Abstract
The family Psathyrellaceae was analysed using phylogenetic and morphological characters. A total of 18,133 sequences (ITS, 5.8S, LSU, ef-1α, β-tubulin), with 45 newly generated, were evaluated from a wide geographic sampling. Special attention was given to the alignment procedures and an iterative multigene guide tree was used to achieve the best possible phylogenetic hypotheses. A new generic system is proposed, which includes the known genera Coprinellus, Coprinopsis, Cystoagaricus, Homophron, Hormographiella, Kauffmania, Lacrymaria, Parasola, Psathyrella and Typhrasa. Six new, monophyletic genera are recognized, viz. Candolleomyces, Britzelmayria, Narcissea, Olotia, Punjabia and Tulosesus, and the corresponding new combinations are proposed. Galerella floriformis is shown to belong to the Psathyrellaceae and the new genus Hausknechtia is erected for it. Psathyrella is subdivided into 18 sections (sections Noli-tangere, Saponaceae, Stridvalliorum, Arenosae, Confusae, Sublatisporae, Sinefibularum are new), and sections Pennatae, Pygmaeae and Pseudostropharia are emended. Coprinellus is divided into nine sections (Disseminati, Aureogranulati, Curti, Hepthemeri and Deminuti are new), and 20 sections are proposed for Coprinopsis (Cinereae, Filamentiferae, Melanthinae, Alopeciae, Xenobiae, Phlyctidosporae, Krieglsteinerorum, Erythrocephalae, Geesteranorum, Mitraesporae, Radiatae, Subniveae and Canocipes are new). And lastly, Parasola is divided into sections Parasola and Conopileae. Many problematic species groups still need revision. A key to the genera based on morphological characters is included.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
When the work of Hopple et al. (1999) and especially Redhead et al. (2001) was published at the dawn of the new millennium, it became apparent that molecular biology techniques would profoundly alter the classical systematics of many dark-spored agarics. Until that time, morphological features were the sole basis for determining family relationships, as in Smith (1972), Romagnesi (1944, 1982), Kühner and Romagnesi (1953), Orton and Watling (1979), Kits van Waveren (1985), Singer (1986), Citérin (1992, 1994) and other authors.
However, the traditional way of working had limits, such as when certain features (e.g. rough spores, a grainy veil, and so on) are not necessarily bound to a systematic position. Extensive comparisons of gene sequences not only led to the splitting of the historical genus Coprinus Pers. (Redhead et al. 2001) and the establishment of the family Psathyrellaceae Vilgalys, Moncalvo & Redhead, but also to various transfers of taxa. For instance, the work of Larsson and Örstadius (2008) showed that Psathyrella conopilea (Br.) A. Pearson & Dennis belongs to the genus Parasola Redhead, Vilgalys & Hopple, while Psathyrella marcescibilis (Britzelm.) Singer and P. pannucioides (J.E. Lange) M.M. Moser were recognized as members of the genus Coprinopsis P. Karst. Moreover, the work of Örstadius et al. (2015) identified further recombinations and the establishment of new genera like Kauffmania Örstadius & E. Larss., Typhrasa Örstadius & E. Larss. and Homophron (Britzelm.) Örstadius & E. Larss. Knowledge of the relationships within the family made rapid progress through the works of Padamsee et al. (2007), Vašutová et al. (2008), Nagy et al. (2009, 2010a, b, 2011a, b, 2012a, b, 2013a, b), Nagy, Vágvölgyi et al. (2013b) and Szarkándi et al. (2017). In the meantime, new taxa described by molecular phylogenetic analyses are often described in the context of just a few related species. Examples are Hazi et al. (2011), Melzer et al. (2017), Yan and Bau (2017), Deschuyteneer et al. (2017), Broussal et al. (2018) and Melzer (2018). If only a few close sequences are used and these are from one or two regions only, there is a risk that the phylogeny will not reflect the truthfulness that would be possible if all the sequences from all regions in the corresponding section were included in the analysis.
This paper presents the Psathyrellaceae in full extent, which is possible with the currently available data, based on a phylogenetic analysis, while also giving regard to the morphology of the taxa. A classification is proposed which is as comprehensive as possible while not unnecessarily complicated, with the ranks of subgenus and subsection being omitted.
Material and methods
Sequence sampling and selection
On March 5, 2018, NCBI GenBank and Unite using PlutoF (Abarenkov et al. 2010) were searched for all nucleotide sequences, concerning the family Psathyrellaceae. From NCBI GenBank, a total of 17,864 raw sequences were exported, further 224 raw sequences from Unite.
All sequences which are classified in the taxonomic group Psathyrellaceae (taxid184208 @ NCBI GenBank) and those that are close to it were collected. Numerous sequences described as “uncultivated” were also analysed, as well as those that were obviously given the wrong taxa names (incorrect determinations). The sequences of the taxonomic group Coprinus (taxid5345 @ NCBI GenBank) were completely exported initially and later critically checked as all others as described below.
From these sources, the basic database with 18,133 single sequences was created. These were first tested for their orientation. The reversed complement sequences were determined and replaced by the generated forward sequences. For the preliminary examination of all sequences, they had to be roughly aligned and sorted. This was done with Mafft using the L-INS-i procedure at default settings. Indels were not coded in the preliminary examination. Partitioning was limited to the ITS1, 5.8S, ITS2, LSU, β-tub, and ef-1α regions.
No MSA filters were applied, but as usual the introns were removed from the protein coding sequences. Using RAxML (model GTR+CAT; final tree optimized by GTR+G) via Cipres, both the preliminary single phylogenies of the regions and the preliminary multigene phylogeny were tested for exclusion of sequences. This was done before the addition of the specially selected outgroup to avoid conflicts.
Sequences which undoubtedly do not belong to the family Psathyrellaceae were excluded. In addition, those without ITS1 and ITS2 regions or only a very short part of them, as well as clearly erroneous sequences that caused incorrect branch lengths or wrong positioning in the topology, were sorted out.
For 100% identical sequences associated to different taxa, the most likely name and sequence ID was chosen. For this, the deposited vouchers and the associated literature were used to ensure the best possible species designation. However, identical sequences for which more than one or different regions were present were kept in the sequence collection in any case.
Sequences with untrue gaps created by the sequence author (deleted regions) were excluded as this would create nonsense gaps in the alignments and errors in the gap matrices. Thus, untrue deviations would have been caused, because the indels were coded and used for the phylogeny. This exclusion did not apply to sequences in which only introns were deleted because they were also removed in the present study (see chapter “The introns of the haploid nuclear genome”).
Note that this was only the preliminary sequence sorting procedure. For the fine selection and error detection, see chapter “Combinability tests of loci, detailed error check of sequence sets from vouchers”. The final “filtered” ingroup sequences are listed in Table S1.01 in Supplement S1. The outgroup sequences are listed in Table 1.
Later available sequences could not be included in the analysis, but are mentioned in the text in important cases. Several times it was necessary to verify determinations by comparison with collections of the authors. The sequencing for these specimens was performed by Pablo Alvarado (ALVALAB, Spain). The sequences were then deposited in NCBI GenBank. See Table S1.01 in Supplement S1 as well.
Reference sampling
Many sequences in the databases have been collected in the context of studies on fungal diversity, mycorrhizal communities, etc. For these studies, exact identifications are often irrelevant. This explains the fact that many sequences in the databases are not designated to a species or have been given an incorrect name.
To avoid confusion, the names are mostly left as they were originally. Additions or corrections of names due to interim recombinations and synonymizations were omitted even if they were clearly possible. Only changes by the same authors in later publications were adopted to ensure better clarity. As a result, different taxon names can sometimes be found in one clade. In the phylograms and in Table S1.01 in Supplement S1, the names of the vouchers or reference IDs were adopted as they are listed in the databases to ensure an unmistakable identification. Therefore, the spelling sometimes does not match the real authors’ names (e.g. LO = Leif Örstadius, Ulje = Uljé).
For this reason, references were selected in which the probability of an exact determination is very high. Of course, type material is most suitable, but it is not always available. Sometimes no type collection exists (e.g. Psathyrella microrhiza) or sequencing is not allowed (e.g. L). The investigation can also fail for older exiccates, which would be an unnecessary consumption of valuable resources. The same purpose is also served by well-described collections from reliable sources, for which an exact determination can be assumed to be certain. A reference voucher (abbreviated as Ref.v.) was therefore selected in each case, accompanied by a comprehensive description of the taxon. Publications by authors with a focus on the taxonomy of Psathyrellaceae were preferred.
In the systematic part, the recognized species with the currently valid name are mentioned. The corresponding reference voucher (abbreviated as Ref.v.), as well as a literature source that allows conclusions on the quality of the determination, is mentioned.
Morphologically studied material
A large number of members of the Psathyrellaceae family have been studied as fresh or dried material to get a more detailed view of the features. The procedure of light microscopy is well known and is not described in detail here. The results are recorded in notes, line drawings or photos and archived by the authors. No voucher is preserved for some small specimens when there is no material left after the examination.
The list in supplement S3 contains all morphologically examined collections as a complete overview, regardless of whether mentioned in the text. Vouchers are stored in the herbaria A. Melzer (AM, AV, CA, HIAS), and D. Wächter (DW); others are designated by name. Samples loaned from the Herbarium GENT are marked with their catalogue number only, because almost all labels are handwritten and sometimes unreadable. The GENT material includes collections from Belgium, France and the Netherlands.
The spore size is simply referred to as “small” (predominantly below 8 μm in length, if remarkably shorter “very small”), medium-sized (about 8–10 μm) and “large” (10–13 μm, if considerably larger “very large”). As the spores of many species are highly variable, this has to be understood as a rough classification.
Phylogenetic analysis
Region selection of molecular phylogenetic markers
Potential and final selected regions
First, a computer-aided determination was made to find out, which of the regions are covered by a sufficient number of sequences (temporarily programmed routines of the authors were used for this step). It turned out that the following regions were considered for the analysis as they were sufficiently covered:
-
From the ribosomal DNA (rRNA), which is a part of the nuclear genome:
-
SSU (small subunit 18S rRNA gene) → later excluded from the analysis (see the following explanation)
-
ITS1 (internal transcribed spacer 1)
-
5.8S (5.8S rRNA gene)
-
ITS2 (internal transcribed spacer 2)
-
LSU (large subunit 28S rRNA gene)
-
From the haploid nuclear genome (these are particularly suitable for the reconstruction of higher phylogenetic relationships):
-
β-tub (β-tubulin gene)
-
ef-1α (translation elongation factor 1-α gene)
All other regions were not used for the analysis because there were obviously too few sequences available. As the following tests have shown, all regions except the SSU region were sufficient and reasonable to be used.
Check whether the sequences of the SSU region are usable
From publications of other authors concerning the family Psathyrellaceae, it was already known as probable that the identified regions do not cause phylogenetic conflicts and are therefore suitable for analysis. As described in the chapter “Combinability tests of loci, detailed error check of sequence sets from vouchers”, this was checked again and confirmed. The only exception was the SSU region, which was not discussed in detail in these studies. Whether the SSU region is meaningful because of its known low phylogenetic information content was therefore not clear at first, especially considering that the phylogenetic analysis covers the entire Psathyrellaceae family plus the genera in the outgroup. There were only 44 sequences from the SSU region (see Table S1.01 in Supplement S1 and Table 1). These are comparatively few. They were aligned with Mafft—with the L-INS-i method. There were no regions that were difficult to align. The available sequences spanned the complete SSU region, i.e. approx. 2180 sites. After trimming the alignment to the inner edges of the terminal gaps, 2099 bp evaluable alignment length remained. With AliView, the sites diverging from the majority rule consensus were first optically checked—this is shown in Fig. 1.
Sites of the SSU alignment diverging from the majority rule consensus; representation from AliView; colours: nucleotides in AliView colour code; coloured sites differ from the majority rule consensus; grey areas are sites matching the consensus; white are indels or missing data; the scale represents the site numbering
Coloured sites in Fig. 1 differ from the consensus. Grey areas are sites matching the consensus. “V” regions (variable regions) of high divergence could not even be identified across the whole family including the outgroup. The phylogenetic content is obviously very low. This was investigated in more detail with Noisy. The following settings for Noisy were used, deviating from the default:
-
--missing ?N—this is necessary because the terminal gaps are filled with “?”.
-
--nogap—this makes sense, because the indels were examined separately for phylogenetic content and an evaluation as 5th state character is not useful—see also chapter “Indel coding method and indel matrices”.
Results:
-
Length of the alignment: 2099 sites
-
Constant sites: 2020
-
Singleton sites: 43
-
Phylogenetically informative sites: 31
-
Phylogenetically very informative sites: 5
-
Sum of phylogenetically informative sites with a reliability score > 80%: 28—corresponds to 1.3%
The result is shown as a visual representation in Fig. 2. The yellow area shows which sites (black lines) could have been used for the study (in addition to the 2020 constant sites).
Visual representation of the phylogenetic content of the alignment of the SSU region and the columns theoretically remaining/removed by the MSA filter. Graphic created with Noisy. Red: phylogenetically uninformative and constant sites; green: phylogenetically informative and very informative sites; black lines in the yellow bar: phylogenetically informative and very informative sites that have a reliability score > 80% and thus would remain in the alignment after application of the filter
Furthermore, the phylogenetically informative indels were analysed in the trimmed SSU alignment, as described in chapter “Indel coding method and indel matrices”.
Results:
-
Sum of gap positions (total): 7
Of those:
-
Informative gap positions: 3
-
Uninformative gap positions: 4
The indels of the SSU region also contain only very little phylogenetic information. The SSU region is covered with too few sequences and has a very low phylogenetic content compared to the sum of all partitions. Therefore, the SSU region was not used for the analysis.
Determination of region boundaries of the SSU, ITS1, 5.8S, ITS2 and LSU regions
One aim of the analysis was to completely remove the SSU region fragments in the ITS1 alignment. Also, the ITS1, 5.8S, ITS2, and LSU regions should be exactly separated for phylogenetic content calculations, best fit model calculations, partitioning, etc. The region boundaries should therefore be determined exactly. The region analysis was performed with ITSx (Bengtsson-Palme et al. 2013) and HMMER including the databases. The alignments showed distinct motifs (in each case majority rule consensus) for the respective regions. These are as follows:
-
SSU end motif:
...GAACCTGCGGAAGGATCATTA
-
ITS1 start motif:
ATGAATATCTATGGC...
-
ITS1 end motif:
...CCTATAAAACAAAAATA
-
5.8S start motif:
CAACTTTCAGCAACGGATCTC...
-
5.8S end motif:
...CCTGTTTGAGTGTCATTA
-
ITS2 start motif:
AATTCTCAACCT...
-
ITS2 end motif:
...GGACAATCTTTTGACA
-
LSU start motif:
ATTTGACCTCAAATCAGG...
