUGAL, Pegões, on nursery plants, June 2017, leg. C. Valente, holotype LISE 96320, ex-type culture MEAN 1313 = CBS 147688; SPAIN, unknown location, on nursery plants, May 2018, leg. C. Valente, culture MEAN 1314 = CBS 147689.
Notes: Neopestalotiopsis iberica is phylogenetically related to N. autralis and N. vitis. These three species are morphologically similar, but N. iberica has shorter apical appendages (N. iberica (11)19.5–22.6(30.7); N. australis (19)21–32(34); N. vitis (14)19–38.6(43)) and a longer single basal appendage (N. iberica 7.5–9.1; N. australis 3–7; N. vitis 2.2–7.2), while N. vitis has one or two basal appendages (Jayawardena et al. 2016, Table 2). Blast searches in GenBank restricted to type species showed that N. iberica differs from N. australis in ITS (KM199348; identities 494/497(99%), 2 gaps (0%)); TUB2 (KM199432; identities 401/405 (99%), no gaps) and TEF1 (KM199537; identities 458/466 (98%), no gaps) and differs from N. vitis ITS (KU140694; identities 419/421(99%), 1 gap (0%)); TUB2 (KU140685; identities 316/319 (99%), no gaps) and TEF1 (KU1406767; identities 370/373 (99%), no gaps).
Neopestalotiopsis longiappendiculata E. Diogo, M. H. Bragança & A.J.L.Phillips sp. nov., Figure 6
MycoBank: MB839460.
Holotype: LISE 96321 (dried culture), ex-type culture MEAN 1315 = CBS 147690.
Etymology: The name reflects the long apical appendages.
Associated with leaves and stem necrosis of Eucalyptus globulus plantlets. Sexual state unknown. Asexual state: Conidiomata in water agar with sterile eucalyptus leaf pieces pycnidial, globose, black, up to 490 µm, scattered, semi-immersed, exuding black globose, glistening, conidial masses. Conidiophores indistinct and reduced to conidiogenous cells. Conidiogenous cells discrete, ampulliform or subcylindrical, smooth, thin-walled, simple, 5.5–16.7 × 2.3–8.4 µm, apex 1.7–3.0 µm diameter. Conidia fusoid to ellipsoid, straight or slightly curved, 4 septate, (20.5)22.4–23.4(26.1) × (5.7)7–7.8(9.6) µm, \(\overline{x }\) ± SD = 22.9 ± 1.5 × 7.4 ± 1.0 µm; basal cell obconic to subcylindrical with a truncate base, hyaline to pale brown, smooth, and thin-walled, 2.8–4.8 µm long, three median cells doliiform, (13.1)14.5–15.2(17.8) µm long, \(\overline{x }\) ± SD = 14.8 ± 1, wall smooth, versicoloured, third septa from the base darker than the cell, second cell from the base pale brown, 4.1–5.9 µm long, third cell honey brown 4.4–6.0 µm long, fourth cell honey brown to brown, 4.2–6.6 µm long, apical cell 3.0–5.0 µm long, hyaline, subcylindrical to conical, smooth and thin-walled; with 2–4, mostly 3 apical appendages arising from the apical crest, unbranched, filiform, flexuous, (17.1)25.7–30.2(42.7) µm long \(\overline{x }\) ± SD = 28 ± 6.3 µm; basal appendage single, filiform, unbranched, centric, 3.3–11.6 µm long.
Culture characteristics: Colony on PDA attaining 80 mm diameter after 7 days at 25 °C, with smooth to slightly undulate edge, pale rosy-buff with sparse aerial mycelium. Conidiomata very rare, scattered. Reverse pale buff.
Habitat: On leaves and stems of Eucalyptus globulus and E. nitens.
Distribution: Furadouro, Paredes de Coura, Portugal.
Specimens examined: PORTUGAL, Furadouro, on Eucalyptus globulus nursery plants, November 2017, leg. A. Reis, holotype LISE 96321, ex-type culture MEAN 1315 = CBS 147690; Paredes de Coura, on young plants of Eucalyptus nitens, March 2016, leg. C. Valente, culture MEAN 1316 = CBS 147691.
