Abstract
The ascomycete genus Diaporthe includes plant pathogens and endophytes on a wide range of hosts including economically important crops. Anamorphs are coelomycetous and reside in the genus Phomopsis. Phomopsis amygdali is the causal agent of twig canker and blight of almonds. In a recent survey of dieback of almonds in Portugal, the most frequent fungi detected were Diaporthe/Phomopsis species. Isolates from almond and other Prunus species were characterised and grouped according to their microsatellite-primed PCR (MSP-PCR) profiles and representatives of the different groups were selected for a phylogenetic study based on the ITS rDNA region (ITS1–5.8S–ITS2). Combining morphological, cultural, molecular and pathogenicity data, three species were distinguished. Phomopsis amygdali was shown to be the main pathogen on almond and is epitypified in the present work. Diaporthe neotheicola is reported for the first time on this host. A third species represented by a single isolate could not be unequivocally identified.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ascomycete genus Diaporthe Nitschke includes plant pathogens and endophytes on a wide range of hosts including economically important crops (Uecker 1988). Species in this genus and their Phomopsis (Sacc.) Bubák anamorphs were characterized largely by host association, which resulted in a proliferation of names (Uecker 1988). Recent studies have revealed that host association is not a reliable character for species definition since the same species can be found on different hosts and several species can occur on the same host (Rehner and Uecker 1994; Mostert et al. 2001). For example, in grapevine (Vitis vinifera), 15 different species of Phomopsis have been reported (Mostert et al. 2001; van Niekerk et al. 2005). Molecular data, such as phylogenies derived from sequences of the internal transcribed spacer (ITS) and other genes have helped to redefine species in this genus (Mostert et al. 2001; Santos et al. 2010; Santos and Phillips 2009; van Niekerk et al. 2005; van Rensburg et al. 2006).
Phomopsis amygdali is the causal agent of twig canker and blight of almonds (Prunus dulcis) and peach (Prunus persica) wherever these hosts are grown. It was first described as Fusicoccum amygdali Delacr. causing cankers on almonds in France (Delacroix 1905). Tuset and Portilla (1989) re-examined the type specimen of F. amygdali and, based on morphology and symptomatology, they considered that it would be best accommodated in Phomopsis as Phomopsis amygdali (Delacr.) J.J Tuset and M.T. Portilla. They also considered Phomopsis amygdalina Canonaco to be a synonym of P. amygdali. Unfortunately, no cultures linked unequivocally to the type exist.
In a recent survey of dieback of almonds in Portugal, the fungi most frequently detected were Diaporthe/Phomopsis species. Although most of the isolates corresponded morphologically with P. amygdali, some were clearly different species. Therefore, the aim of this work was to use morphological, molecular and pathogenicity data to clarify the identity of the Diaporthe/Phomopsis species that occur on almond in Portugal, and to select a suitable specimen as epitype of P. amygdali.
Materials and methods
Isolations
Branches from almond trees showing symptoms of cankers or dieback were collected in the two major production areas of Portugal. Six orchards in the northern region of Trás-os-Montes were sampled while in the south of the country two orchards in Algarve and one in Alentejo were sampled. Isolations were made from small pieces of host tissue taken from the margin of the cankers. Tissue pieces of about 3 mm2 were surface disinfected in 3% sodium hypochlorite for 5 min, rinsed in sterile distilled water and plated on potato dextrose agar (PDA, Difco Laboratories, Detroit, MI, USA.) amended with 100 ppm of streptomycin sulphate (Sigma-Aldrich, St. Louis, MO, USA) to suppress bacterial growth. The resulting cultures were induced to sporulate and single spore isolations were made for all isolates. Isolates were maintained on 2% water agar (WA) slants with a piece of sterile alfalfa stem at 4°C (Farr et al. 1999). The isolates used in this study are summarised in Table 1. For comparison, isolates from other Prunus hosts were included. Reference isolates were deposited in the public culture collection of the Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands.