At the same time, the exact length of the 5.8S region could also be determined—it is exactly 158 sites long for almost all sequences. A few sequences of the genus Parasola contain a deletion, so that the 5.8S region of these sequences is only 157 base pairs long. Some of them also contain some more indels, but most of them are probably stutter sites. See also chapter “MSA of the 5.8S region” and Fig. 14.
Sequence selection for the outgroup and root branch validation
It goes without saying that a meticulous outgroup selection is indispensable for a large phylogenetic study like this one. Extensive pre-tests were conducted with different compositions of outgroups, different outgroup sizes, different divergences to the ingroup and different loci. Among other things, the following important findings were obtained:
-
As not uncommon in large phylogenetic studies, the root node of the Psathyrellaceae family is mathematically unstable and, if the outgroup was selected unfavourably, shifts to an extreme position in the tree where it does not belong. This is favoured by a too small outgroup, too few loci, and too high divergence from outgroup to ingroup.
-
The higher phylogeny of critical genera and/or species close to the family Psathyrellaceae can only be solved mathematically with an outgroup selection specifically dedicated to this problem. Especially when only rapidly evolving loci are present for the critical species. These genera/species are Mythicomyces, Stagnicola perplexa and Pachylepyrium. All 3 do not belong to the family Psathyrellaceae, but fall into it in case of an unfavourable outgroup selection. Details can be found in the results chapter “Outgroup and critical genera”.
-
A too large outgroup is not directly harmful, but should also be avoided because of the high influence on the overall length of the partitions, since the already enormous computing time of the MCMCMC (several months—even using supercomputer clusters as used in this study) would be unnecessarily extended.
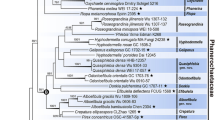
Mainly for these reasons, special care was taken to choose an outgroup as close to the family as possible, containing all critical taxa/genera, and if possible only sets with all loci. Special attention was also paid to the ability of root branch stabilization. The final outgroup found to be optimal can be taken from the phylogram in Fig. 39. The corresponding sequences can be found in Table 1. “Seq.-ID” is the voucher number or the most useful reference number if no voucher number was given by the original authors. The resulting stable position of the root branch was confirmed by various tests and probably currently provides the most accurate possible rooting of the family Psathyrellaceae. See also chapter “Plausibility check using a “HLPGT” (high-level phylogeny guide tree)”.
MSA of the problematic ITS1 and ITS2 regions
Alignment strategy for the ITS1 and ITS2 regions
The alignment of the ITS region posed a major problem due to the many rapidly evolving, indel-rich areas in these regions. An alignment of these regions without an iterative multigene guide tree, which was also calculated from conserved regions of the other loci, cannot achieve the truthfulness like an alignment with such a guide tree. In the course of the study, it was even shown that such an alignment procedure is indispensable in order to obtain a low-conflict and thus best truthful phylogram. Other authors Nagy et al. (2013a) and Tóth et al. (2013) have already found that the accuracy of the guide tree is the decisive factor for a truthful alignment of the ITS1 and ITS2 regions. At this point, it should again be mentioned that the exclusion of divergent regions (filtering with Gblocks (Castresana 2000) and similar programs) also leads to a distortion of the phylogeny—see chapter “MSA filter for divergent regions (not applied)”. Therefore, it was necessary to apply an iterative multigene guide tree refinement method for the ITS region, which includes the conserved regions (5.8S, LSU, ef-1α and β-tub) and also the indel partitions in the guide tree. Originally, the resulting best tree from a Bayesian analysis should be used as guide tree. However, this idea had to be discarded as it would have required an exorbitant amount of time. Therefore, after each alignment step, an ML analysis with RAxML via Cipres (model: GTRCAT, refinement under GTR+G for DNA, GTR2 + G with acquisition bias correction according to Lewis (2001) for the indel partitions) with the partitions as described in chapter “Partitioning of alignments and indel matrices/model selection for DNA alignments” was carried out instead and the resulting best tree was used as guide tree for the next refinement of the ITS1 respectively ITS2 alignment. Prank requires a rooted binary tree (i.e. straight bifurcating). Each tree therefore had to be rooted (root branch identification see chapter “Sequence selection for the outgroup and root branch validation”). This step was done with Treegraph. The alignment result was again the basis for a further ML analysis with RAxML via Cipres (models as described before). The resulting best tree was again used as a guide tree for the next refinement step of the ITS MSAs. This was done in a loop until no significant change in the best tree from the previous one was detectable respectively no significant change in the recorded values (see Table 3) occurred. The trees were evaluated using Treegraph and Node Integrator (authors’ tool) with which the number of nodes of ≥ 75% bootstrap as well as the average bootstrap values of all branches and the “weighted cumulative node reliability S” (according to Wächter) were calculated. The computational comparison of the trees was also performed with Node Integrator (authors’ tool), which determines the percentage of equal nodes from the new tree to the previous tree. Also the Robinson-Foulds distances (RF), the weighted Robinson-Foulds distances (WRF) and the weighted Robinson-Foulds distances incl. support consideration of common bipartitions (WRF2) were recorded. These were calculated with RAxML.
Selection of the alignment method for the iteration loops of the guide tree
The initial alignment of the ITS region was carried out with Mafft using the FFT-NS-2 method, as it turned out to be advantageous after some preliminary tests. The gap matrices of the initial alignment were coded with SeqState (Müller 2005) as described in chapter “Indel coding method and indel matrices”. The initial alignments including gap matrices were combined with the alignments and gap matrices of the other regions. With RAxML (settings as described before), the best tree for the complete phylogeny was calculated. This tree was rooted with Treegraph and served as multigene guide tree for the first alignment step. The quality values from this tree were recorded as described above.
It had to be checked which alignment method with guide tree is most advantageous for the alignment steps. For this purpose, the ITS1 region was again aligned with 4 different probabilistic methods—this time with guide tree—and the results were evaluated. The following 4 tests were performed with Prank. The switch -once was set to disable the automated iteration of Prank.
-
Test 1:
settings: Leave Gappy Regions (+F set); rest: default settings
-
Test 2:
settings: +F disabled; rest: default settings
-
Test 3:
settings: Leave Gappy Regions (+F set); -uselogs set; rest: default settings
-
Test 4:
settings: +F disabled; -uselogs set; rest: default settings
The results were evaluated as previously described by comparing the single phylogeny of the ITS1 alignments including gap matrices (also calculated with RAxML via Cipres, settings as described before) with the total phylogeny. In addition, the sum of the informative sites with a reliability score > 80% was calculated with Noisy. Table 2 shows the test results.
The diagram in Fig. 3 shows the weighted cumulative node reliability S(b) (according to Wächter) from tests 1 to 4.
Test 2 turned out to be the best method; therefore, this method (i.e. Prank—with default setting) was used for all subsequent iteration steps of the ITS1 and ITS2 regions. The method with the function +F (Leave Gappy Regions) was clearly worse in both cases. For the ITS2 region, it was assumed that the procedure from test 2 was also best choice.
Following alignment loop of the ITS1 and ITS2 regions with iterative multigene guide tree over all regions
After each iteration step, it was necessary to analyse how far the respective alignment was still from the optimum. After the respective alignment, which including the gap matrices was reintegrated into the overall partition, another ML bootstrap analysis with RAxML via Cipres (settings as described before) was performed and the best tree calculated from this was compared with the previous tree and evaluated as described above and below.
Among others, the following values were traced:
-
The length of the ITS1 alignment
-
The gap matrix length of the ITS1 alignment
-
The length of the ITS2 alignment
-
The gap matrix length of the ITS2 alignment
-
The number of nodes of the total phylogeny with ML bootstrap values ≥ 75%
-
The average ML bootstrap values of the total phylogeny
-
The percentage of identical nodes to the previous phylogeny with a ML bootstrap value ≥ 75%. Node Comparator (author’s tool) was used to calculate the percentage of identical nodes of the total phylogram resulting from the new iteration in relation to nodes with ≥ 75% reliability of the previous total phylogram
-
The “weighted cumulative node reliability S” according to Wächter (see below)
-
The -logLikelihood score of the total phylogeny from RAxML (model: GTRCAT; refinement under GTR+G for DNA; GTR2+G with acquisition bias correction according to Lewis (2001) for Indel partitions)
-
The Robinson-Foulds distances (RF)
-
The weighted Robinson-Foulds distances (WRF)
-
The weighted Robinson-Foulds distances including support consideration of common bipartitions (WRF2)
Weighted cumulative node reliability S
A new method was developed to analyse the calculated trees qualitatively. This is briefly presented here. To analyse the level of support of the trees, the bootstrap values were counted according to their size from b = 0 to b = 100 (the percentage sign of bootstrap values is not used in the following formulas for simplification). The numbers n were weighted linearly according to the bootstrap values b and summed up. This sum was divided by the number of all nodes in order to obtain a percentage ratio in the diagram from which the function.
- S :
-
= weighted cumulative node reliability S
- b :
-
= magnitude of the bootstrap value
- n :
-
= number of the respective bootstrap value size
- i :
-
= control variable
From this curve, the AUC (area under curve) was determined by the formula
AUCS = area under curve S—the area under curve S in general
respectively for the complete bootstrap range from 0 to 100 with
AUCStotal = the area under the complete curve S
In order to make the values of the iteration steps comparable, the relation to an ideal tree was used, which would have only bootstrap values of 100, from which the formula
or simply, since AUCStotal max = 100·100
AUCStotal% = the area under the complete curve S as a ratio to an ideal 100% tree
This formula would give 100% if all nodes in the tree had the bootstrap value 100%, and correspondingly less the smaller they become (where the numbers n are weighted). The formulas were implemented in a tool (Node Integrator). One criticism of this procedure could be that the tree with the highest bootstrap values does not have to be the most truthful one. However, the goal was not to optimize or increase the bootstrap values of the iterative multigene guide tree alignments. The fact that the bootstrap values increase is a positive side effect which can be controlled quickly and easily with this method, but the purpose was to record the change of the bootstrap values with this method to determine the end of the iteration loop.
The curves S(b) in the diagram Fig. 4 show the result. A strong increase was clearly visible after the first iteration step. After that, especially poorly supported nodes were slightly improved. After 4 iteration steps, no significant change was visible. After the 5th iteration step, the LSU region was also aligned using the guide tree from iteration step 5 and included in the evaluation (see chapter “MSA of the LSU region and range selection”). The ITS1 and ITS2 alignments and gap matrices of the last step were used for the final tree inferences. After completion of the final phylogeny, the values of the final ML tree from RAxML were additionally included in Table 3 and diagram Fig. 4. The final calculation of the ML phylogeny is explained in chapter “Final maximum likelihood (ML) estimation and bootstrapping”.
Table 3 shows the test results.
It can be seen that already with the first iteration step, there was a substantial leap-like improvement in the alignments. After that, the improvement was small but steady. Only a few more iteration steps had to be done until no significant improvement could be detected. Step 6 is not qualitatively comparable to step 5 as far as the previous phylogeny is concerned; since the LSU region was added, therefore the corresponding values are entered in brackets in Table 3. The best values of the important tracing values are shown in italics.
The final ITS1 and ITS2 alignments were then trimmed with Mega (Tamura et al. 2013) to the exact boundaries of the regions using the motifs (see chapter “Determination of region boundaries of the SSU, ITS1, 5.8S, ITS2 and LSU regions”).
Checking the final ITS1 alignment
Figure 5 shows the final MSA of the ITS1 region. Figure 6 shows the sites differing from the majority rule consensus. Both alignment graphics are already in phylogenetic order of the final total tree.
Sites of the ITS1 alignment diverging from the majority rule consensus; representation from AliView; colours: nucleotides in AliView colour code; coloured sites differ from the majority rule consensus; grey areas are sites matching the consensus; white are indels or missing data; the scale represents the site numbering
The phylogenetically informative content was analysed with Noisy. However, Noisy was not used for MSA filtering (see chapter “MSA filter for divergent regions (not applied)” for reason). The following settings for Noisy were used, deviating from the default:
-
--missing? N—this is necessary because the terminal gaps are filled with “?”.
-
--nogap—this makes sense, because the indels were examined separately for phylogenetic content and an evaluation as 5th state character is not useful—see also chapter “Indel coding method and indel matrices”.
Results:
-
Length of the alignment: 933 sites
-
Constant sites: 413
-
Singleton Sites: 61
-
Phylogenetically informative sites: 77
-
Phylogenetically very informative sites: 382
-
Sum of phylogenetically informative sites with a reliability score > 80%: 404—corresponds to 43.3%
The result as graphical representation shows Fig. 7.
Visual representation of the phylogenetic content of the alignment of the ITS1 region and the columns theoretically remaining/removed by the MSA filter. Graphic created with Noisy. Red: phylogenetically uninformative and constant sites; green: phylogenetically informative and very informative sites; black lines in the yellow bar: phylogenetically informative and very informative sites that have a reliability score > 80% and thus would remain in the alignment after application of the filter
Checking the final ITS2 alignment
Figure 8 shows the final MSA of the ITS2 region. Figure 9 shows the sites differing from the majority rule consensus. Both alignment graphics are also already in phylogenetic order of the final total tree.
Sites of the ITS2 alignment diverging from the majority rule consensus; representation from AliView; colours: nucleotides in AliView colour code; coloured sites differ from the majority rule consensus; grey areas are sites matching the consensus; white are indels or missing data; the scale represents the site numbering
The phylogenetic information content was again analysed with Noisy (settings as described above).
Results:
-
Length of the alignment: 700 sites
-
Constant sites: 288
-
Singleton Sites: 44
-
Phylogenetically informative sites: 63
-
Phylogenetically very informative sites: 305
-
Sum of phylogenetically informative sites with a reliability score > 80%: 337—corresponds to 48.1%
The result as graphical representation shows Fig. 10.
Visual representation of the phylogenetic content of the alignment of the ITS2 region and the columns theoretically remaining/removed by the MSA filter. Graphic created with Noisy. Red: phylogenetically uninformative and constant sites; green: phylogenetically informative and very informative sites; black lines in the yellow bar: phylogenetically informative and very informative sites that have a reliability score > 80% and thus would remain in the alignment after application of the filter
The ITS alignments thus prepared were the basis for the indel coding (see chapter “Indel coding method and indel matrices”) model determination and partitioning (see chapter “Partitioning of alignments and indel matrices/model selection for DNA alignments”).
MSA of the LSU region and range selection
Testing and initial alignment of the LSU region
Only LSU sequences reaching at least up to the right end of domain D1 were used and some shorter ones as exceptions which seemed important for rare taxa. After sorting, 745 LSU sequences were available (see sequence Table S1.01 in Supplement S1). For the LSU region, it was initially unclear whether there were areas with indels that were difficult to align (gappy regions). Therefore, indels and also the change of phylogeny when using a multigene guide tree were analysed in more detail. It turned out that the LSU region also contains indels that are difficult to align and must be aligned with a multigene guide tree. This was performed and evaluated in the course of the iteration loop of the ITS1 and ITS2 alignments after the 5th iteration step as final refinement step of the LSU alignment (see chapter “MSA of the problematic ITS1 and ITS2 regions”).