Notes: See notes under N. hispanica.
Neopestalotiopsis lusitanica E. Diogo, M. H. Bragança & A. J. L. Phillips sp. nov., Figure 7
MycoBank: MB839461.
Holotype: LISE 96322 (dried culture), ex-type culture MEAN 1317 = CBS 147692. Etymology: Named after the ancient Roman province of Lusitania that later became Portugal, the country where it was first detected.
Associated with leaf and stem necrosis of Eucalyptus globulus seedlings. Sexual state not observed. Asexual state: Conidiomata in culture on PDA pycnidial, globose, black, covered by white mycelia, up to 580 µm, aggregated or scattered, semi-immersed, exuding black globose, glistening, conidial masses. Conidiophores indistinct and reduced to conidiogenous cells. Conidiogenous cells discrete, ampulliform to lageniform, rarely cylindrical to subcylindrical, smooth, thin-walled, simple, 5.2–13.7 × 2.5 5.7 µm, apex 1.6–2.9 µm diameter. Conidia fusoid to ellipsoid, straight, rarely slightly curved, 4 septate, (27.5)30.6–32.5(38.7) × (8.2)9.6–10.3(12.5) µm, \(\overline{x }\)± SD = 31.6 ± 2.6 × 9.9 ± 1.0 µm; basal cell obconic with a truncate base, hyaline to very pale brown, smooth, and thin-walled, 4.5–7.9 µm long, three median cells doliiform, (17)18.9–20.4(24.1) µm long, \(\overline{x }\)± SD = 19.6 ± 2.1, wall smooth and slightly thick-walled, sometimes very slightly constricted at the septa, versicoloured, septa darker than the cell, second cell from the base pale brown, 5.4–9.6 µm long, third cell honey brown 4.7–7.6 µm long, fourth cell honey brown to brown, 4.7–7.6 µm long, apical cell 3.2–7.6 µm long, hyaline, conical to subcylindrical, rugose and thin-walled; with 3–4 apical appendages, rarely 2, arising from the apical crest, unbranched, filiform, flexuous, (8.2)12.5–14.7(19.7) µm long \(\overline{x }\) ± SD = 13.6 ± 3.1 µm; basal appendage single, filiform, unbranched, centric, 3.8–15.0 µm long.
Culture characteristics: Colony on PDA attaining 85 mm diameter after 7 days at 25 °C with 12 h of near UV light, with smooth to slightly undulate edge, pale rosy-buff with sparse white aerial mycelium. Conidiomata scattered, isolated or aggregated with concentric rings of more abundant conidiomata. Reverse pale buff.
Habitat: Associated with leaves and stem necrosis of Eucalyptus globulus.
Distribution: Pegões, Portugal.
Specimens examined: PORTUGAL, Pegões, on Eucalyptus globulus nursery plants, June 2017, leg. C. Valente, holotype LISE 96322, ex-type culture MEAN 1317 = CBS 147692; Pegões, on Eucalyptus globulus nursery plants, December 2012, leg. C. Valente, culture MEAN 1318 = CBS 147692, MEAN 1319; Pegões, on Eucalyptus globulus nursery plants, March 2013, leg. C. Valente, culture MEAN 1321.