DNA isolation, MSP-PCR and sequence analysis
DNA was isolated from mycelia scraped from the surface of a PDA plate. DNA was extracted with the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, USA), following the manufacturer’s instructions. Instead of using ground lyophilized mycelia, fresh mycelium was used and was disrupted by adding approx. 50 μl of glass beads (425–600 μm diam.) to the extraction buffer and vortexing for 2 min. MSP-PCR profiles were generated following the protocol of Uddin and Stevenson (1997) using the primers (GTG)5, (GGA)7, (ACAC)5 and phage M13 core sequence. (Meyer et al. 1993). Amplicons were separated by electrophoresis at 5 V.cm−1 and visualised as described in Santos and Phillips (2009). The isolates were clustered on the basis of their profiles in a consensus dendrogram built with GelCompar™ version 4.1 (Applied Maths BVBA, Kortrijk, Belgium) using Pearson’s correlation coefficient and UPGMA (Fig. 1). These experiments were repeated once.
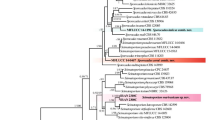
Consensus dendrogram from M13, (GTG)5, (GGA)7 and (ACAC)5 MSP-PCR profiles performed in GelCompar™ using Pearson’s correlation coefficient and UPGMA. Isolates shown in bold were selected for the phylogenetic analysis. Scale bar represents percentage of similarity. Groups I, II, III and IV are referred to in the text
A set of 19 isolates representing each MSP-PCR cluster was selected for a phylogenetic analysis of the ITS region following the protocol of Alves et al. (2004) and increasing the amount of primers to 25 pmol each and of Taq polymerase to 1.25 U. The cycling conditions were: initial denaturation of 10 min at 95°C, 35 cycles of 1 min at 95°C, 1 min at 55°C and 30 sec at 72°C, and a final extension step of 10 min at 72°C. The amplicons were purified and sequenced as in Santos and Phillips (2009). Sequences were edited and used in a phylogenetic analysis following the protocol of Santos and Phillips (2009). The sequence of Phomopsis theicola CBS 187.27 was retrieved from TreeBASE (S1506, M2708, van Rensburg et al. 2006). Sequences obtained from GenBank are listed by their taxon names and accession numbers in the tree (Fig. 2), while newly generated sequences are listed by their isolate number. Newly generated sequences have been deposited in GenBank (Table 1) and the alignment and phylogeny in TreeBASE (S10334, M5013).
One of seven equally parsimonious trees resulting from the alignment of 504 characters of the ITS rDNA region. Length = 256; consistency index (CI) = 0.813; retention index (RI) = 0.952; homoplasy index (HI) = 0.188; rescaled consistency index (RC) = 0.774. Newly generated sequences are listed in bold. Bootstrap values with 1000 replications are shown above the branches for Maximum Parsimony and below the branches for Neighbour-Joining (NJ). Branches marked with * where not present in NJ tree. Thickened lines indicate branches that were present in the strict consensus tree. Bar represent 10 changes. Isolates followed by § are ex-type/ex-epitype isolates. Valsa ceratosperma (DQ241769) and Leucostoma persoonii (DQ996042) were included as outgroups. Clades A, B, C and D are referred to in the text. Phylogeny deposited in TreeBASE (S10334)
Morphology
Cultures were grown on 2% WA with a sterile alfalfa stem (Farr et al. 1999) at 22°C with 12 h of near UV light per day. Morphological characters were studied and recorded as in Santos and Phillips (2009). Growth rates were determined on PDA plates as described by Santos and Phillips (2009). Colony diameters were measured after 3 days of incubation.
Pathogenicity
One year old almond twigs cv. Ferragnès, about 30 cm long, were inoculated with 4 isolates of P. amygdali, 4 of D. neotheicola and the single isolate of the unidentified Phomopsis species. A 3 mm2 flap of epidermis was lifted, colonised agar plugs of about the same size were placed underneath the epidermis and the wounds sealed with parafilm. The twigs were kept in an upright position with their lower ends immersed in jars of water in a controlled environment at 25°C with 14 h of light per day. The twigs were covered with a plastic bag during the first 7 days to keep a moist environment. The water was changed every 3 days. Ten twigs per isolate were used. A negative control was treated in the same way with a sterile agar plug. Lesion lengths were measured 24 days after inoculation and analysed by analysis of variance. Means were compared by least significant difference test (LSD) (Table 2).