The initial LSU alignment which was used for the alignment tests described below was carried out with Mafft using the iterative refinement method “E-INS-i”. The terminal gaps and the projecting ends of the ITS2 region were removed. The initial LSU motif was the one described in chapter “Determination of region boundaries of the SSU, ITS1, 5.8S, ITS2, and LSU regions”.
For an alignment test with different software or methods, all gaps were removed from this alignment, re-aligned and evaluated using different methods. The following 9 tests were performed:
-
Test 1:
Software: Mafft (on cbrc.jp); version: 7.372; method: L-INS-i
Settings: default
-
Test 2:
Software: Mafft (on cbrc.jp); version: 7.372; method: L-INS-i
Settings: Leave Gappy regions; rest: default
-
Test 3:
Software: Mafft (on cbrc.jp); version: 7.372; method: E-INS-i
Settings: default
-
Test 4:
Software: Mafft (on cbrc.jp); version: 7.372; method: E-INS-i
Settings: Leave Gappy regions; rest: default
-
Test 5:
Software: Probalign (Roshan and Livesay 2006) (over Cipres); version: 1.4
Settings: default
-
Test 6:
Software: Prank (local); version: 140603
Settings: Leave Gappy Regions (+F set); rest: default settings
-
Test 7:
Software: Prank (local); version: 140603
Settings: +F disabled; rest: default settings
-
Test 8:
Software: Prank (local); version: 140603
Settings: Leave Gappy Regions (+F set); -uselogs set; rest: default settings
-
Test 9:
Software: Prank (local); version: 140603
Settings: +F disabled; -uselogs set; rest: default settings
The -logLikelihood score for the model GTR+G (Generalised Time-Reversible—Tavaré 1986) was calculated from the alignments resulting from these tests using JModelTest (Darriba et al. 2012) (via Cipres), since this model was determined to be the best fit model for the LSU region (see chapter “Partitioning of alignments and indel matrices/model selection for DNA alignments”). Table 4 shows the relevant test results.
The method used in test 3 turned out to be the best for the initial alignment; therefore, this method (i.e. Mafft—method: E-INS-i) was used.
Refinement of the LSU alignment using the iterative multigene guide tree
As mentioned above, refinement was performed using the multigene guide tree from iteration step 5 using Prank during the ITS1 and ITS2 MSA iteration loop (for statistical results, see chapter “MSA of the problematic ITS1 and ITS2 regions”). The alignment improved significantly in that process. So, the final alignment method of the LSU region used for the tree inference was Prank with +F disabled, -uselogs and -once set, using the multigene guide tree from iteration step 5.
The left end of the alignment was then exactly trimmed with Mega (Tamura et al. 2013) based on the motif (consensus) ATTTGACCTCAAATCAGG...—see chapter “Determination of region boundaries of the SSU, ITS1, 5.8S, ITS2 and LSU regions”. The frayed right ends were trimmed to a meaningful length. The alignment thus prepared was the basis for the indel coding of the LSU region (see chapter “Indel coding method and indel matrices”) and the partitioning (see chapter “Partitioning of alignments and indel matrices/model selection for DNA alignments”).
Figure 11 shows the 1499 sites long MSA of the LSU region in phylogenetic order of the final tree.
The LSU alignment was analysed in more detail for areas of phylogenetic informative content in order to define the area to be used. This was first checked optically with AliView. The domains could be identified by the sites deviating from the majority rule consensus, as Fig. 12 illustrates by the already final alignment.
Sites of the LSU alignment diverging from the majority rule consensus in phylogenetic order of the final total tree with the left end domains; representation from AliView; colours: nucleotides in AliView colour code; coloured sites differ from the majority rule consensus; grey areas are sites matching the consensus; white are indels or missing data; Yellow areas are divergent “D” areas of the domains. “C” areas are the conserved regions of the domains; the scale represents the site numbering
The phylogenetically informative content in the D regions was clearly recognizable, but pi-positions (parsimony informative positions) were also present in the C regions. This was analysed with Noisy more detailed. Again, Noisy was not used for MSA filtering (reason see chapter “MSA filter for divergent regions (not applied)”). The following settings for Noisy were used, deviating from the default:
-
--missing ?N—this is necessary because the terminal gaps are filled with “?”.
-
--nogap—this makes sense, because the indels were examined separately for phylogenetic content and an evaluation as 5th state character is not useful—see also chapter “Indel coding method and indel matrices”.
Results:
-
Length of the alignment: 1499 sites
-
Constant sites: 946
-
Singleton Sites: 180
-
Phylogenetically informative sites: 201
-
Phylogenetically very informative sites: 172
-
Sum of phylogenetically informative sites with a reliability score > 80%: 285—corresponds to 19%
The result as graphical representation shows Fig. 13.
Visual representation of the phylogenetic content of the alignment of the LSU region and the columns theoretically remaining/removed by the MSA filter. Graphic created with Noisy. Red: phylogenetically uninformative and constant sites; green: phylogenetically informative and very informative sites; black lines in the yellow bar: phylogenetically informative and very informative sites that have a reliability score > 80% and thus would remain in the alignment after application of the filter
The LSU alignment thus prepared was the basis for the indel coding of the LSU region (see chapter “Indel coding method and indel matrices”), model determination and partitioning (see chapter “Partitioning of alignments and indel matrices/model selection for DNA alignments”).
MSA of the 5.8S region
All 5.8S sequences were embedded in between the ITS sequences. These are listed in Table S1.01 in Supplement S1. Within the 5.8S alignment, there were no regions difficult to align and no phylogenetically informative indels were expected. Nevertheless, indels were analysed more detailedly (see chapter “Indel coding method and indel matrices”).
Since the alignment was part of the initial ITS alignment, the 5.8S region only had to be trimmed with Mega (Tamura et al. 2013) and visually checked. The alignment was impeccable, so there was no need for further testing with other alignment procedures or refinement. The final alignment in phylogenetic order of the final total tree is shown in Fig. 14, which clearly shows the conservation even across the entire family.
However, there were also quite clear uniform divergences, for example in section Spadiceogriseae, in which 2 sites deviated unanimously from the whole family, or a deletion that occurs only in clades within Parasola (this is for example a phylogenetically informative indel). Both can clearly be seen in Fig. 15, which shows the sites in 5.8S alignment that deviate from the majority rule consensus.
Sites of the 5.8S alignment diverging from the majority rule consensus; representation from AliView; colours: nucleotides in AliView colour code; coloured sites differ from the majority rule consensus; grey areas are sites matching the consensus; white are indels or missing data; the scale represents the site numbering
The phylogenetically informative content of the 5.8S region was analysed with Noisy. The following settings for Noisy were used, deviating from the default:
-
--missing ?N—this is necessary because the terminal gaps are filled with “?”.
-
--nogap—this makes sense, because the indels were examined separately for phylogenetic content and an evaluation as 5th state character is not useful—see also chapter “Indel coding method and indel matrices”.
Results:
-
Length of the alignment: 167 sites
-
Constant sites: 84
-
Singleton Sites: 39
-
Phylogenetically informative sites: 16
-
Phylogenetically very informative sites: 28
-
Sum of phylogenetically informative sites with a reliability score > 80%: 41—corresponds to 24.6%
Although it is not obvious at first glance, the 5.8S region contains 24.6% phylogenetically informative sites. This is even higher than at the LSU region, which has only 19% informative content (see chapter “MSA of the LSU region and range selection”). The result as a visual representation is shown in Fig. 16.
Visual representation of the phylogenetic content of the alignment of the 5.8S region and the columns theoretically remaining/removed by the MSA filter. Graphic created with Noisy. Red: phylogenetically uninformative and constant sites; green: phylogenetically informative and very informative sites; black lines in the yellow bar: phylogenetically informative and very informative sites that have a reliability score > 80% and thus would remain in the alignment after application of the filter
The use of the 5.8S region was therefore found to be very reasonable.
The 5.8S alignment thus prepared was the basis for the indel coding of the 5.8S region (see chapter “Indel coding method and indel matrices”), model determination and partitioning (see chapter “Partitioning of alignments and indel matrices/model selection for DNA alignments”).
MSA of the β-tub region and its exon extraction
From the β-tubulin region, 297 sequences were present after sorting out (see chapter “Sequence sampling and selection”; see Table S1.01 in Supplement S1). The intron regions were removed from the alignment after the usability study (see chapter “The introns of the haploid nuclear genome”). The alignment did therefore not make any special demands. The sequences were aligned with Mafft using the iterative refinement method “L-INS-i”. This was also tested with the “E-INS-i” method which, however, yielded a worse result. After the removal of some stutter sites, the exons and introns were identifiable.
Figure 17 shows the translation of the complete 621-bp-long nucleotide alignment including the introns in phylogenetic order of the final tree. The pink areas were removed from the alignment. These were the beginning and the end in the area of the terminal gaps and the 2 introns. The numbering of the exons and introns corresponds to the interpretation of Russo et al. (1992).
Translation of the β-tub alignment including introns in phylogenetic order of the final tree; representation from AliView; colours: codons in ClustalX colour code (Larkin et al. 2007)
All introns started with the donator GT and ended with the acceptor AG, which made the extraction of the exons very easy. The amino acid sequence over exon 6 to 7 is divided intron-overstretching.
Figure 18 shows the final β-tub alignment after removal of the introns as codon representation which is 384 sites (128 amino acids) long.
Final β-tub alignment as codon representation; representation from AliView; colours: codons in ClustalX colour code (Larkin et al. 2007)
An ORF that spanned over the range of bp 9 to beyond the end of the consensus sequence out of the alignment is a small part of the total OFR that forms the β-tubulin protein. The match of this ORF was tested with SWISS-MODEL. As expected, this section corresponds to a fragment of the β-tubulin protein models—e.g. SMTL ID 5fnv.1.D (Yang et al. 2016)—which was used as a template for a purely informative calculation of the section model of the consensus amino acid sequence over the alignment. Figure 19 shows this section model.
The β-tubulin alignment is consistently phylogenetically informative, as can already be seen from the presentation of the sites deviating from the majority rule consensus in Fig. 20.
Sites of the β-tubulin alignment diverging from the majority rule consensus; representation from AliView; colours: nucleotides in AliView colour code; coloured sites differ from the majority rule consensus; grey areas are sites matching the consensus; white are missing data; the scale represents the site numbering
With Noisy, the phylogenetically informative fractions of the codon positions were analysed in relation to the respective codon partition. The following settings for Noisy were used deviating from the default:
-
--missing ?N—this is necessary because the terminal gaps are filled with “?”.
-
--nogap—this makes sense because the few gaps present are most likely sequencing errors and cannot be considered phylogenetically informative—see also chapter “Indel coding method and indel matrices”.
The result as a visual representation is shown in Fig. 21. The black bars in the yellow marked area are the sites that would remain in alignment if Noisy was used for filtering the MSA—but MSA filtering was generally not applied (reason see chapter “MSA filter for divergent regions (not applied)”).
Visual representation of the phylogenetic content of the 3 β-tubulin-codon partitions and the columns theoretically remaining/removed by the MSA filter. Graphics created with Noisy. Red: phylogenetically uninformative and constant sites; green: phylogenetically informative and very informative sites; black lines in the yellow bars: phylogenetically informative and very informative sites that have a reliability score > 80% and thus would remain in the alignment after application of the filter
Results:
β-tubulin codon 1 partition (128 sites):
-
Constant sites: 87
-
Singleton Sites: 8
-
Phylogenetically informative sites: 22
-
Phylogenetically very informative sites: 11
-
Sum of phylogenetically informative sites with a reliability score > 80%: 30—corresponds to 23.4%
β-tubulin codon 2 partition (128 sites):
-
Constant sites: 104
-
Singleton Sites: 14
-
Phylogenetically informative sites: 6
-
Phylogenetically very informative sites: 4
-
Sum of phylogenetically informative sites with a reliability score > 80%: 7—corresponds to 5.5%
β-tubulin codon 3 partition (128 sites):
-
Constant sites: 7
-
Singleton Sites: 6
-
Phylogenetically informative sites: 36
-
Phylogenetically very informative sites: 79
-
Sum of phylogenetically informative sites with a reliability score > 80%: 109—corresponds to 85.2%
The results show that codon position 3 has a multiple information content compared to codon positions 1 and 2. See also diagram Fig. 29.
MSA of the ef-1α region and its exon extraction
From the ef-1α region, 185 sequences were present after sorting out (reason see chapter “Sequence sampling and selection”; see Table S1.01 in Supplement S1). Since the intron regions were also removed from the alignment (see chapter “The introns of the haploid nuclear genome”), this alignment also had no special requirements. The sequences were also aligned with Mafft using the iterative refinement method “L-INS-i”, for testing purposes also with the “E-INS-i” method, which also yielded a slightly worse result here. After the removal of some stutter sites the exons and introns were identifiable.
Figure 22 shows the translation of the complete 1338-bp-long nucleotide alignment including the introns in phylogenetic order of the final tree. The pink areas were removed from the alignment. These were the start and the end in the area of the terminal gaps, and the 4 introns and the exon at the end, as Fig. 23 shows.
Translation of the ef-1α alignment including introns in phylogenetic order of the final tree; representation from AliView; colours: codons in ClustalX colour code (Larkin et al. 2007)
Final ef-1α alignment as codon representation; representation from AliView; colours: codons in ClustalX colour code (Larkin et al. 2007)
All introns also started with the donator GT and ended with the acceptor AG. The amino acid sequence over exons 2 to 3 and 3 to 4 are divided intron-overstretching.
Figure 23 shows the final ef-1α alignment after removal of the introns as codon representation which is 993 sites (331 amino acids) long.
An ORF that spanned over the range of bp 10 to beyond the end of the consensus sequence out of alignment is a section of the entire OFR that forms the ef-1α protein. The match of this ORF was checked with SWISS-MODEL. As expected, the section corresponds to a fragment of the elongation factor 1α proteins—e.g. SMTL ID 2b7c.1 (Pittman et al. 2006)—which was used as a template for a calculation of the section model of the consensus amino acid sequence over the ef-1α alignment. Figure 24 shows this section model of the complete ORF.
The ef-1α alignment is consistently phylogenetically informative as can already be seen from the presentation of the sites deviating from the majority rule consensus in Fig. 25.