Notes: Neopestalotiopsis lusitanica forms a well-resolved and well-supported clade (Fig. 2), sister to a large clade containing N. brasiliense, N. macadamiae, N. asiatica, N. umbrinospora, N. chrysea and N. surimanemsis. However, it can be distinguished from all these species by the larger conidia and longer basal appendage (Table 2). Blast searches in GenBank restricted to type species showed that N. lusitanica differs from N. brasiliensis in ITS (MG686469; identities 449/452(99%), no gaps); TUB2 (MG692400; identities 383/385(99%), no gaps) and TEF1 (MG692402; identities 455/474(96%), 2 gaps(0%)); from N. asiatica in ITS (NR_120181; identities 482/484 (99%), 2 gaps (0%)); TUB2 (JX399018; identities 399/403 (99%)), no gaps) and TEF1 (JX399049; identities 457/477 (96%), 7 gaps(1%)); from N. umbrinospora in ITS (NR_111783; identities 482/484 (99%), 2 gaps (0%)); TUB2 (JX399019; identities 398/403 (99%)), no gaps) and TEF1 (JX399050; identities 458/474 (97%), 2 gaps (0%)); from N. chrysea in ITS (NR_111784; identities 482/484 (99%), 2 gaps (0%)); TUB2 (JX399020.; identities 398/403 (99%)), no gaps) and TEF1 (JX399051; identities 456/474 (96%), 2 gaps(0%)) and from N. surinamensis TUB2 (KM199465; identities 388/390 (99%), no gaps) and TEF1 (KM199518; identities 442/455 (97%), 2 gaps (0%)); from N. macadamiae in ITS (NR_161002; identities 457/463 (99%), no gaps); TUB2 (KX186654; identities 390/395 (99%), no gaps) and TEF1 (9 pb difference and two gaps, verified in an 213 pb alignment with MEAN 1317, since the TEF1 sequence of the type species of N. macadamiae is very short and is not available for blast searches in Genbank).
Discussion
In this study, twenty-three isolates of Neopestalotiopsis species derived from diseased young plants of E. globulus and E. nitens were characterised in terms of morphology and phylogeny based on combined ITS, TEF1 and TUB2 sequence data. Two isolates clustered with the N. hadrolaeliae/N. honoluluana/N. zimbabwana complex, and four clustered in a poorly resolved clade sister to N. eucalyptorum. Fourteen isolates were resolved into five well-supported clades separate from all other species in Neopestalotiopsis and were described and introduced as new species. These are the first reports of Neopestalotiopsis species associated with Eucalyptus in Portugal.
Morphological identification of pestalotioid species is unreliable due to overlapping morphological characteristics used to define species and the plasticity of these characters that change with culture conditions, age of cultures and subculturing (Hu et al. 2007). In a phylogenetic study of diversity of endophytic Pestalotiopsis species obtained from Pinus armandii and Ribes sp., Hu et al. (2007) found that ITS was not sufficiently informative to segregate species in this genus and proposed β-tubulin as an additional gene. Later, Maharachchikumbura et al. (2012) tested ten different genes (act, cal, gs, gapdh, ITS, LSU, 18 S nrDNA, rpb1, TEF1 and TUB2) and selected ITS, TEF1 and TUB2 as the most suitable to resolve species in Pestalotiopsis.
Neopestalotiopsis was introduced by Maharachchikumbura et al. (2014b) who segregated Pestalotiopsis into three genera, namely, Pestalotiopsis, Neopestalotiopsis and Pseudopestalotiopsis, based on morphology and phylogenetic analysis. Accordingly, Neopestalotiopsis can be distinguished from Pestalotiopsis s.s. by versicoloured conidial cells and indistinct conidiophores that are often reduced to the conidiogenous cells; Pestalotiopsis species have concolourous conidial cells and conspicuous conidiophores, while Pseudopestalotiopsis species have darker concolourous median cells and indistinct conidiophores. Morphologically, all the strains isolated in this study fit within Neopestalotiopsis, having versicoloured conidial cells and indistinct conidiophores.
Following the work of Maharachchikumbura et al. (2014b), phylogenetic analyses of Neopestalotiopsis have been based on combined ITS, TEF1 and TUB2, and numerous species have been introduced in the genus (see Table 1). In our analysis, not all strains could be identified, but five clades received good support and were considered to represent previously unknown species that are introduced here as new species. Two isolates clustered in a clade with three known species that are not well-resolved. For this reason, we decided not to give them a name until that clade has been resolved. This is in accordance with recent studies where poor resolution was obtained for some species in Neopestalotiopsis (Belisário et al. 2020; Gerardo-Lugo et al. 2020; Liu et al. 2017).