Results
MSP-PCR and sequence analysis
Based on the MSP-PCR profiles the isolates clustered in four groups (Fig. 1). Group I included isolates from almond and peach, obtained from all the regions sampled. Group II included isolates from almond and apricot, all from orchards in Alentejo and Algarve except those from apricot with unknown origin. The two isolates in group III were obtained from plum leaves, and isolate 58A, from almond in Algarve, was the sole isolate in group IV. Almond isolates representative of each group and geographic origin within each group were selected for phylogenetic analysis.
Amplification products and sequences ranged from 578 to 590 bp. Sequences selected from a BLAST search in GenBank were added to the sequences generated in this study together with additional GenBank sequence of Phomopsis amygdali and P. theicola. The dataset consisted of 39 ingroup taxa and two outgroup taxa and the alignment contained 504 characters including alignment gaps. Of the 504 characters, 113 were parsimony informative and included in the analysis resulting in seven equally parsimonious trees. One of these trees is represented in Fig. 2. Neighbour-Joining analysis resulted in a tree with the same topology as the MP tree except for three branches on terminal clades indicated by * in Fig. 2. Four major clades could be distinguished. Clade A (with 100% support in both NJ and MP) included sequences from P. amygdali isolates. Within this clade a subclade (with bootstrap values of 99% for NJ and 95% for MP) included isolates from almond in the three sampled geographic regions. Sequences of P. amygdali from almonds in Portugal were identical to sequences of isolates from almonds in Italy (AF102994) and Spain (AF102997) and peach in USA (U86406, AF102995 and AF102996). Clade B (with bootstrap values of 100% for NJ and 99% for MP) included the ex-epitype isolate of Diaporthe viticola and the ex-type isolate of Diaporthe australafricana. Clade C (with 100% bootstrap support for both NJ and MP) included almond isolates collected only in the South of Portugal that clustered with ex-type isolates of P. theicola and Diaporthe neotheicola. Clade D (with bootstrap values of 100% for NJ and 99% for MP) included only one almond isolate that clustered close to a reference isolate of Diaporthe eres. This clade also included sequences from other isolates assigned to different species.
Morphology
According to Tuset and Portilla (1989) the type specimen of Fusicoccum amygdali (basionym of P. amygdali) was deposited in herbarium PC. In the present study, we attempted to study this specimen in detail. Unfortunately, the herbarium PC was closed for renovation during the time period of this work. Therefore, we based our morphological comparisons between P. amygdali and the Portuguese isolates found on almond on published descriptions of this species.
Overall, the morphology of the Portuguese P. amygdali isolates, especially that of isolate CBS 126679 (= 3B), correlated very well with the original description of F. amygdali given by Delacroix (1905) as well as with the detailed description of its type specimen provided by Tuset and Portilla (1989). The pycnidial diameter range of our isolates falls within the range given by Tuset and Portilla (1989). Moreover, conidiophores have the same shape, are rarely branched and exhibit the same dimensions as described by the same authors. Alpha conidia are ovoid-ellipsoid, matching the shape and dimensions given by Delacroix (1905) and Tuset and Portilla (1989). The absence of beta conidia in the Portuguese isolates correlates with their absence in Delacroix (1905) and their scarcity reported by Tuset and Portilla (1989). In fact, these last authors did not see beta conidia in the type specimen of F. amygdali nor in any in vitro culture. These spores were only seen in a pycnidium on an almond leaf.
The D. neotheicola isolates obtained in the present study were morphologically identical to the type isolates and type specimen of this species as described by Santos and Phillips (2009). Pycnidia are globose to subglobose, having the same dimensions as in Santos and Phillips (2009). Conidiophores are cylindrical, septate, ranging from 9 to 30 μm in length. Conidiogenous cells are phialidic, 7–21 × 1.5–3 μm. Alpha conidia are fusoid, 5.5–10.5 × 2–3 μm, while beta conidia are filiform, 21–32 × 1–1.5 μm.
Pathogenicity
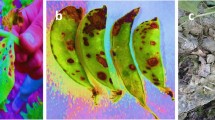
The mean length of lesions observed on almond twigs inoculated with the isolates in this study are given in Table 2. The lesions observed on twigs inoculated with isolates identified as P. amygdali and isolate 58A were significantly longer than those caused by D. neotheicola, except for isolate 17B, as determined by the LSD test. All isolates identified as P. amygdali produced lesions in all inoculated almond twigs. Some of the twigs inoculated with D. neotheicola isolates did not show any lesions.