Sites of the ef-1α alignment diverging from the majority rule consensus; representation from AliView; colours: nucleotides in AliView colour code; coloured sites differ from the majority rule consensus; grey areas are sites matching the consensus; white are indels or missing data; the scale represents the site numbering
Here too, Noisy was used to analyse the phylogenetically informative proportions of the codon positions in relation to the respective codon partition. The same settings as for the β-tubulin alignment were used.
The results are shown as an optical representation in Fig. 26. The black bars in the yellow marked area are the sites that would remain in alignment if Noisy was used for filtering the MSA—but MSA filtering was generally not applied (reason see chapter “MSA filter for divergent regions (not applied)”).
Visual representation of the phylogenetic content of the 3 ef-1α codon partitions and the columns theoretically remaining/removed by the MSA filter. Graphics created with Noisy. Red: phylogenetically uninformative and constant sites; green: phylogenetically informative and very informative sites; black lines in the yellow bars: phylogenetically informative and very informative sites that have a reliability score > 80% and thus would remain in the alignment after application of the filter
Results:ef-1α codon 1 partition (331 sites):
-
Constant sites: 213
-
Singleton Sites: 42
-
Phylogenetically informative sites: 36
-
Phylogenetically very informative sites: 40
-
Sum of phylogenetically informative sites with a reliability score > 80%: 53—corresponds to 16%
ef-1α codon 3 partition (331 sites):
-
Constant sites: 228
-
Singleton Sites: 45
-
Phylogenetically informative sites: 28
-
Phylogenetically very informative sites: 30
-
Sum of phylogenetically informative sites with a reliability score > 80%: 45—corresponds to 13.6%
ef-1α codon 3 partition (331 sites):
-
Constant sites: 33
-
Singleton Sites: 7
-
Phylogenetically informative sites: 97
-
Phylogenetically very informative sites: 194
-
Sum of phylogenetically informative sites with a reliability score > 80%: 254—corresponds to 76.7%
The results show that in the ef-1α gene, the information content of codon position 3 is several times higher than that in the codon positions 1 and 2. See also diagram Fig. 29.
The introns of the haploid nuclear genome
It was investigated whether the introns of the β-tubulin and the ef-1α region are usable for phylogeny. Therefore, only the completely present introns were extracted from the alignment with Mega (Tamura et al. 2013). Without a guide tree, the introns cannot be aligned at all. Therefore, the alignment had to be done at a later time. The alignment was done with the last guide tree from the ITS alignment iteration loop (see chapter “MSA of the problematic ITS1 and ITS2 regions”) with Prank. The settings of Prank were +F disabled; -uselogs set because these settings gave the best results in the test; additionally, -prunetree was set because the guide tree contained all sequence sets. Furthermore, -once was set to switch off the further iterations at Prank.
The result is it turned out that all introns can be aligned on a lower clades level, but because of the extreme divergence, they are ambiguous and therefore unsafe and not usable for phylogeny. The introns were therefore not used for this study.
MSA filter for divergent regions (not applied)
Two reasons are often cited why divergent areas should be excluded from the alignments by MSA filters. On the one hand, the computing time becomes shorter. On the other hand, if the difference between the sequences in the divergent areas is so distinctive that a false alignment occurs, the phylogeny is distorted accordingly. By excluding these “gappy regions”, the phylogeny would be more accurate. This is correct in principle, but only if there is a false alignment in these “gappy regions”. If an expansion of the sequences is present only, but all bases in the “gappy regions” are aligned correctly, then exactly the opposite happens: instead of removing incorrectly aligned areas, phylogenetic information is removed. This exactly was found out in the study “Current Methods for Automated Filtering of Multiple Sequence Alignments Frequently Worsen Single-Gene Phylogenetic Inference” Tan et al. (2015). This study has also shown that, at the current state of the art, no filtering of divergent areas should be used at all.
Our own results also clearly showed the high phylogenetic information content of the indels (as explained below). For these reasons, the ITS and LSU regions relevant in this respect were aligned in our study with an iterative multigene guide tree method, without applying any filters for divergent regions. Similarly, no MSA filters were applied to the other regions. One exception are the intron regions, which do not allow proper alignment. They had to be excluded as usual.
Indel coding method and indel matrices
Selection of the indel coding method
The best known methods for indel coding are “5th-state coding”, SIC = “simple indel coding” (Simmons and Ochoterena 2000) and MCIC = “modified complex indel coding” (Müller 2006). As it was recognized in the study “The relative performance of indel-coding methods in simulations” Simmons et al. (2007), 5th state coding is the coding that contains the highest apparent phylogenetic information compared to other methods. The authors of study “Re-Mind the Gap! Insertion – Deletion Data Reveal Neglected Phylogenetic Potential of the Nuclear Ribosomal Internal Transcribed Spacer (ITS) of Fungi” Nagy et al. (2012) rightly state, however, that this method evaluates each multiple indel as a multiple biological event and is therefore rather unsuitable. Simmons et al. (2007) come to the conclusion that SIC and MCIC are superior to all other methods and approximately equivalent in performance. However, since MCIC is more critical in terms of coding bias correction (see chapter “Models for the indel partitions”), the SIC procedure was chosen for the present study. SeqState (Müller 2005) was selected as coding software.
Calculation of the number of phylogenetically informative gaps
For all statistical evaluations, the number of phylogenetically informative gaps of the matrices was calculated by

with
- I:
-
number of informative gaps in the matrix
- G:
-
gap position (column)
- n:
-
number of the gap position G
- x:
-
total number of gap positions G in the matrix
- C:
-
information (0/1) of gap position G with number n
- s:
-
sequence
- A:
-
gap is absent
- P:
-
gap is present
Gap matrices of ITS1, 5.8S and ITS2 regions
The gap matrices of the ITS1 and ITS2 regions are shortened by the MSA refinement using the iterative multigene guide tree and gradually approach the length that corresponds to optimal truthfulness. Figure 27 shows as an example the gap matrices of the ITS1, 5.8S and ITS2 regions in a row after the last iteration step in phylogenetic order of the final tree.
Results ITS1 region:
-
Sum of gap positions (total): 900
Of those:
-
Informative gap positions: 676—corresponds to 75.1%
-
Uninformative gap positions: 224—corresponds to 24.9%
Results 5.8S region:
-
Sum of gap positions (total): 18
Of those:
-
Informative gap positions: 3—corresponds to 16.7%
-
Uninformative gap positions: 15—corresponds to 83.3%
Results ITS2 region:
-
Sum of gap positions (total): 838
Of those:
-
Informative gap positions: 625—corresponds to 74.6%
-
Uninformative gap positions: 213—corresponds to 25.4%
The astonishingly high values of ITS1 and ITS2 prove the high information content and thus the importance of the indels, but also that MSA filters should not be used. See also diagram Fig. 29.
Gap matrix of the LSU region
Figure 28 shows the gap matrix of the final LSU alignment, also in phylogenetic order of the final total tree. The LSU region also contains distinct indels, especially at the level of higher phylogeny, as can be clearly seen in Fig. 28.
Results:
-
Sum of gap positions (total): 218
Of those:
-
Informative gap positions: 94—corresponds to 43.1%
-
Uninformative gap positions: 124—corresponds to 56.9%
Gap matrices of the β-tub and ef-1α regions
It was to be expected that hardly any phylogenetically informative indels were present in these regions. Nevertheless, this was investigated.
Result for the β-tub gap matrix:
-
Sum of gap positions (total): 11
Of those:
-
Informative gap positions: 4—corresponds to 36.4%
-
Uninformative gap positions: 7—corresponds to 63.6%
-
Gap positions—divisible by 3: 0
Result for the ef-1α gap matrix:
-
Sum of gap positions (total): 24
Of those:
-
Informative gap positions: 2—corresponds to 8.3%
-
Uninformative gap positions: 22—corresponds to 91.7%
-
Gap positions—divisible by 3: 3—corresponds to 12.5%
-
Informative gap positions—divisible by 3: 1—corresponds to 4.2%
Since there were no usable indels in these regions, no indel partitions were created for the β-tub and ef-1α regions.
Proportion of information content of the regions
After completion of the last iteration step, the approximate information content of all final alignments and gap matrices in relation to the total length of all partitions was examined on the basis of the informative sites or binary positions. Table 5 summarizes the lengths and informative positions of all regions and gap matrices. The phylogenetically informative sites with a reliability score > 80% were used for DNA alignments. For the gap matrices (indels), the number of informative gaps was used. Since the significance of the gap matrices (dual system ➔ base 2) is 21 and that of the nucleotides (quadral system ➔ base 4) is 41, only 50% of the significance of the gap matrices was used for the calculation of information content.
The diagram in Fig. 29 shows the approximate percentages of the information content of all alignments and gap matrices used in this study, relative to the total length of all partitions.
Approximate percentage of the information content of all alignments and gap matrices used in this study, relative to the total length of all partitions; red font: indels; colour marking as in Table 5
Of course, this description only applies to the case examined in the present study and is only approximate. In addition, it was not taken into account that many sequence sets lacked sequences outside the ITS1 and ITS2 regions.
Partitioning of alignments and indel matrices/model selection for DNA alignments
Partitioning method and software
The alignments of the individual regions were not sub-partitioned in smaller parts, since there is still no properly functioning algorithm for it. The k-means algorithm sometimes used for this purpose is no longer recommended by the authors of PartitionFinder except for morphology matrices, as it has been proven (see e.g. Baca et al. 2016) that it generates bad inference for the following phylogenetic analysis. Therefore, the biologically logical pre-partitioning of the individual DNA alignments, codon position alignments and indel matrices was only used as previously described. However, in order to avoid over-partitioning, all alignments and gap matrices were analysed with PartitionFinder for the best partitioning scheme.
Partitioning was performed in 2 steps. The first level could only be carried out without a guide tree for the first and intermediate alignments. The second level was performed after the multigene guide tree iteration, with the final alignments and also with guide tree. Attempts with software which included codon models failed—obviously because of the amount of data.
Settings in PartitionFinder:
The following settings were used in PartitionFinder:
-
User tree: not possible for the first partitioning step and for the intermediate partitioning steps. Used for final partitioning.
-
Branch length linking between partitions: The phylogeny software used in this study (MrBayes and RAxML) support both unlinked branch lengths. However, it was assumed as usual that the branch lengths between the partitions evolve in equal rates, therefore “branchlengths = linked” was used.
-
Evolution models for the nucleotide partitions: all models supported by MrBayes were chosen except those involving proportions of invariable sites. MrBayes supports the following models: JC, K80, SYM, F81, HKY, GTR, JC+G, K80+G, SYM+G, F81+G, HKY+G, GTR+G, JC+I, K80+I, SYM+I, F81+I, HKY+I, GTR+I, JC+I+G, K80+I+G, SYM+I+G, F81+I+G, HKY+I+G, GTR+I+G. To avoid the ping-pong effect described by Rannala (2002), Nylander et al. (2004) and later Stamatakis (2006), which occurs when gamma distribution and proportion of invariant sites (+I) are applied simultaneously, the +I option was not used, although in many publications this is not the case. Based on this, the following models were included: JC, K80, SYM, F81, HKY, GTR, JC+G, K80+G, SYM+G, F81+G, HKY+G, GTR+G. RAxML does not support all of these models, but RAxML was used as a secondary phylogeny software.
-
Evolution model for the indel partitions: for the reason described in the following chapter “Models for the indel partitions”, acquisition bias correction (Lewis 2001) should be enabled. For this purpose, PartitionFinder provides the model “BINARY+G+A”, which was used.
-
Information Criteria: always 2 runs were started. One with the corrected Akaike information criterion (AICc) and one with the Bayesian information criterion (BIC). As the tests of the authors of PartitionFinder showed, a significant difference between the result when using BIC and AICc is rarely to be expected. However, in the study “The relative performance of AIC, AICc and BIC in the presence of unobserved heterogeneity” Brewer et al. (2016), the authors showed that the BIC often produces better results for data sets with high heterogeneity. Our tests showed that in the data set used in the present study, the BIC score was always more constant in the results between different calculation programs. The results of the two runs were compared and evaluated—see below.
-
Search algorithm: “all” was used for all analyses, with the exception for the DNA total phylogenies, where “greedy” was used, since “all” would have caused an exorbitantly high calculation time.
-
Search software: only PhyML (Guindon et al. 2010) could be used for DNA partitions, since RAxML does not provide the required models. For the indel partitions, however, only RAxML could be used, since only this provides the BINARY+G+A model.
First and intermediate partitions of the overall phylogeny
The previously described 4 DNA alignments, 6 codon position alignments and 4 indel matrices were programmed as input in PartitionFinder. These were always those from the previous iteration step, respectively the first alignments/first indel matrices at the first partitioning. The partitions of the individual iteration steps were combined according to the respective results and used for the next iteration step.
Final partitioning of the overall phylogeny
After the multigene guide tree iteration, the resulting final 4 DNA alignments, 6 codon position alignments and 4 indel matrices were programmed as input in PartitionFinder. The final guide tree from the multigene guide tree iteration was programmed for the final partitioning as described above.
Result for the nucleotide alignments:
The results were different for both information criteria:
-
Result according to the BIC information criterion: all 10 DNA partitions as single partitions
-
Result according to the AICc information criterion: 5 partitions combined by the following data blocks (ITS1, ITS2) (LSU, BET1) (BET2, ALP1, ALP2) (BET3, ALP3)
Note that “BET” is used as a shortcut for the β-tub codon alignments and “ALP” is used for the ef-1α alignments.
Since over-partitioning is less critical than under-partitioning (see e.g. Brown and Lemmon 2007) and because partitioning according to the BIC information criterion produced better convergence values in the pre-tests, the BIC information criterion was chosen. The 10 DNA partitions were therefore used as single partitions.
Result for the indel matrices:
The results were different for the BIC and AICc information criteria as well. Note that “IND” is used as a shortcut for “indel matrix”.
-
Result according to the BIC information criterion: 3 partitions combined by the following data blocks: (IND_ITS1, IND_ITS2) (IND_58S) (IND_LSU)
-
Result according to the AICc information criterion: 1 partition combined by all data blocks: (IND_ITS1, IND_58S, IND_ITS2, IND_LSU)
The partitioning scheme was chosen according to the BIC information criterion as well. The 3 indel partitions therefore were combined.
Final partitioning scheme of the total phylogeny and models for the DNA partitions for MrBayes
The partitioning schemes determined were combined to form the following final partitioning scheme of the total phylogeny:
-
ITS1 = 1-933;
-
58S = 934-1100;
-
ITS2 = 1101-1800;
-
LSU = 1801-3299;
-
BETcodon1 = 3300-3683\3;
-
BETcodon2 = 3301-3683\3;
-
BETcodon3 = 3302-3683\3;
-
ALPcodon1 = 3684-4676\3;
-
ALPcodon2 = 3685-4676\3;
-
ALPcodon3 = 3686-4676\3;
-
IND_ITS1_ITS2 = 4677-5576 5595-6432;
-
IND_58S = 5577-5594;
-
IND_LSU = 6433-6650;
This was used for the final phylogeny.