Pestalotiopsis s. l. is generally considered to be endophytes or weak pathogens causing disease only when the plants are under stress (Maharachchikumbura et al. 2011). However, a new Pestalotiopsis species, P. pini, was reported recently in Portugal causing shoot blight and trunk necrosis on Pinus pinea (Silva et al. 2020). Furthermore, a common disease in nurseries in Brazil has been attributed to an unidentified species of Pestalotiopsis. It was regarded as a minor disease associated with the conditions required for the propagation process. This includes wounds, high humidity, and long periods of leaf wetness (Alfenas et al. 2009; Santos et al. 2020). Therefore, this species has been regarded as a secondary and opportunistic pathogen. Recently, Belisário et al. (2020) demonstrated that N. rosae and N. protearum as well as other unidentified species can infect unwounded leaves of Eucalyptus under long periods of leaf wetness. They also tested the pathogenicity of six other species from different hosts and found that all tested species caused similar symptoms. Santos et al. (2020) also identified N. rosae, N. australis and N. eucalypti causing disease in old cuttings in conditions less favourable to infection. On the pathogenicity tests, in addition to the injury on the cutting stem at the inoculation point, Santos et al. (2020) observed the presence of the fungus in the leaf petiole above the inoculation point, suggesting that the fungus may migrate through the vascular system of the plant and cause lesions at points distant from the site of inoculation. In the present study, we also found lesions in plants recently transplanted into the field suggesting that the fungus may remain latent in the nursery plants but then cause symptoms during the transplantation stress. It is well-known that some fungi that are considered endophytic may became pathogenic when their hosts became stressed (Stergiopoulos and Gordon 2014). Therefore, it is important to monitor and control the presence of these fungi in nurseries.
Although pathogenicity of the species described in this study has not been determined, according to pathogenicity tests previously done on Eucalyptus (Belisário et al. 2020; Santos et al. 2020), several species of Neopestalotiopsis proved to be pathogenic and cannot be regarded simply as endophytes or opportunistic pathogens. Since all isolates obtained in this study were obtained from diseases plants, it likely that they are pathogenic. In addition, several recently described Neopestalotiopsis species have been associated with plant diseases. Solarte el al. (2018) showed that guava scab in Colombia is caused by several species of Pestalotiopsis and Neopestalotiopsis. Neopestalotiopsis rosicola causes rose stem canker in China (Jiang et al. 2018), and Neopestalotiopsis vitis is responsible for leaf spots of Vitis vinifera also in China (Jayawardena et al. 2016). Various Neopestalotiopsis species were also isolated in association with grapevine trunk diseases in France (Maharachchikumbura et al. 2016). Neopestalotiopsis macadamiae causes dry flower disease of Macadamia in Australia (Akinsanmi et al. 2017). Chen et al. (2018) reported that Neopestalotiopsis clavispora and other pestalotioid species cause grey blight disease on Camellia sinensis in China. Furthermore, other pestalotioid fungi (e.g., Pestalotiopsis biciliata) have already been proven to be pathogenic on Eucalyptus species (Morales-Rodríguez et al. 2019).
Pathogenicity, host specificity and geographic distribution of the new species remain unknown, and these are issues that should be considered in future studies. To our knowledge this is the first report of Neopestalotiopsis species causing leaf and stem necrosis on Eucalyptus globulus and E. nitens in Portugal.
Data availability
Strains are available at MEAN (https://www.iniav.pt/recursos-geneticos-microbianos) and CBS (https://wi.knaw.nl/page/Collection) culture collections. Sequences are available at GenBank (https://www.ncbi.nlm.nih.gov/genbank) and alignments and trees are deposited in TreeBase (https://treebase.org).