Taxonomy
The isolates in clade A (Fig. 2) were morphologically identical to F. amygdali as described by Delacroix (1905) and P. amygdali as described by Tuset and Portilla (1989). Furthermore, they were phylogenetically indistinguishable from isolates from Italy, Spain and USA. One specimen (CBS-H 20420) was selected and designated herein as epitype.
Phomopsis amygdali (Delacr.) J.J. Tuset and M.T. Portilla, Can J Bot 67: 1280 (1989).
(Fig. 3)
MycoBank MB 518643
Basionym: Fusicoccum amygdali Delacr., Bull Trimest Soc Mycol Fr, 21 (3): 184 (1905).
≡ Phomopsis amygdalina Canonaco, Riv Pat Veget 26: 157 (1936).
Teleomorph: Unknown.
Conidiomata eustromatic, subglobose to ampuliform, subepidermal, erumpent, dark brown to black, 240–390 μm wide × 140–160 μm tall on host. In culture 160–220 μm wide × 120–300 μm tall. Conidia exuding from the pycnidia in white to cream drops. Conidiophores subcylindrical, hyaline, seldom branched, 7.4–36.3 × 1.5–3.2 μm, \( \overline x \) ± S.D. = 14.5 ± 4.16 × 2.3 ± 0.36 μm (n = 380). Conidiogenous cells phialidic, cylindrical, tapering toward the apex, periclinal thickening and collarette present, 5.0–20.0 × 1.5–3.2 μm, \( \overline x \) ± S.D. = 9.8 ± 2.29 × 2.3 ± 0.36 μm (n = 380). Alpha conidia ovoid-ellipsoid, mostly with one end obtuse and the other acute, 0–2 guttulate, (4.18–)6.27–6.32(–9.64) × (1.63–)2.36–2.38(–3.31) μm, \( \overline x \) ± S.D. = 6.3 ± 0.63 × 2.37 ± 0.22 μm (n = 2100). Beta conidia not seen.
Colonies on PDA reaching a diameter of 43.5 mm after 3 days at 25°C in the dark. Colonies white, cottony, with raised margins, becoming pale brown, reverse pale brown with brown patches.
Habitat
On branches and fruits of Prunus armeniaca (Garofalo 1973), P. dulcis (Adaskaveg et al. 1999), P. persica (Farr et al. 1999) and Vitis vinifera (Mostert et al. 2001).
Known distribution
France (Delacroix 1905), Greece (Pantidou 1973), Italy (Canonaco 1936; Garofalo 1973), Japan (Kanematsu et al. 1999), Portugal (Dias et al. 1982), South Africa (Mostert et al. 2001), Spain (Tuset and Portilla 1989), Tunisia (Trigui 1968) and USA (Adaskaveg et al. 1999; Farr et al. 1999).
Material examined
PORTUGAL: Trás-os-Montes, Mirandela, September 2005, E. Diogo, (CBS-H 20420, EPITYPE designated herein; culture ex-epitype 3B = CBS 126679). See Table 1 for other isolates studied.
Notes
Beta conidia were not seen in any of the isolates. Tuset and Portilla (1989) observed beta conidia in pycnidia on the host but never saw them in culture. However, these authors were unable to find beta conidia in the type specimen. This species differs from Diaporthe neotheicola in having consistently shorter alpha conidia and white colonies becoming pale brown instead of olivaceous green.
Discussion
Sequence analyses based on the ITS rDNA region revealed three species of Diaporthe and Phomopsis on almonds in Portugal. These three species correlated perfectly with the groups revealed in the MSP-PCR profiles. Although ITS is the locus most commonly used to infer phylogenies in fungi (Nilsson et al. 2008) it was recently demonstrated that this region of the ribosomal gene cluster shows high within species variation in some Diaporthe/Phomopsis species (Santos et al. 2010). Nevertheless, ITS can still be used to distinguish species in this genus provided the phylogenies are interpreted with care. Indeed, as shown in this paper, there was little within species variation in P. amygdali (group I in Fig. 1 and clade A in Fig. 2).