The calculated best fit models proposed by PartitionFinder for the final partitioning scheme using the guide tree were used for MrBayes. These were:
-
Partition ITS1: GTR+G
-
Partition 58S: K80+G
-
Partition ITS2: GTR+G
-
Partition LSU: GTR+G
-
Partition BETcodon1: SYM+G
-
Partition BETcodon2: GTR+G
-
Partition BETcodon3: GTR+G
-
Partition ALPcodon1: HKY+G
-
Partition ALPcodon2: SYM+G
-
Partition ALPcodon3: GTR+G
Partitioning for the individual phylogenies (β-tubulin and ef-1α alignments)
For the partitioning of the single phylogenies for the combinability tests and detailed error check (see chapter “Combinability tests of loci, detailed error check of sequence sets from vouchers”), only the alignments from the haploid nuclear genome had to be examined, since the individual regions were not partitioned in smaller parts as described above. For the single phylogeny of ITS1+5.8S+ITS2+indels and for LSU+indels, the final partitioning scheme as described above was reduced to the respective regions (ITS or LSU). For the β-tubulin alignment and the ef-1α alignment, a single calculation was performed in each case. The settings for PartitionFinder were also as described above.
Partitioning of the β-tubulin alignment for the single phylogeny:
The results were different for the BIC and AICc information criteria:
-
Result according to the BIC information criterion: Codon positions 1, 2, and 3 as separate partitions
-
Result according to the AICc information criterion: Codon positions 1, 2, and 3 as one partition
Again, the partitioning scheme was chosen according to the BIC information criterion.
Partitioning of the ef-1α alignment for the single phylogeny:
There was no difference between the BIC and AICc information criteria.
Result according to the BIC and AICc information criteria: Codon position 1, 2, and 3 as separate partitions.
The partitions were combined accordingly.
Models for the indel partitions
The programs used for the tree inferences (MrBayes and RAxML) do not provide methods that consider indels using realistic stochastic models. However, both programs provide alternatives to include indel partitions in the calculation. For indel partitions and other binary or multi-state partitions, models with “acquisition bias correction” should be used, for example, the two-parameter model “Mkv” (Lewis 2001), which is often proposed and available in both programs. In MrBayes, a model similar to the F81 model (Felsenstein 1981) was implemented especially for restriction sites and binary partitions, which is also proposed for indel partitions by the authors of MrBayes. Therefore, it was chosen for the analyses with MrBayes. Since version 8, RAxML includes a two parameter model (two state time-reversible model) for binary partitions, which was chosen for the indel partitions processed in RAxML. RAxML also provides the acquisition bias correction according to the method of Lewis (2001), which was used.
Settings at MrBayes for the indel partitions
The following settings were used:
-
Data type: the data type “Restriction” was used for the reason described above.
-
Model: the model described above is selected automatically by MrBayes as soon as data type “Restriction” has been set and does not need to be programmed.
-
State Frequencies: since gap matrices are not matrices with arbitrary state labels, the stationary state frequencies were left according to the default setting, i.e. estimated according to the Dirichlet function.
-
Across-Site Rate Variation: a gamma distribution was assumed.
-
Coding Bias: since the determination of gaps is made by the sequence length change, neither always present nor always absent states can be recorded. Thus, the setting “coding=variable” is the correct one for this partition and was set.
Settings in RAxML for the indel partitions
The following settings were used:
-
Data type: BIN
-
Correction for acquisition bias (ASC_): yes
-
Acquisition bias correction type: Lewis
Bayesian tree inference of the phylogeny and Bayesian posterior probabilities
The main part of Bayesian MCMCMC analysis (Metropolis-coupled Markov Chains with Monte Carlo simulation) was done via Cipres with MrBayes 3.2.6 64-bit, as parallel version on 8 processors of the Cipres cluster at the San Diego Supercomputer Center.
The previously mentioned 1744 taxon sets were programmed as input. The total length of the alignments and matrices was 6650 characters, with 4676 for the DNA alignments and 1974 for the gap matrices. The selected data type was “mixed”. The missing characters were programmed as described above with “?”, the gaps with “-”.
MrBayes commands:
-
dimensions ntax=1744 nchar=6650;
-
Format datatype=mixed(dna:1-4676,restriction:4677-6650) missing=? gap=- interleave=no;
The 13 partitions from PartitionFinder described in chapter “Partitioning of alignments and indel matrices/model selection for DNA alignments” were programmed.
MrBayes commands:
-
charset ITS1 = 1-933;
-
charset 58S = 934-1100;
-
charset ITS2 = 1101-1800;
-
charset LSU = 1801-3299;
-
charset BET1 = 3300-3683\3;
-
charset BET2 = 3301-3683\3;
-
charset BET3 = 3302-3683\3;
-
charset ALP1 = 3684-4676\3;
-
charset ALP2 = 3685-4676\3;
-
charset ALP3 = 3686-4676\3;
-
charset IND_ITS1_ITS2 = 4677-5576 5595-6432;
-
charset IND_58S = 5577-5594;
-
charset IND_LSU = 6433-6650;
-
partition favored= 13: ITS1, 58S, ITS2, LSU, BET1, BET2, BET3, ALP1, ALP2, ALP3, IND_ITS1_ITS2, IND_58S, IND_LSU;
-
set partition=favored;
The models resulting from PartitionFinder described under “Partitioning of alignments and indel matrices/model selection for DNA alignments” were also programmed for the DNA partitions and the model described under “Models for the indel partitions” was programmed for the indel partitions.
MrBayes commands:
-
lset applyto=(1) nucmodel=4by4 nst=6 rates=gamma; [GTR+G model for ITS1 Partition]
-
lset applyto=(2) nucmodel=4by4 nst=2 rates=gamma; prset applyto=(2) statefreqpr=fixed(equal); [K80+G model for 58S Partition]
-
lset applyto=(3) nucmodel=4by4 nst=6 rates=gamma; [GTR+G model for ITS2 Partition]
-
lset applyto=(4) nucmodel=4by4 nst=6 rates=gamma; [GTR+G model for LSU Partition]
-
lset applyto=(5) nucmodel=4by4 nst=6 rates=gamma; prset applyto=(5) statefreqpr=fixed(equal); [SYM+G model for BET1 Partition]
-
lset applyto=(6) nucmodel=4by4 nst=6 rates=gamma; [GTR+G model for BET2 Partition]
-
lset applyto=(7) nucmodel=4by4 nst=6 rates=gamma; [GTR+G model for BET3 Partition]
-
lset applyto=(8) nucmodel=4by4 nst=2 rates=gamma; [HKY+G model for ALP1 Partition]
-
lset applyto=(9) nucmodel=4by4 nst=6 rates=gamma; prset applyto=(9) statefreqpr=fixed(equal); [SYM+G model for ALP2 Partition]
-
lset applyto=(10) nucmodel=4by4 nst=6 rates=gamma; [GTR+G model for ALP3 Partition]
-
lset applyto=(11) coding=variable rates=gamma; [settings for IND_ITS1_ITS2 Partition]
-
lset applyto=(12) coding=variable rates=gamma; [settings for IND_58S Partition]
-
lset applyto=(13) coding=variable rates=gamma; [settings for IND_LSU Partition]
-
No outgroup was programmed. The model parameters over the partitions were set to be unlinked and “ratepr” was set to “variable” to allow the partitions to evolve at different rates.
MrBayes command:
-
unlink statefreq=(all) revmat=(all) shape=(all) pinvar=(all) tratio=(all);
-
prset applyto=(all) ratepr=variable;
The remaining parameters were left at default settings. This resulted in 39 active parameters which were monitored with Tracer.
One problem (concerning the flood of data) was the sample frequency and the diagnostic frequency. After a test run, which took 18 days computing time, these values could be calculated more closely. According to this estimation, about 100 million generations were necessary, but for safety reasons, the sample frequency was calculated for 120 million generations with a maximum of 2GB sampled trees and diagnostic data. So the following final values were set:
-
Sample frequency of Markov chains and print frequency: 5000 generations
-
Diagnostic frequency: 50000 generations
A stop rule was programmed to deviation of split frequency = 0.01, but this was done just routinely, as a very long estimated computing time (120 to 160 days) and continuous monitoring would have meant that it could have been stopped manually. The number of generations was gradually increased, while continuous monitoring the parameters with Tracer. Topology convergence diagnostic was enabled. Two independent analyses were used (anyway, the stop rule needs at least two analyses to compare the split frequencies between two runs). One cold chain and three incremental heated Markov chains were programmed, i.e. a total of 8 MCMCMC. For the convergence diagnostics, the first 25% of the runs were discarded.
MrBayes command:
-
mcmc ngen=xxx samplefreq=5000 printfreq=5000 diagnfreq=50000 nruns=2 stoprule=YES stopval=0.01 mcmcdiagn=YES;
“ngen=xxx” stands here for the number of generations used continuously for the next statistics. The increment was 2 million, so that after about each 3 days, an intermediate result could be stored and statistically evaluated. The estimated computing time including statistics and programming breaks was 120 to 160 days. The actual computing time was 150 days.
The average standard deviation of split frequency was recorded continuously and visualized as a diagram. For an analysis of this size, MrBayes converged surprisingly quickly and the ASDSF had a good steepness until the end of the calculation—see diagram Fig. 30.
It took 105.4 million generations to reach ASDSF = 0.01. A total of 42,162 trees were sampled. After removal of the trees within the 25% burn-in phase, the remaining 31,622 trees were used to calculate the 50% majority rule consensus trees and the Bayesian posterior probabilities.
All following phylograms and cladograms follow the 50% majority consensus rule. Branch support values on the branches in blue are MrBayes posterior probabilities; those under the branches are ML bootstrap support values. A -lnL score of 146,382 was reached.
No manual collapsing of the phylograms and cladograms were made (except the desired collapsing to triangle leaves).
Final maximum likelihood (ML) estimation and bootstrapping
As described in chapter “MSA of the problematic ITS1 and ITS2 regions”, a final ML analysis with the final partitioning scheme (see chapter “Partitioning of alignments and indel matrices/model selection for DNA alignments”) was performed after the multigene guide tree iteration. This ML analysis is described in this chapter, while all previous ML analyses have already been described in the corresponding chapters.
The final maximum likelihood analysis was performed using Cipres with RAxML 8.2.10 as parallel version on 12 processors/48 cores. The partitions were programmed according to chapter “Partitioning of alignments and indel matrices/model selection for DNA alignments”:
-
DNA, ITS1 = 1-933
-
DNA, 58S = 934-1100
-
DNA, ITS2 = 1101-1800
-
DNA, LSU = 1801-3299
-
DNA, BET1 = 3300-3683\3
-
DNA, BET2 = 3301-3683\3
-
DNA, BET3 = 3302-3683\3
-
DNA, ALP1 = 3684-4676\3
-
DNA, ALP2 = 3685-4676\3
-
DNA, ALP3 = 3686-4676\3
-
ASC_BIN, IND_ITS1_ITS2 = 4677-5576, 5595-6432
-
ASC_BIN, IND_58S = 5577-5594
-
ASC_BIN, IND_LSU = 6433-6650
No outgroup was programmed.
For all DNA partitions, the GTR substitution matrix (Tavaré 1986) was used under the CAT model of RAxML as this was found to be the best combination in all previous tests. The final optimization was computed under gamma distribution. For the CAT model, 25 distinct rate categories were used (this does not affect the 4 rate categories for the gamma approximation).
For the binary partitions, the “two state time-reversible model” as described in chapter “Models for the indel partitions” was used, with acquisition bias correction, according to the procedure of Paul Lewis (2001).
At the pre-test, 300 ML bootstrap inferences were required up to convergence according to the MRE-based (majority rule) bootstrapping criterion. However, 1000 ML bootstrap inferences were used. One thousand trees were sampled of these and the best tree was labelled with the ML bootstrap support values. This was merged with the Bayesian tree using Treegraph.
A final -lnL score of 146,052 was reached. In all following phylograms, ML bootstrap values are given in black numbers under the branches. Conflict values, i.e. those where the branches were located at a different position than in the Bayesian tree, are shown in square brackets [xx], where the value in the square brackets shows the ML bootstrap value, the fictitious common branch would have had in the ML phylogram. Even if unusual, none of the low ML bootstrap values were deleted.
Combinability tests of loci, detailed error check of sequence sets from vouchers
It was examined whether the different loci may be combined for phylogeny. Additionally, any existing differences between the topology positions of all sequence sets in the single phylogenies and the total phylogeny were checked and, if necessary, the conflicted sequence set discarded. A congruence test of all single topologies to the total topology was performed as well.
For the loci ef-1α, β-tub, LSU, LSU+indels and ITS1+5.8S+ITS2+indels, a ML bootstrap analysis with RAxML was performed in each case and the result was compared with the total phylogeny. Nodes with high support values (> 70%), which differed in position at the individual phylogenies compared to the total phylogeny, were considered a conflict. In such cases, appropriate measures have been taken (exclusion of the corresponding sequence or exclusion of the complete sequence set). Furthermore, for quality assurance purposes, a preliminary tree was calculated with MrBayes without removing the conflict causing sets, while the convergence values and other quality values were recorded. After 2 months of calculation and analysis time, which was necessary to eliminate the conflict causing sequence sets, there were no more conflicts in the higher and lower phylogeny detectable. Likewise, there was no longer any doubt about the combinability of the loci (including gap matrices). Please note that this procedure is not intended to sort out sequences in order to obtain a “desired beautiful result”. Quite the opposite was the goal, namely to keep strongly divergent sequences or sequence sets in the analysis. The goal of this method was to find out those sequences from sets that most likely could not belong together or were faulty due to their different positioning. MrBayes was only able to achieve a useful convergence after the main part of the sequence sets identified as erroneous were removed. This procedure is time-consuming but crucial for the high quality of the final phylogram. After completion of the sorting work, all nodes of the higher phylogeny exhibited excellent probability values (for final result, see e.g. collapsed total phylogram—Fig. 42).
Plausibility check using a “HLPGT” (high-level phylogeny guide tree)
Purpose of the HLPGT
The HLPGT was used to control and ensure the truthfulness of the higher phylogeny from the initial alignment of the ITS1 and ITS2 regions until the final tree inference was done. This was reached by continuously comparing the HLPGT with the alignment guide trees until the final tree was inferred. Please note that the HLPGT was not used as a guide tree for the alignments at all.