References
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Automat Contr 19:716–723. https://doi.org/10.1109/TAC.1974.1100705
Akinsanmi OA, Nisa S, Jeff-Ego OS et al (2017) Dry flower disease of macadamia in australia caused by Neopestalotiopsis macadamiae sp. nov. and Pestalotiopsis macadamiae sp. nov. Plant Dis 101:45–53. https://doi.org/10.1094/PDIS-05-16-0630-RE
Alfenas AC, Zauza EAV, Mafia RG, Assis TF (2009) Clonagem e doenças do eucalipto, 2nd edition. Editora UFV, Viçosa
Almeida FA, Araújo E, Gonçalves Junior H et al (2003) Diagnóstico e quantificação de doenças fúngicas da acerola no Estado da Paraíba. Fitopatol Bras 28:176–179. https://doi.org/10.1590/s0100-41582003000200010
Ayoubi N, Soleimani Pari S (2016) Morphological and molecular identification of Neopestalotiopsis mesopotamica causing tomato fruit rot. J Plant Dis Prot 123:267–271. https://doi.org/10.1007/s41348-016-0042-z
Bakry M, Bussières G, Lamhamedi MS et al (2011) A first record of Pestalotiopsis clavispora in argan mass cutting propagation: prevalence, prevention and consequences for plant production. Phytoprotection 90:117–120. https://doi.org/10.7202/045780ar
Barradas C, Phillips AJL, Correia A et al (2016) Diversity and potential impact of Botryosphaeriaceae species associated with Eucalyptus globulus plantations in Portugal. Eur J Plant Pathol 146:1–13. https://doi.org/10.1007/s10658-016-0910-1
Belisário R, Aucique-Pérez CE, Abreu LM et al (2020) Infection by Neopestalotiopsis spp. occurs on unwounded eucalyptus leaves and is favoured by long periods of leaf wetness. Plant Pathol 69:194–204. https://doi.org/10.1111/ppa.13132
Bezerra JDP, Machado AR, Firmino AL et al (2018) Mycological diversity description I. Acta Bot Brasilica 32:656–666. https://doi.org/10.1590/0102-33062018abb0154
Bragança H, Simões S, Onofre N et al (2007) Cryphonectria parasitica in Portugal: diversity of vegetative compatibility types, mating types, and occurrence of hypovirulence. For Pathol 37:391–402. https://doi.org/10.1111/j.1439-0329.2007.00513.x
Bragança H, Diogo ELF, Neves L et al (2016) Quambalaria eucalypti a pathogen of Eucalyptus globulus newly reported in Portugal and in Europe. For Pathol 46:67–75. https://doi.org/10.1111/efp.12221
Branco M, Bragança H, Sousa E, Phillips AJL (2014) Pests and diseases in Portuguese forestry: current and new threats. In: Reboredo F (ed) Forest Context and Policies in Portugal-Present and Future Challenges World Forests 19. Springer International Publishing Switzerland, Switzerland, pp 117–154
Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous fungi. Mycologia 91:553–556
Chen Y, Zeng L, Shu N et al (2018) Pestalotiopsis-like species causing gray blight disease on Camellia sinensis in China. Plant Dis 102:1–28. https://doi.org/10.1094/PDIS-05-17-0642-RE
Conforto C, Lima NB, Silva FJA et al (2019) Characterization of fungal species associated with cladode brown spot on Nopalea cochenillifera in Brazil. Eur J Plant Pathol 155:1179–1194. https://doi.org/10.1007/s10658-019-01847-3
Crous PW, Gams W, Stalpers JA et al (2004) MycoBank: an online initiative to launch mycology into the 21st century. Stud Mycol 50:19–22 (https://doi.org/citeulike-article-id:9861703)
Crous PW, Summerell BA, Swart L et al (2011) Fungal pathogens of Proteaceae. Persoonia 27:20–45. https://doi.org/10.3767/003158511X606239
Crous PW, Wingfield MJ, Le RJJ et al (2015) Fungal planet description sheets: 371–399. Persoonia 35:264–327
Crous PW, Wingfield MJ, Chooi Y-H et al (2020) Fungal planet description sheets: 1042–1111. Persoonia - Mol Phylogeny Evol Fungi 44:301–459. https://doi.org/10.3767/persoonia.2020.44.11
Diogo ELF, Santos JM, Phillips AJL (2010) Phylogeny, morphology and pathogenicity of Diaporthe and Phomopsis species on almond in Portugal. Fungal Divers 44:107–115. https://doi.org/10.1007/s13225-010-0057-x