By far the largest number of isolates from almonds resided in a clade with isolates previously identified as P. amygdali. These isolates were collected from widely separate geographical regions including Georgia and South Carolina (Southern USA), Italy and Spain. Isolates in this clade were the most virulent ones in the pathogenicity tests and morphologically all the isolates in this clade that we studied corresponded with the original description of F. amygdali (Delacroix 1905) and the detailed description of the type specimen given by Tuset and Portilla (1989). Although the type of F. amygdali is housed in PC, it was not available during the course of this work. However, this specimen was examined by Tuset and Portilla (1989) who provided a detailed description. Since no ex-type cultures of P. amygdali exist we proposed CBS-H 20420 as epitype with CBS 126679 as ex-epitype culture. Ideally an epitype should be derived from the same host and same locality as the holotype (Hyde & Zhang 2008). However, since ITS sequences of P. amygdali from almonds in Italy, Portugal and Spain are identical, and the isolates from Portugal are indistinguishable morphologically from the type we felt justified in proposing the specimen from Portugal as epitype. In a similar way, Phillips et al. (2006) found that ITS sequences of two isolates of Botryosphaeria corticis (Demaree & M.S. Wilcox) Arx & E. Müll. from North Carolina (the type locality of this species) were identical to isolates from New Jersey. Since this latter collection from New Jersey was morphologically identical to the type it was designated epitype.
Phomopsis amygdali is not restricted to peach and almond but has been isolated, albeit infrequently, from grapevines in South Africa (van Niekerk et al. 2005). ITS sequences of the two isolates from grapevines, and a further isolate from peach from Arkanas, USA, difffered slightly from the typical isolates of P. amygdali. These differences would suggest that recombination had ocurred at some time further suggesting that a sexual state of this species may exist although none has been reported for P. amygdali in nature. However, Kanematsu et al. (2000) reported succesful mating between Diaporthe G-type and P. amygdali isolates in culture. Furthermore, Santos et al. (2010) showed that these two taxa may be the same phylogenetic species. Taking together these two observations it seems that compatible isolates of P. amygdali are able to mate, although, for some reason, this rarely happens in nature.
Diaporthe neotheicola (group II in Fig. 1 and clade C in Fig. 2) was identified by association with the ex-type isolate CBS 123208 as well as with the ex-type isolate of Phomopsis theicola (CBS 187.27). As far as we know, this is the first report of D. neotheicola on Prunus dulcis and Prunus armeniaca. This species was first described as P. theicola on Camellia sinensis in Italy (Curzi 1927) but has since been isolated from several different hosts including Vitis vinifera, Protea, Pyrus (Mostert et al. 2001; van Niekerk et al. 2005), Aspalathus linearis (van Rensburg et al. 2006) and recently from Foeniculum vulgare in Portugal (Santos and Phillips 2009). The pathogenicity results showed that D. neotheicola is probably a weak pathogen on P. dulcis, which confirms previous studies with different hosts (van Niekerk et al. 2005; van Rensburg et al. 2006; Santos and Phillips 2009). The results obtained in the present work further confirm the previous findings that host affiliation is an unreliable character for identification of Diaporthe/Phomopsis species (Farr et al. 2002; Mostert et al. 2001; Rehner and Uecker 1994).