Creation and application of the HLPGT
Before the alignment of all loci was done, a phylogenetic tree was calculated from an alignment containing only the sequence sets containing at least one ef-1α and/or β-tubulin sequence (see Table S1.01 in Supplement S1 and Table 1). This “high-level phylogeny guide tree” offers the maximum possible truthfulness for a first alignment concerning higher phylogeny, since it has the lowest missing data level. The areas of the ITS1 and ITS2 regions that are difficult to align have little harmful effects and are of little importance for a high-level phylogeny guide tree, since it is used to detect and control the higher phylogeny before the iterative alignment. The HLPGT is therefore accurate in the higher phylogeny, but still inaccurate in the lower phylogeny. So, this guide tree was not used as a guide tree for the iteration. Such a high-level phylogeny guide tree is particularly advantageous if, as in the case of this study, there are only relatively few sequences that determine the higher phylogeny (LSU, β-tubulin, ef-1α) but many that determine the lower phylogeny (ITS1, ITS2) respectively additionally areas that are difficult to align such as in the ITS1 and ITS2 regions. Several HLPGTs were computed and evaluated using different alignment, partitioning, and calculation methods. Different alignment combinations and alignment software were also investigated and evaluated. The best result was obtained with the following composition:
-
ITS1 to ITS2 aligned with Mafft using the FFT-NS-2 method
-
LSU aligned with Mafft using the E-INS-i method
-
β-tubulin and ef-1α see chapters “MSA of the β-tub region and its exon extraction” and “MSA of the ef-1α region and its exon extraction”
The gap matrices were coded with SeqState (Müller 2005). The best partitioning scheme was determined by using PartitionFinder. The following 11 partitions were found as best scheme: (ITS2+ITS1) (58S) (LSU) (BET1) (BET2) (BET3) (ALP1) (ALP2) (ALP3), (IND_ITS1, IND_ITS2) (IND_LSU, IND_58S). The HLPGT was calculated with RAxML 8.2.10. The HLPGT essentially showed the topology as in Fig. 42, i.e. the same as the final topology; therefore, the representation of the HLPGT as a graphic is omitted at this point.
Comparison with other phylogenetic studies
In addition to several others, the studies Padamsee et al. (2007); Larsson and Örstadius (2008); Vašutová et al. (2008); Nagy et al. (2009, 2011a, 2013a); Nagy, Urban et al. (2010a); Hazi et al. (2011); Nagy, Walther et al. (2011b); Nagy, Vágvölgyi et al. (2013b); Tóth et al. (2013); Örstadius et al. (2015); and Szarkándi et al. (2017) were mainly used for comparison and control purposes of the phylogenetic analysis. Within these studies, some species and sections already appeared at different positions. Even whole genera are shifted (example: Coprinellus—see Örstadius et al. (2015) to Nagy, Urban et al. (2010a) or Nagy, Vágvölgyi et al. (2013b) to Tóth et al. (2013)). The contradictions in these studies were analysed to the best of our ability. However, a comparison with other studies is only meaningful if there were at least qualitatively similar conditions as in this study. Therefore, the phylogeny influencing factors used in the previously mentioned studies were recorded, for example regions used, alignment strategy, indel coding, models, partitions, phylogeny software and others. However, the detailed comparison would go beyond the scope of this study and will not be elaborated.
The rough result
Only 2 studies, namely Nagy et al. (2013a) and Tóth et al. (2013), use an iterative guide tree alignment procedure. In some cases, the higher phylogeny could also be used as a comparison, whereby in Tóth et al. (2013) the phylogenetic tree which described by the authors as uploaded to TreeBASE was missing there and therefore was not comprehensible at all. From all other studies listed above, only the lower phylogeny of some genera, sections or clades could be used for comparison. The comparison was carried out with the best possible precision. Some essential differences between the studies listed above and the present one could be explained by the unfavourable phylogenetic influencing factors chosen in some studies (see chapter “Summary of phylogenetic findings concerning the workflow”).
Summary of phylogenetic findings concerning the workflow
High support values of the final 1744 taxa containing consensus tree were achieved across the entire Psathyrellaceae family using an iterative multigene guide tree alignment procedure for an alignment set consisting of ITS1, 5.8S, ITS2, LSU, β-tub and ef-1α regions and the indel matrices from ITS1, 5.8S, ITS2 and LSU alignments. The high phylogenetic information content of the indels within the ITS1 and ITS2 regions was determined and with it the necessity to include them in the phylogenetic analysis using an iterative multigene guide tree alignment instead of removing difficult alignable areas using MSA filters.
As can be seen from the final phylogram, there are some clades where the internal branches are short, while the terminal branches are relatively long. For example, this is the case with genus Psathyrella—sections Noli-tangere and Spadiceogriseae or genus Candolleomyces. This is a serious phylogenetic problem and phylogenetic analyses are often difficult to reproduce. However, the main problem with Psathyrellaceae is the higher phylogeny, which strongly depends on the workflow, alignment procedure, regions used, partitioning and use of indel matrices, but less on the phylogeny software used to reconstruct the tree. The high phylogenetic content of all regions, but also of the indel matrices, is shown as a percentage in diagram Fig. 29. It should be noted that this diagram applies only to the specific case of this analysis and not to the Psathyrellaceae family in general, but it clearly shows that omitting regions or gap matrices means that phylogenetic information is withheld in such analysis. And this automatically must lead to another, in some cases to a more or less distorted topology. An exception, however, is if only one clade is studied in which one or more of the regions are conserved. For example, in one of the above examples—Psathyrella spadiceogrisea—the LSU region (at least in the range of domains described in the chapter “MSA of the LSU region and range selection”) is completely irrelevant if the clade is studied alone, but of high importance if the clade is studied within other clades with diverging LSU regions. It must therefore be checked at each analysis whether regions can be omitted without problems.
A serious problem is the correct alignment of the ITS region, which cannot be solved without an iterative multigene guide tree, which also includes conserved regions. This fact will also cause a distorted phylogeny, if ignored. For the correct calculation of the root position, the choice of sequence sets for the outgroup is a crucial, important point.
The following points summarize the important factors in large, accurate phylogenetic reconstruction analyses like the present one:
-
Region selection—see chapter “Region selection of molecular phylogenetic markers”.
-
Data quality: ensured by the selection method applied as described above (chapter “Sequence sampling and selection”) and sorting out erroneous sequence sets (chapter “Combinability tests of loci, detailed error check of sequence sets from vouchers”).
-
Sufficient and overlapping alignment length—see chapter “Sequence sampling and selection”.
-
Selection of sequence sets for the outgroup—see chapter “Sequence selection for the outgroup and root branch validation”.
-
Correct alignment of indel-rich areas—see chapters “MSA of the problematic ITS1 and ITS2 regions” to “MSA of the LSU region and range selection”.
-
Using MSA Filters should be omitted—see chapter “MSA filter for divergent regions (not applied)”.
-
Use of indels as gap matrices—see chapter “Indel coding method and indel matrices”.
-
Phylogenetic content of the data—see chapter “Proportion of information content of the regions”.
-
Best possible model selection—see chapters “Partitioning of alignments and indel matrices/model selection for DNA alignments” and “Models for the indel partitions”. Over-fitting of the models to the data leads to a reduction in support values, while under-fitting leads to high support values for a false topology. Ideal is the model that is least complex but sufficient to model the data. Normally, this selection is done by a model test software. But what if the results of the programs are different? In such cases of doubt, the over-fitting model was chosen to avoid the previously described effect, i.e. the correct topology was given priority.
-
Sufficient partitioning with simultaneous avoidance of over-partitioning—see chapter “Partitioning of alignments and indel matrices/model selection for DNA alignments”.
-
Conflict-free combinability of regions—see chapter “Combinability tests of loci, detailed error check of sequence sets from vouchers”.
Phylogenetic results and discussion
Overview
As expected, the genera Coprinopsis, Cystoagaricus, Homophron, Kauffmania, Lacrymaria, Parasola and Typhrasa could be shown to be monophyletic. In contrast, Coprinellus is not uniform and forms two clades. The species group within the clade /patouillardii does not belong to Coprinopsis, as previously thought. It does not belong to Coprinellus sensu stricto either (which is labelled as the clade /Coprinellus A, since that is where the type of the genus belongs). The clade /Coprinellus B deviates morphologically as well as phylogenetically from /Coprinellus A.
Within the historical genus Psathyrella, /candolleana and /supernula are clearly separated, as is /codinae. Psathyrella is thus paraphyletic. Surprisingly, the results show that Galerella floriformis Hauskn. belongs to the Psathyrellaceae. Coprinellus pakistanicus does not belong to Coprinellus but constitutes another separated clade.
There is some residual uncertainty in the higher phylogeny because not as many sequences of the ef-1α- and β-tubulin-region (and LSU region) were available than was the case for ITS sequences. However, despite these shortcomings, this phylogenetic analysis is considered to be relatively robust, as well as being the most accurate that is possible to achieve with the existing data.
The present study shows that the available sequence data of family Psathyrellaceae form different distinct clades. After comparing all available morphologic data, the 16 coloured clades in Fig. 31 were detected as different genera, nine of which already exist and seven new as proposed below. The evolution takes place in two main directions: on one hand in the direction of the crown group /Coprinopsis, on the other hand in the direction of the crown groups /Psathyrella and /Coprinellus.
All in all, the result of the phylogenetic analysis fits well with the main morphological characteristic groups of fungi within the clades. The systematic and nomenclatorial consequences are discussed below.
Coprinellus
As previously mentioned, the historical classification of the taxa which have been assigned to the genus Coprinellus produces a diphyletic system. However, /patouillardii positions itself in between with relatively high divergence to the neighbour clades, as shown by the 360° radial consensus phylogram in Fig. 32.
/Coprinellus A
This clade was found to comprise 15 confirmed and several unclear taxa, which are distributed over 9 subclades. These are shown in the 310° radial consensus phylogram in Fig. 33.
Pileocystidia may be absent or present and are then utriform or lageniform, partially capitate; sclerocystidia do not occur. The spores mostly have a smooth surface, although some species have rough spores or a perisporium. The most important common feature is the veil which is always present, at least partially, and consists of globose to subglobose elements. In addition, chains of subcylindrical cells often appear, which are sometimes thick-walled and encrusted.
The name for /Coprinellus A must be Coprinellus P. Karst., because the type of the genus is included here: Coprinellus deliquescens (Bull.) P. Karst., Bidr Känn Finl Nat Folk 32:542, 1879, designated by Earle (1909:384). Readhead et al. (2001) accepted this typification with explicit reference to the representation in Horak (1968). The synonymization with Coprinus silvaticus Peck is wrong, because Peck (1872) described and drew a fungus that is reminiscent of Parasola (see Melzer 2017). In the possible event that C. deliquescens is rejected because of ambiguity, then Coprinellus tardus (P. Karst.) P. Karst. would be the next valid type.
For the differentiation in sections, the available names are Coprinellus (= /deliquescens), Micacei (Fr.) D.J. Schaf. (= /micaceus), Domestici (Singer) D.J. Schaf. (= /domesticus) and Flocculosi Citérin (= /flocculosus). The remaining clades will be considered below as further sections and the following names will be proposed for them: Aureogranulati (= /aureogranulatus), Curti (= /curtus), Deminuti (= /deminutus), Disseminati (= /disseminatus), and Hepthemeri (= /hepthemerus).
/patouillardii
This clade is extremely sensitive to the applied phylogenetic technique, since inappropriate techniques can result in a very different positioning of this clade. It must be emphasized that in the present work, no manual influence was exerted on the multigene guide tree and no constraints or other leads were programmed, so the calculated positions only resulted mathematically from the described alignment technique under use of an iterative multigene guide tree. The ITS region can be incorrectly aligned with the ITS of the genus Psathyrella, resulting in phylograms where the clade falls within Psathyrella; for example Nagy, Urban et al. (2010a) position the clade /patouillardii close to Psathyrella fagetophila Örstadius & Enderle and Psathyrella umbrina Kits van Wav. In contrast, Örstadius et al. (2015) show /cordisporus (= /patouillardii) to be far from Psathyrella.
Coprinus patouillardii Quél. and Coprinus cordisporus Gibbs were transferred to the genus Coprinopsis (Moreno and Manjón 2010 resp. Krieglsteiner and Gminder 2010), because the pileipellis is supposedly similar. Keirle et al. (2004) have also examined the pileipellis and found “...a cutis of somewhat inflated to cylindrical, radially arranged hyphae…”. Still, it might not be a pure cutis, but more detailed studies are needed here. The statement by Uljé and Noordeloos (1993) “Pileipellis made up of (sub)globose to ellipsoid elements, smooth to granular, up to 50 μm wide” is certainly an editorial mistake because this description clearly refers to the veil.
Rejinders (1979) has analysed the veil elements in detail and commented on this “…immediately over the cap, the hyphae are divided into short cells. There is no sharp boundary between veil and pileus trama, and this is also the case in older stages”.
The phylogenetic results make it clear that /patouillardii cannot be placed in Coprinopsis (see Fig. 31). Proximity to /Coprinellus A is indicated by the structure of the veil; there also are globose to subglobose elements mixed with chains of subcylindrical cells. But there are important morphological differences. In addition to the pileipellis, these are the polygonal, strongly flattened spores. The establishment of a genus is therefore justified and Narcissea will be proposed as the name (see below).
/Coprinellus B
In this clade, 30 unique taxa were found, which always have lageniform (to subutriform) pileocystidia; some also have sclerocystidia. A veil is often lacking; if present, it usually consists of chains of slightly diverticulate cells. Only the species with polygonal spores have a veil of globose to subglobose cells.
The separation of Coprinellus is recommended for phylogenetic and morphological reasons; the name Tulosesus will be proposed below. At first sight, the morphological features do not give any apparent reason for a division into sections. No sharp phylogenetic separation can be recognized between the species with and without a veil. The differences between them are very delicate, but nevertheless they exist.
/supernula
The two previously known species of this clade, Psathyrella supernula (Britzelm.) Örstadius & Enderle and Psathyrella multipedata (Peck) A.H. Sm., are usually found in small to very large clusters of fruiting bodies and have a long pseudorrhiza and green deposits on the cystidia. They show similarities to Coprinellus christianopolitanus Örstadius & E. Larss. regarding the shape of the pileocystidia and presence of green deposits, while the LSU of C. christianopolitanus is almost identical to the LSU of the clade /supernula. The morphological and phylogenetic characteristics allow the clade /supernula to be considered a separate genus. The name Britzelmayria is proposed below. The resulting phylogenetic position of /supernula is shown in the radial phylogram in Fig. 31.
/Psathyrella s. str
The revised genus as refined by this study shows 18 distinct subclades (see Fig. 34). Some of these can be assigned to existing taxonomic categories which can still stand as sections: Cystopsathyra (Singer) Kits van Wav. (= /kellermanii), Pseudostropharia A.H. Sm. (= /caput-medusae), Hydrophilae Romagn. ex Singer (= /piluliformis), Pygmaeae Romagn. (= /pygmaea), Obtusatae (Fr.) Singer (= /obtusata), Spadiceogriseae Kits van Wav. (= /spadiceogrisea), Atomatae Romagn. ex Singer (= /prona), Microrhizae Romagn. ex Singer (= /microrhiza), Lutenses Kits van Wav. (= /lutensis), Pennatae Romagn. ex Romagn. (= /fibrillosa) and Psathyrella Kits van Wav. (= /corrugis).