Espinoza JG, Briceño EX, Keith LM, Latorre BA (2008) Canker and twig dieback of blueberry caused by Pestalotiopsis spp. and a Truncatella sp. in Chile. Plant Dis 92:1407–1414. https://doi.org/10.1094/PDIS-92-10-1407
FAO (2001) Mean annual volume increment of selected industrial forest & plantation species by L. Ugalde & D. Pérez, forest plantation thematic papers, Working Paper 1. Forest Resources Development Service, Forest Resources Division. FAO, Rome, 27 pp. http://www.fao.org/3/a-ac121e.pdf. Accessed 8 Feb 2021
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution (NY) 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Ferreira MC, Ferreira GW, Fonseca N (1994). Manual de Sanidade dos Viveiros Florestais, IEADR, Lisboa
Freitas EFS, Da Silva M, Barros MVP, Kasuya MCM (2019) Neopestalotiopsis hadrolaeliae sp. nov., a new endophytic species from the roots of the endangered orchid Hadrolaelia jongheana in Brazil. Phytotaxa 416:211–220. https://doi.org/10.11646/phytotaxa.416.3.2
Gerardo-Lugo SS, Tovar-Pedraza JM, Maharachchikumbura SSN et al (2020) Characterization of Neopestalotiopsis species associated with mango grey leaf spot disease in Sinaloa, Mexico. Pathogens 9:1–17. https://doi.org/10.3390/pathogens9100788
Glass NL, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330
Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174. https://doi.org/10.1007/BF02101694
ICNF (2019) 6,º Inventário Florestal Nacional (IFN6) - 2015 Relatório Final. Instituto de Conservação da Natureza e Florestas, Lisboa. www2.icnf.pt/portal/florestas/ifn/ifn6. Accessed 16 Mar 2021
Hopkins KE, McQuilken MP (2000) Characteristics of Pestalotiopsis associated with hardy ornamental plants in the UK. Eur J Plant Pathol 106:77–85. https://doi.org/10.1023/A:1008776611306
Hu H, Jeewon R, Zhou D et al (2007) Phylogenetic diversity of endophytic Pestalotiopsis species in Pinus armandii and Ribes spp.: evidence from rDNA and β-tubulin gene phylogenies. Fungal Divers 24:1–22
Huannaluek N, Jayawardena RS, Maharachchikumbura SSN, Harishchandra DL (2021) Additions to pestalotioid fungi in Thailand: Neopestalotiopsis hydeana sp nov Pestalotiopsis hydei sp. nov. Phytotaxa 479:23–43. https://doi.org/10.11646/phytotaxa.479.1.2
Ismail AM, Cirvilleri G, Polizzi G (2013) Characterisation and pathogenicity of Pestalotiopsis uvicola and Pestalotiopsis clavispora causing grey leaf spot of mango (Mangifera indica L.) in Italy. Eur J Plant Pathol 135:619–625. https://doi.org/10.1007/s10658-012-0117-z
Jayawardena RS, Liu M, Maharachchikumbura SSN et al (2016) Neopestalotiopsis vitis sp. nov. causing grapevine leaf spot in China. Phytotaxa 258:63–74. https://doi.org/10.11646/phytotaxa.258.1.4
Jayawardena RS, Hyde KD, Jeewon R et al (2019) One stop shop II: taxonomic update with molecular phylogeny for important phytopathogenic genera: 26–50 (2019). Fungal Divers 94:41–129. https://doi.org/10.1007/s13225-019-00418-5
Jiang N, Bonthond G, Fan XL et al (2018) Neopestalotiopsis rosicola sp. nov. causing stem canker of Rosa chinensis in China. Mycotaxon 133:271–283. https://doi.org/10.5248/133.271
Jiang N, Fan X, Tian C (2021) Identification and characterization of leaf-inhabiting fungi from Castanea plantations in China. J Fungi 7:64. https://doi.org/10.3390/jof7010064
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. https://doi.org/10.1093/bib/bbx108
Kumar V, Cheewangkoon R, Gentekaki E et al (2019) Neopestalotiopsis alpapicalis sp. Nov. a new endophyte from tropical mangrove trees in Krabi province (Thailand). Phytotaxa 393:251–262. https://doi.org/10.11646/phytotaxa.393.3.2
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Liu F, Hou L, Raza M, Cai L (2017) Pestalotiopsis and allied genera from Camellia, with description of 11 new species from China. Sci Rep 7:866. https://doi.org/10.1038/s41598-017-00972-5