References
Adaskaveg JE, Forster H, Connell JH (1999) First report of fruit rot and associated branch dieback of almond in California caused by a Phomopsis species tentatively identified as P. amygdali. Plant Dis 83:1073
Alves A, Correia A, Luque J, Phillips AJL (2004) Botryosphaeria corticola sp. nov. on Quercus species, with notes and description of Botryosphaeria stevensii and its anamorph Diplodia mutila. Mycologia 96:598–613
Canonaco A (1936) Il seccume dei rameti di mandorlo in relazione ad alcuni micromiceti. Riv Patol Veget 26:145–164
Curzi M (1927) De novis theae micromycetibus pathogenis. Atti Ist Bot Univ Pavia 3:59–72
Delacroix G (1905) Sur une maladie des amandiers en Provence. Bull Trimest Soc Mycol Fr 21:180–185
Dias MRS, Lucas MT, Lopes MC (1982) Fungi Lusitaniae XXIX. Agron Lusit 41:175–192
Farr DF, Castlebury LA, Pardo-Schultheiss R (1999) Phomopsis amygdali causes peach shoot blight of cultivated peach trees in the southeastern United States. Mycologia 91:1008–1015
Farr DF, Castlebury LA, Rossman A (2002) Morphological and molecular characterization of Phomopsis vaccinii and additional isolates of Phomopsis from blueberry and cranberry in the eastern United States. Mycologia 94:494–504
Garofalo F (1973) L’Albicocco “Tonda di Costigliole”, nuovo ospite di Fusicoccum amygdali Del. Informatore Fitopatologico 23:13–15
Hyde KD, Zhang Y (2008) Epitypification: should we epitypify? J Zhejiang Univ Sci B 9:842–846
Kanematsu S, Yokoyama Y, Kobayashi T (1999) Taxonomic reassessment of the causal fungus of peach Fusicoccum canker in Japan. Ann Phytopathol Soc Jpn 65:531–536
Kanematsu S, Minaka N, Kobayashi T, Kudo A, Ohtsu Y (2000) Molecular phylogenetic analysis on ribosomal DNA internal transcribed spacer regions and comparison of fertility in Phomopsis isolates from fruit trees. J Gen Plant Pathol 66:191–201
Meyer W, Mitchell TG, Freedman EZ, Vilgalys R (1993) Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans. J Clin Microbiol 31:2274–2280
Mostert L, Crous PW, Kang JC, Phillips AJL (2001) Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: morphological, cultural, molecular and pathological characterization. Mycologia 93:146–167
Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson K-H (2008) Intraspecific ITS variability in the kingdom fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol Bioinformatics 4:193–201
Pantidou ME (1973) Fungus-host index for Greece. Benaki Phytopathol Inst, Kiphissia, Athens
Phillips AJL, Oudemans PV, Correia A, Alves A (2006) Characterization and epitypification of Botryosphaeria corticis, the cause of blueberry cane canker. Fungal Divers 21:141–155
Rehner SA, Uecker FA (1994) Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete Phomopsis. Can J Bot 72:1666–1674
Santos JM, Phillips AJL (2009) Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculum vulgare in Portugal. Fungal Divers 34:111–125
Santos JM, Correia VG, Phillips AJL (2010) Primers for mating-type diagnosis in Diaporthe and Phomopsis: their use in teleomorph induction in vitro and biological species definition. Fungal Biol 114:255–270
Trigui A (1968) Sur la présence en Tunisie de Fusicoccum amygdali Delacroix sur Amandier. Bull ENSAT 18/19:65–68
Tuset JJ, Portilla MT (1989) Taxonomic status of Fusicoccum amygdali and Phomopsis amygdalina. Can J Bot 67:1275–1280
Uddin W, Stevenson KL (1997) Pathogenicity of a species of Phomopsis causing a shoot blight on peach in Georgia and evaluation of possible infection courts. Plant Dis 81:983–989
Uecker FA (1988) A world list of Phomopsis names with notes on nomenclature, morphology and biology. Mycologia Memoir 13:1–231
van Niekerk LM, Groenewald JZ, Farr DF, Fourie PH, Hallen F, Crous PW (2005) Reassessment of Phomopsis species on grapevine. Australas Plant Pathol 34:27–39
van Rensburg JCJ, Lamprecht SC, Groenewald JZ, Castebury LA, Crous PW (2006) Characterization of Phomopsis spp. associated with die-back of rooibos (Aspalathus linearis) in South Africa. Stud Mycol 55:65–74
Acknowledgements
This work was financed by the European Regional Development Fund and Fundação para a Ciência e a Tecnologia (FCT) under the project PTDC/AGR-AAM/67064/2006 and J. Santos was supported by a research grant within the project. A. Phillips was supported by grant number SFRH/BCC/15810/2006 from FCT. João Inácio (Laboratório Nacional de Investigação Veterinária, Instituto Nacional de Recursos Biológicos, I.P.) helped with the construction of the MSP-PCR dendrogram, and Cecília Rego (Instituto Superior de Agronomia, Laboratório de Patologia Vegetal “Veríssimo de Almeida”) provided bibliographic material.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Diogo, E.L.F., Santos, J.M. & Phillips, A.J.L. Phylogeny, morphology and pathogenicity of Diaporthe and Phomopsis species on almond in Portugal. Fungal Diversity 44, 107–115 (2010). https://doi.org/10.1007/s13225-010-0057-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-010-0057-x