The remaining subclades will be considered below as further sections and the following names will be proposed for them: Arenosae (= /arenosa), Confusae (= /gordonii), Jacobssoniorum (= /jacobssonii), Noli-tangere (= /noli-tangere), Saponaceae (= /saponacea), Stridvalliorum (= /stridvalii) and Sinefibularum (= /vinosofulva).
It should be noted that few of these clades have striking characteristics found in all species. The only clades which are defined relatively well in morphological terms are /Cystopsathyra with globose veil elements, /vinosofulva without clamps, /lutensis with striking deposits on the cystidia, and /spadiceogrisea with predominantly clavate and sphaeropedunculate marginal cells. Of course, no general statement can be usefully made for clades with only a few or even a single species.
The clade /fibrillosa represents a very fanned-out crown group. It could be further split into four sections, but their morphology would overlap; therefore, this is refrained from for the time being.
/candolleana
This clade can be subdivided into 13 distinct subclades, which could be established as sections from a phylogenetic point of view. The radial phylogram (Fig. 35) illustrates the crown group /candolleana s. str., in which the neotype of Psathyrella candolleana (Fr.) Maire is located. All other subclades are therefore not considered /candolleana. Noteworthy is the extraordinary accumulation of indefinite (or unnamed) species, which have a very similar morphology despite apparent molecular biological separability. The only branch that has an unusual length is that of /typhae; this subclade comprises an indeterminate collection and Psathyrella typhae (Kalchbr.) A. Pearson & Dennis. This species can be recognized by its ecology, growing on aquatic plants just above the water level. In principle, the section name Typhicolae (Romagn.) Singer ex Singer, Sydowia 15:67, 1962 “1961”, could be used for /typhae. However, taking into account the incomplete state of knowledge, a further subdivision of the entire species complex is avoided for the time being as the sections emerging phylogenetically are not sufficiently understood and morphologically distinct.
The search for morphological properties that are valid for each species is difficult and has not been completed for a long time. Members of the candolleana group are not often thoroughly examined. The main criterion which differentiates /candolleana from Psathyrella could be the complete absence of pleurocystidia. Psathyrella probably always has pleurocystidia, although they can be extremely rare and only found after a long search. However, the final tests have not yet been carried out to find exceptions. A second may be the veil, with sphaerocysts with slightly thickened walls being observed for Psathyrella candolleana, P. sulcatotuberculosa (J. Favre) Einhell., P. aberdarensis A. Melzer, Kimani & R. Ullrich and P. bivelata Contu (hence the name). However, further examinations are necessary here. So far, the fleeting veil has rarely been closely observed. Already the phylogenetic results justify the transferring of all the taxa in /candolleana to a separate genus; the name Candolleomyces will be proposed (see below).
/floriformis
In the original description of Galerella floriformis (Hausknecht and Contu 2003), young basidiomata are compared with Coprinus and it is described as “... a very spectacular member of the genus”. Tóth et al. (2013) also draw attention to the special phylogenetic position. It is impossible to place this species in the genus Galerella Earle, and an assignment into Candolleomyces does not seem sensible due to the deliquescent lamellae and the absence of cheilocystidia. Hausknechtia will be proposed below as the name of the new genus.
/codinae
Psathyrella codinae Deschuyteneer, A. Melzer & Pérez-De-Gregorio has no morphological features that would otherwise preclude it from belonging to Psathyrella (compare Deschuyteneer et al. 2018). The phylogenetic result, however, is clear; the species must be transferred to a new genus, to be named as Olotia (see below).
/pakistanicus
The “Coprinellus” specimen MEL 2382843A in the National Herbarium of Victoria is preserved with the find data: Australia, Northern Territory, Darwin, Casuarina coastal Reserve, Darwin surf club 1st parking area, mulch on garden bed, 2014-01-22, leg. G. M. Bonito. The voucher was sequenced (Bonito G, Barrett M, Udovicic F, Lebel T, Fleshy Macrofungi of Northern Tropical Australia: Filling in the sampling gap, unpublished) and filed under GenBank accession number KP012718.1. Within the same clade are two uncultured Psathyrella-species (vouchers 2.54E, 2.67E) from the unpublished work Gouveia GV, Yano de Melo AM, da Costa MM, Gouveia J: Soil microbial diversity of a Brazilian semiarid region. From a phylogenetic point of view, these three collections belong to the same species and form a separate genus. No statement could be made about their characteristics because further information was not available. It is noteworthy that the taxa were determined as both Coprinellus and Psathyrella.
A surprising turn came when the work of Hussain et al. (2018a, 2018b) was published and Coprinellus pakistanicus Usman & Khalid was described. The sequence MH366735 (voucher LAH35322) proved to be 100% identical to KP012718.1, the sequence of the Australian voucher. In fact, there was a detailed description (pages 53–55 and colour photograph on page 45) of the taxon at this time, which allows proposing the new genus Punjabia (see Fig. 82 as well).
/Cystoagaricus
The genus Cystoagaricus Singer emend. Örstadius & E. Larss. includes only lignicolous, rather large species with a fibrous-scaly pileus. The spores are never dominantly ellipsoid in shape, generally being subtriangular, rounded-angular, broadly ellipsoid, mitriform or even irregular in outline instead.
/Typhrasa
The unique feature of the genus Typhrasa Örstadius & E. Larss. is the presence of cystidia with large refractive globules.
/Kauffmania
Kauffmania larga (Kauffman) Örstadius & E. Larss., the only species in this monotypic genus, is a relatively stately fungus with pale spores with no or only an indistinct germ pore. There is no single morphological feature to distinguish it from Psathyrella at first glance.
/Coprinopsis
Of the historical genera analysed in this phylogenetic study, Coprinopsis P. Karst. is the most heterogeneous, in terms of habit and micro-features. Phylogenetically, there are 20 subclades detectable, which are considered here as sections. A system of sections and subsections can be conceived as illustrated by Fig. 36. However, this is avoided as it would be complicated and some apparent gaps could be closed by an increase of the available data (sharpening the boundaries of some taxonomic units) and the discovery of further species.
Usable names are Coprinopsis (= /friesii), Picacei Penn. in Kauffman (= /picacea), Atramentariae (Fr.) D.J. Schaf. (= /atramentaria), Narcotici (Uljé & Noordel.) D.J. Schaf. (= /narcotica), Lanatulae (Fr.) D.J. Schaf. (= /lagopus) and Nivei (Citérin) D.J. Schaf. (= /nivea).
The remaining subclades will be considered below as further sections and the following names will be proposed for them: Alopeciae (= /alopecia), Canocipes (= /canoceps), Cinereae (= /cinerea), Subniveae (= /cortinata), Erythrocephalae (= /erythrocephala), Filamentiferae (= /filamentifera), Geesteranorum (= /geesterani), Krieglsteinerorum (= /krieglsteineri), Melanthinae (= /melanthina), Mitraesporae (= /mitraespora), Phlyctidosporae (= /phlyctidospora), Quartoconatae (= /marcescibilis), Radiatae (= /radiata), and Xenobiae (= /xenobia).
The subclades are each more or less homogeneous in their morphology, although they often lack exclusive characteristics, making it difficult to offer a precise morphological description in some cases. However, relatively clear morphological differentiation is possible for /friesii (veil consisting of strongly diverticulate cells which are often thick-walled), /nivea (non-ellipsoid spores; main element of the veil being cells which are globose, smooth to scattered warty and sometimes incrusted), /atramentaria (robust species with a persistent veil), /melanthina (lignicolous species with very pale spores) and /phlyctidospora (rough to warty spores), and partly for /narcotica (mostly globose, densely warty cells as the main element of the veil). For other subclades, the simplest and most appropriate description is problematic, for example in /alopecia, some species have warty spores and others have smooth spores.
/Lacrymaria
The genus Lacrymaria Pat. is well characterized according to current knowledge. A very rich veil and rough to warty spores are characteristic.
/Homophron
All species of the genus Homophron (Britzelm.) Örstadius & E. Larss. lack a veil; the cystidia are at least partially thick-walled and carry crystals; the spores are pale with an indistinct or absent germ pore.
/Parasola
The subdivision into sections Parasola and Auricomi can no longer be maintained from a morphological and phylogenetic point of view. On the basis of the morphological features, a new classification is proposed, which was supported by the phylogeny (see Fig. 37).
Only Parasola conopilea (Fr.) Örstadius & E. Larss. contrasts with other members of the genus. It is not ephemeral and the spores are always ellipsoid in front as well as lateral view (not being much flattened). The habit is also distinctly different. The phylogenetic results also indicate differences, making it appropriate to establish a section for this species, to be named Conopileae (see below).
The phylogenetic tree, marks, symbols and labels of leaves
Because of the size of the entire tree, the result is initially collapsed at the genus level (Fig. 42) and section level (Fig. 43), then expanded on the following pages to species level. That means, from the large and coarse overview to the detail.
Figure 38 shows a rectangular phylogram as an example to describe all possible marks.
For all subsequent phylograms, the following applies to the marks, symbols and labels of the leaves shown in sample phylogram Fig. 38:
-
a)
Blue numbers on branches: posterior probabilities resulting from the Bayesian tree inference.
-
b)
Black numbers below the branches: ML bootstrap support values, taken from the maximum likelihood analysis in %. Black numbers below the branches in rectangular brackets [xx] are conflict ML values, those in which the position of the node in the ML tree deviated from the Bayesian tree.
-
c)
Short black triangle symbols without length specification represent a general collapse of a clade. The length and height of the triangle have no meaning.
-
d)
Black stretched triangles represent the clades as collapsed groups, whereas the length of the triangle represents the longest path length within the clade. The height of the triangles has no meaning.
-
e)
Green numbers on black triangles are the longest path length (thus the length of the triangle) in expected changes/site.
-
f)
Clamps with clade labels represent, as usual, the intended association of the taxa of a clade. Black arrows on clamps (not shown in example) and on vertical lines of rectangular phylogram parts mean: If a phylogram part of the complete phylogram is split in more than one part (since it was too large to fit on one page), the arrows show the linking of the parts.
-
g)
The labelling of the leaves will be explained by the following example.
Example for the label key:
ILBA Psathyrella piluliformis-type - XYZ
The first letters in the label explain which loci were used for the leave in the analysis: I—ITS region (including the 5.8S region); L—LSU region; B—β-tubulin region; A—ef-1α region. This key follows the original taxon name of the original author(s) of the sequence or sequence set. Note that in many cases, different taxon names were used by the authors for one set, or in the database another name was used as in the corresponding voucher. In this case, the latest or most logical name was selected. The species names as well as the genera were always advertised, never abbreviated to avoid confusion between Coprinellus, Coprinus and Coprinopsis. Terms like “cf.”, “aff.”, “var.” and “f.” have always been printed. If “-type” or similar is printed, the sequence or sequence set was explained by the original authors to be from the type (or similar). Also some important remarks were added to the taxon name separated by a “-”. The last entry “- XYZ” in the example is the voucher number or the most useful reference number if no voucher number was given by the original authors. These are called “Seq.-ID” in Table S1.01 in Supplement S1 and in Table 1. Use these tables to find out the accession numbers of the sequence or sequence set which was/were used for the analysis.
All rectangular phylograms were edited in Treegraph. All radial phylogram and cladogram skeletons were calculated with Dendroscope (Huson and Scornavacca 2012).
Outgroup and critical genera
The outgroup includes three species originally assigned to the Psathyrellaceae. The detailed outgroup is shown in Fig. 39. The used sequences of the outgroup can be found in Table 1. The outgroup position within the entire tree can be seen in Fig. 42 and Fig. 43.
Phylogram part of the outgroup; position in tree see Fig. 42
Örstadius et al. (2015) already discovered that Psathyrella ornatispora M. Villarreal & Esteve-Rav. does not belong to the Psathyrellaceae, but the phylogenetic position was left unclear. The phylogenetic analysis in the present study revealed the position shown in Fig. 39, in close proximity to Agaricaceae.
At the time the alignments were created, there were no sequences from the ITS region of Stagnicola perplexa available. In the course of the work, Karl Soop fortunately provided us a Stagnicola perplexa exsiccate so that the sequencing and phylogenetic positioning could be performed as a separate analysis. The genomic DNA was extracted from the dried fruiting bodies and the ITS region and part of the LSU region was sequenced. The same molecular phylogenetic markers were used for the additional analysis as described above. The analysis, of which the details are not further discussed in this publication, showed that Stagnicola perplexa undoubtedly does not belong to the Psathyrellaceae family. Stagnicola perplexa is closely related to Mythicomyces corneipes. The sequences were deposited in GenBank under accession numbers MK045203.1 (ITS) and MK045260.1 (LSU). Shortly thereafter, in June 2019, an article by Vizzini et al. (2019) was released, which confirmed the findings of the authors. Vizzini et al. (2019) even established the new family Mythicomycetaceae for Stagnicola and Mythicomyces Redhead & A.H. Sm., established in Redhead and Smith (1986) on the basis of Mythicomyces corneipes (Fr.) Redhead & A.H. Sm. We can confirm the findings of Vizzini et al. (2019) (see also phylogram part of the outgroup in Fig. 39). The distinct phylogenetic distance from Mythicomyces to Psathyrellaceae is clearly visible in Fig. 39 and in the phylogram provided by Vizzini et al. (2019). However, the ITS sequence of Mythicomyces corneipes can be well aligned with sequences of the family Psathyrellaceae. If a bad workflow or technique is used, Mythicomyces corneipes phylogenetically shifts into the middle of the family Psathyrellaceae. Detailed representations of Mythicomyces corneipes are also contained in Huhtinen and Vauras (1992), Strittmatter and Obenauer (2013).
Proposed classification and nomenclatural novelties
As already mentioned, based on the molecular phylogenetic evidence and considering the morphological characteristics, the family Psathyrellaceae is divided into 16 genera, as new are proposed and characterized below the genera Narcissea, Tulosesus, Britzelmayria, Candolleomyces, Hausknechtia, Olotia and Punjabia. The radial phylogram in Fig. 40 and the collapsed rectangular phylogram in Fig. 42 show the positions in the context of the existing genera Coprinellus, Psathyrella, Kauffmania, Typhrasa, Cystoagaricus, Parasola, Homophron, Lacrymaria and Coprinopsis. The radial cladogram in Fig. 41 illustrates the numbers and the relationships of the taxa within the genera. Figure 43 shows an overview of both the genera and the sections.
Genus Coprinellus P. Karst.