Liu F, Bonthond G, Groenewald JZ et al (2019) Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Stud Mycol 92:287–415. https://doi.org/10.1016/j.simyco.2018.11.001
Lin HF, Chen TH, Da LS (2011) The antifungal mechanism of Bacillus subtilis against Pestalotiopsis eugeniae and its development for commercial applications against wax apple infection. African J Microbiol Res 5:1723–1728. https://doi.org/10.5897/AJMR10.169
Ma X-Y, Maharachchikumbura SSNN, Chen B-W et al (2019) Endophytic pestalotiod taxa in Dendrobium orchids. Phytotaxa 419:268–286. https://doi.org/10.11646/phytotaxa.419.3.2
Maharachchikumbura SSN, Guo L-D, Chukeatirote E et al (2011) Pestalotiopsis —morphology, phylogeny, biochemistry and diversity. Fungal Divers 50:167–187. https://doi.org/10.1007/s13225-011-0125-x
Maharachchikumbura SSN, Guo L-DD, Cai L et al (2012) A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Divers 56:95–129. https://doi.org/10.1007/s13225-012-0198-1
Maharachchikumbura SSN, Guo L-D, Chukeatirote E, Hyde KD (2014a) Improving the backbone tree for the genus Pestalotiopsis; addition of P. steyaertii and P. magna sp. nov. Mycol Prog 13:617–624. https://doi.org/10.1007/s11557-013-0944-0
Maharachchikumbura SSN, Hyde KD, Groenewald JZ et al (2014b) Pestalotiopsis revisited. Stud Mycol 79:121–186. https://doi.org/10.1016/j.simyco.2014.09.005
Maharachchikumbura SSN, Larignon P, Hyde KD et al (2016) Characterization of Neopestalotiopsis, Pestalotiopsis and Truncatella species associated with grapevine trunk diseases in France. Phytopathol Mediterr 55:380–390
Metz AM, Haddad A, Worapong J et al (2000) Induction of the sexual stage of Pestalotiopsis microspora, a taxol-producing fungus. Microbiology 146(Pt 8):2079–2089
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE). IEEE, New Orleans, pp 1–8. https://doi.org/10.1109/GCE.2010.5676129
Mishra Y, Singh A, Batra A, Sharma MM (2014) Understanding the biodiversity and biological applications of endophytic fungi: a review. J Microb Biochem Technol s8:1–11. https://doi.org/10.4172/1948-5948.S8-004
Morales-Rodríguez C, Dalla Valle M, Aleandri MP, Vannini A (2019) Pestalotiopsis biciliata, a new leaf pathogen of Eucalyptus spp. recorded in Italy. For Pathol 49:1–7. https://doi.org/10.1111/efp.12492
Norphanphoun C, Jayawardena RS, Chen Y et al (2019) Morphological and phylogenetic characterization of novel pestalotioid species associated with mangroves in Thailand. Mycosphere 10:531–578. https://doi.org/10.5943/mycosphere/10/1/9
Nylander JAA (2004) MrModeltest v2. program distributed by the author. Evolutionary Biology Centre, Uppsala University. https://www.abc.se/_nylander/mrmodeltest2/mrmodeltest2.html
O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci 95:2044–2049. https://doi.org/10.1073/pnas.95.5.2044
Old KM, Lee SS, Sharma JK, Yuan ZQ (2000) A manual of diseases of tropical acacias in Australia, South-East Asia and India. Center for International Forestry Research (CIFOR). Jakarta, Indonesia
Pirralho M, Flores D, Sousa VB et al (2014) Evaluation on paper making potential of nine Eucalyptus species based on wood anatomical features. Ind Crops Prod 54:327–334. https://doi.org/10.1016/j.indcrop.2014.01.040
Qi M, Xie C-X, Chen Q-W, Yu Z-D (2021) Pestalotiopsis trachicarpicola, a novel pathogen causes twig blight of Pinus bungeana (Pinaceae: Pinoideae) in China. Antonie Van Leeuwenhoek 114:1–9. https://doi.org/10.1007/s10482-020-01500-8
Rannala B, Yang Z (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol 43:304–311. https://doi.org/10.1007/BF02338839
Rayner RW (1970) A mycological color chart. Commonwealth Mycological Institute, Kew
Ronquist F, Teslenko M, van der Mark P et al (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Santos GS, Mafia RG, Aguiar AM et al (2020) Stem rot of eucalyptus cuttings caused by Neopestalotiopsis spp. in Brazil. J Phytopathol 168:311–321. https://doi.org/10.1111/jph.12894
Silva MRC, Diogo E, Bragança H et al (2015) Teratosphaeria gauchensis associated with trunk, stem and foliar lesions of Eucalyptus globulus in Portugal. For Pathol 45:224–234. https://doi.org/10.1111/efp.12160
Silva AC, Diogo E, Henriques J et al (2020) Pestalotiopsis pini sp. nov., an emerging pathogen on stone pine (Pinus pinea L.). Forests 11:1–17. https://doi.org/10.3390/f11080805
Silvério ML, de Cavalcanti MA, Q, Silva GA da, et al (2016) A new epifoliar species of Neopestalotiopsis from Brazil. Agrotópica 28:151–158
Solarte F, Muñoz CG, Maharachchikumbura SSN, Álvarez E (2018) Diversity of Neopestalotiopsis and Pestalotiopsis spp., causal agents of guava scab in Colombia. Plant Dis 102:49–59. https://doi.org/10.1094/PDIS-01-17-0068-RE
Song YU, Geng KUN, Zhang BIN et al (2013) Two new species of Pestalotiopsis from Southern China. Phytotaxa 126:22–30. https://doi.org/10.11646/phytotaxa.126.1.2
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Stergiopoulos L, Gordon TR (2014) Cryptic fungal infections: the hidden agenda of plant pathogens. Front Plant Sci 5:10–13. https://doi.org/10.3389/fpls.2014.00506
Stone JK, Polishook JD, White JF (2004) Endophytic fungi. In: Biodiversity of Fungi. Elsevier, Burlington, pp 241–270
Swofford DL (2002) Phylogenetic analysis using parsimony. Sinauer Associates, Sunderland
Tejesvi MV, Kini KR, Prakash HS et al (2007) Genetic diversity and antifungal activity of species of Pestalotiopsis isolated as endophytes from medicinal plants. Fungal Divers 24:37–54
Tibpromma S, Hyde KD, McKenzie EHC et al (2018) Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Divers 93:1–160. https://doi.org/10.1007/s13225-018-0408-6
Vaidya G, Lohman DJ, Meier R (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27:171–180. https://doi.org/10.1111/j.1096-0031.2010.00329.x
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols. Academic Press, San Diego, pp 315–322
Xu J, Yang X, Lin Q (2014) Chemistry and biology of Pestalotiopsis-derived natural products. Fungal Divers 66:37–68. https://doi.org/10.1007/s13225-014-0288-3
Acknowledgements
The authors would like to thank to Joana Henriques for help on molecular biology, to Isabel Lourenço and Florinda Medeiros for assistance in the Mycology laboratory. We also thank Ana Reis, from Altri Florestal, for providing one sample and Saowaluck Tibpromma, Kunming Institute for Botany, China, for sending sequences before they were made available in GenBank. Alan JL Phillips acknowledges the support from UIDB/04046/2020 and UIDP/04046/2020 Centre grants from FCT, Portugal (to BioISI).
Funding
This work was funded by a collaborative protocol, “Estudo das Doenças do eucalipto – prospecção e controlo” established between INIAV, the RAIZ Institute and Altri Florestal.
Author information
Authors and Affiliations
Contributions
H.B., E.D., C.I.G. and C.V. designed the experiments; E.D. and A.C.S. performed morphological and phylogenetic analysis; C.I.B and E.D. wrote the first draft of the manuscript; H.B and A.J.L.P. reviewed and edited the manuscript. HB and A.J.L.P. supervised the study. All authors have read, edited and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Section editor: Marc Stadler
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Diogo, E., Gonçalves, C.I., Silva, A.C. et al. Five new species of Neopestalotiopsis associated with diseased Eucalyptus spp. in Portugal. Mycol Progress 20, 1441–1456 (2021). https://doi.org/10.1007/s11557-021-01741-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-021-01741-5