Overview
Figure 44 first shows the 360° radial phylogram of the complete historical genus Coprinellus, divided into the proposed new system. The genus Tulosesus represented the basal group in the historical system.
Nine sections of genus Coprinellus were possible to identify with the currently available sequence data and morphological features. Figure 45 shows the nine sections of the genus Coprinellus as a radial phylogram. The angle is closed approx. 50° for a better view. The radial cladogram in Fig. 46 illustrates the numbers and the relationships of the taxa in the sections. Figure 47 shows an overview in a phylogram collapsed to section level and serves above all for further orientation; the red brackets refer to the detailed phylograms.
Coprinellus sect. Disseminati Wächter & A. Melzer, sect. nov. MB 831453 (Fig. 48)
Phylogram part of the section Disseminati; position in tree see Fig. 47
Description: Basidiomata small, lignicolous or terrestrial, in groups or caespitose, lamellae never deliquescent. Veil sparse, consisting of chains of often somewhat thick-walled and pigmented subcylindrical and globose cells. Spores are medium-sized, in front view more or less fusiform to ovoid with a central germ pore. Basidia 4-spored. Marginal cells of the lamellae edge clavate, utriform, subcylindrical. Pleurocystidia always absent. Pileocystidia very large, utriform. Clamps absent.
Type species: Coprinellus disseminatus (Pers.) J.E. Lange, Dansk bot. Ark. 9(6): 93, 1938.
Representative:
Coprinellus disseminatus (Pers.) J.E. Lange; Ref.v: SZMC-NL-0786 (Nagy et al. 2011)
Remarks:
The separate subclades clearly reflect the origin of the material. Ko et al. (2001) described a similar phenomenon for Hawaii and East Asia. Either they are geographical varieties or they are closely related species. Coprinellus disseminatus-similis S. Hussain also belongs in this section (Hussain et al. 2018b).
Coprinellus sect. Micacei (Fr.) D.J. Schaf., Field Mycology 11(2):50, 2010 (Fig. 49)
Phylogram part of the section Micacei; position in tree see Fig. 47
Description: Basidiomata medium to large-sized, mostly lignicolous. Lamellae deliquescent or withering. Veil at first distinct, granular, consisting of globose cells with thin connection hyphae, often slightly thick-walled and pigmented. Spores medium-sized with a central germ pore. Basidia 4-spored. Marginal cells of the lamellar edge clavate, sphaeropedunculate, ellipsoid. Pleurocystidia usually present, voluminous. Pileocystidia absent. Clamps present, absent (or overlooked) in Coprinellus truncorum.
Type species: Coprinus micaceus (Bull.) Fr., Epicr Syst Mycol:247, 1838 ≡ Coprinellus micaceus (Bull.) Vilgalys, Hopple & Jacq. Johnson, Taxon 50(1):234, 2001, designated by Lange (1915:38).
Representatives:
Coprinellus micaceus (Bull.) Vilgalys, Hopple & Jacq. Johnson; Ref.v.: SZMC-NL-2744 (Nagy et al. 2011a)
Coprinellus saccharinus (Romagn.) P. Roux, Guy García & Dumas; Ref.v.: SZMC-NL-3888 (Hazi et al. 2011)
Coprinellus truncorum (Scop.) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-1101 (Nagy et al. 2011b)
Remarks:
Section Micacei is the only paraphyletic group in the entire Psathyrellaceae family. Somewhat aside E14512I (Rundell et al. 2015) and 2Di102-1 (Held and Blanchette, 2017) can be found. The identity of these two species is not satisfactorily clarified; the material comes from South America and the Antarctis. There is no information about the morphology.
During the study, it was examined whether Disseminati and Micacei form a common section. All species grow on wood or on pieces of wood, usually in clusters or large groups, the veil and the marginal cells consist mainly of globose elements, the spores tend to be mitri- or fusiform. However, C. disseminatus has pileocystidia, while pleurocystidia are missing. Because of these striking morphological differences, also the very wide phylogenetic distance Micacei was considered a separate paraphyletic group.
Coprinellus campanulatus S. Hussain & H. Ahmad belongs also in this section (Hussain et al. 2018a). It is strange that no pleurocystidia were detected. The identity of C. saccharinus was verified by a comparative study with the own voucher from Germany: Saxony, Kyhna, 29.VI.2009, A. Melzer (AM1265). The sequence is deposited at GenBank as MG696612.1. The question of whether C. pallidissimus (Romagn.) P. Roux, Guy Garcia & S. Roux (≡ Coprinus palllidissimus Romagn.) and Coprinus rufopruinatus Romagn. belong here as suspected by Romagnesi (1976) could only be answered by examining the types.
There are several clades of C. micaceus and C. saccharinus of which material differs in geographical origin. Ko et al. (2001) once again found something similar. Whether those are independent species or a beginning separation due to geographic isolation cannot be assessed here. Presumably, the varieties (or species) of C. micaceus and C. saccharinus as well as C. truncorum can hardly be differentiated morphologically. Therefore, a sequence key was created for the separation. Figure 50 shows the phylogenetic differences across the entire ITS1 to ITS2 region for the complete section Micacei (without the upper two “/sp”-branches), especially the non-consensus sites and the indels. The optimal key area extends from site about 105 to about 160 of the ITS1 region. This area with the prominent key points (outlined in yellow) is shown in Fig. 51. The difference between Coprinellus saccharinus “Europe” and “America” is minimal but clear at this point. Thus, the 5 varieties (or species) of C. micaceus and C. saccharinus as well as C. truncorum are clearly separable.
ITS alignment of the /micaceus, /saccharinus, and /truncorum clades; coloured sites (in AliView colour code) represent the difference from majority rule consensus. Grey areas are sites which match the consensus. White sites are gaps. The yellow area highlights the sites which were used for the sequence key. Presentation from AliView; the scale represents the site numbering
Coprinellus sect. Aureogranulati Wächter & A. Melzer, sect. nov. MB 831454 (Fig. 52)
Phylogram part of the sections Aureogranulati and Coprinellus; position in tree see Fig. 47
Description: Basidiomata medium-sized, terrestrial, lignicolous. Lamellae deliquescent. Ozonium present at the base of the stipe. Veil well developed, brown, granular, consisting of chains of subglobose and subcylindrical, often thick-walled, encrusted, yellow-brown pigmented cells. Spores small to medium in size, phaseoliform in side view, with a central germ pore. Basidia 4-spored. Marginal cells of the lamellar edge clavate, lageniform to subutriform. Pleurocystidia present, but rare. Pileocystidia subutriform. Clamps absent.
Type species: Coprinellus aureogranulatus (Uljé & Aptroot) Redhead, Vilgalys & Moncalvo, Taxon 50(1):232, 2001.
Representative:
Coprinellus aureogranulatus (Uljé & Aptroot) Redhead, Vilgalys & Moncalvo; Ref.v.: CBS973.95 (Nagy et al. 2011)
Remarks:
The presence of an ozonium indicates a transition to the Section Domestici (see below), but the presence of pileocystidia is an important difference.
Coprinellus sect. Coprinellus (Fig. 52)
Description: Basidiomata small to medium-sized, terrestrial or lignicolous. Lamellae slowly deliquescent. Veil sparse, granular, consisting of slightly thick-walled and encrusted globose, subglobose or subcylindrical cells. Spores large-sized, smooth or rough with a perisporium and a central germ pore. Basidia 4-spored. Marginal cells of the lamellar edge lageniform or clavate. Pleurocystidia present or absent. Pileocystidia present. Clamps present or absent.
Representatives:
Coprinellus deliquescens (Bull.) P. Karst.; Ref.v.: LO172-08 (Örstadius et al. 2015).
Coprinellus verrucispermus (Joss. & Enderle) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-2146 (Nagy et al. 2011)
Coprinellus sect. Domestici (Singer) D.J. Schaf., Field Mycology 11(2):51, 2010 (Fig. 53)
Phylogram part of the section Domestici; position in tree see Fig. 47
Description: Basidiomata medium-sized to large, terrestrial or lignicolous. Lamellae deliquescent. Ozonium often present at the base of the stipe. Veil initially strongly developed, small-grained to floccose, consisting of subglobose elements and chains of subcylindrical, often thick-walled, encrusted and brownish pigmented cells. Spores medium-sized, laterally partially phaseoliform, germ pore slightly eccentric. Basidia 4-spored. Marginal cells of the lamellar edge clavate and utriform. Pleurocystidia present. Pileocystidia and clamps absent.
Type species: Coprinus domesticus (Bolton) Gray, Nat Arr Brit Pl 1:635, 1821 ≡ Coprinellus domesticus (Bolton) Vilgalys, Hopple & Jacq. Johnson, Taxon 50(1):233, 2001, designated by Singer (1948:36).
Representatives:
Coprinellus domesticus (Bolton) Vilgalys, Hopple & Jacq. Johnson; Ref.v.: SZMC-NL-2933 (Nagy et al. 2011)
Coprinellus radians (Desm.) Vilgalys, Hopple & Jacq. Johnson; Ref.v.: SZMC-NL-1373 (Nagy et al. 2011)
Coprinellus xanthothrix (Romagn.) Vilgalys, Hopple & Jacq. Johnson; Ref.v.: SZMC-NL-3417 (Nagy et al. 2011)
Remarks:
Descriptions of the sequenced vouchers are hard to find. One of the few exceptions is Yagame et al. (2013) with a very detailed description of a fungus identified as C. domesticus from the rhizome of Cremasta appendiculata (D. Don) Makino. The associated vouchers AF1-1 and AF1-1B are located in a subclade within /domesticus. The report of C. radians from Hawaii by Keirle et al. (2004) is C. domesticus (SFSU DEH1026 and SFSU DEH1765), recognizable by the slender, phaseoliform spores with a central germ pore as well as exclusively clavate to sphaeropedunculate marginal cells of the lamellar edge.
There are great uncertainties in the classification of species, especially for C. radians and C. xanthothrix, which can be found in several different subclades. Both species are very similar and also very variable. The only reliable difference are the spores; those of C. radians are slightly larger, much darker and hardly phaseoliform. For this reason, two own collections of C. radians were examined: Germany: Saxony-Anhalt, Landsberg, 1.IX.2006, A. Melzer (AM783); and Germany: Saxony, Kyhna, 8.X.2006, A. Melzer (AM804). Sequences are deposited at GenBank as MK072830.1 and MK072830.1. The comparative analysis showed 100% compliance with SZMC-NL-1373; consequently, this voucher is the only certain C. radians in the phylogram. In other clades, C. radians appears frequently and in different positions; possibly, these are morphologically extremely similar species. Equally ambiguous is C. xanthothrix; three vouchers (SZMC-NL-3417, SZMC-NL-128-08, TOK12808) are in the immediate vicinity of C. radians. It is hard to imagine that an error in the determination exists. Because C. radians and C. xanthothrix differ in principle only by the spores, they could also be close phylogenetic neighbours. If this interpretation does not apply and the true C. xanthothrix is placed in one of the upper clades, this does not change the membership of the section Domestici. Another member would be C. ellisii (P.D. Orton) Redhead, Vilgalys & Moncalvo, if the proof for an independent species is provided, and most likely also C. albidofloccosus (Locq.) Gminder & Manz. The latter species is extensively characterized in Moreau et al. (2002). In addition, at least four, probably even six, previously unknown taxa are hidden in the section. The entire species complex needs a thorough investigation.
Singer (1948) chose C. domesticus sensu Lange as the type; however, according to a modern view, this is C. xanthothrix; a subsequent correction does not seem desirable.
Note the anamorph of Coprinellus domesticus is Hormographiella verticillata (see Table 6).
Coprinellus sect. Flocculosi (Citérin) Wächter & A. Melzer, comb. & stat. nov. MB 831798 (Fig. 54)
Phylogram part of the sections Flocculosi, Curti, Hepthemeri and Deminuti; position in tree see Fig. 47
Basionym: Coprinus subsect. Flocculosi Citérin, Docums Mycol 22(86):19, 1992
Description: Basidiomata medium-sized to large, terrestrial, lignicolous, herbiolous or rarely fimicolous. Lamellae deliquescent. Veil strongly developed, visible as white to brownish patches, consisting of chains of subcylindrical and subglobose, hyaline or brownish pigmented cells. Spores very large in size with a strikingly eccentric germ pore. Basidia 4-spored. Marginal cells of the lamellar edge clavate, sphaeropedunculate. Pleurocystidia present. Pileocystidia and clamps absent.
Type species: Coprinus flocculosus (DC.) Fr., Epicr Syst Mycol:245, 1838 ≡ Coprinellus flocculosus (DC.) Vilgalys, Hopple & Jacq. Johnson, Taxon 50(1):233, 2001, designated by Citérin (1992:19).
Representative:
Coprinellus flocculosus (DC.) Vilgalys, Hopple & Jacq. Johnson; Ref.v.: SZMC-NL-1567 (Nagy et al. 2011)
Remarks:
C. flocculosus was included in section Domestici, but the much larger and darker spores with a strongly eccentric, often almost dorsal germ pore differentiate it significantly from the other members of this section.
Coprinellus sect. Curti Wächter & A. Melzer, sect. nov. MB 831455 (Fig. 54)
Description: Basidiomata tiny to small, fimicolous. Lamellae deliquescent. Veil brown, granular, consisting of subglobose, thick-walled, incrusted elements. Spores medium-sized to large with an eccentric germ pore. Basidia 4-spored. Marginal cells of the lamellar edge clavate, sphaeropedunculate. Pleurocystidia absent. Pileocystidia and clamps present.
Type species: Coprinellus curtus (Kalchbr.) Vilgalys, Hopple & Jacq. Johnson, Taxon 50(1):233, 2001.
Representative:
Coprinellus curtus (Kalchbr.) Vilgalys, Hopple & Jacq. Johnson; Ref.v.: SZMC-NL-1023 (Nagy et al. 2012a)
Remarks:
The study by Hussain et al. (2018) shows that C. tenuis S. Hussain belongs in this section.
With high probability, there is another species Coprinus curtusoides Bogart (inval.) with smaller spores. Especially the presentation of C. curtus in Keirle et al. (2004) is suspicious, because the spores reach only up to 10 (10.5) μm in length. For a review, the voucher Germany: Nordrhein-Westfalen, Mönchengladbach, 07.VIII.2014, H. Bender (HB20140807A) was sequenced and deposited at GenBank as MK070111.1. The match is excellent; all characteristics fit very well with Bogart’s description.
The vouchers FDBC47 and FDBC45 have their origin in Central America (Romero-Olivares et al. 2013) and represent another independent species.
After the sequence collection for the present study was already completed, a new sequence of Hormographiella candelabrata MH862273.1, strain CBS 517.91 Vu et al. (2019), was uploaded to GenBank. With a separate phylogeny, it was possible to verify that this sequence is identical to the sequences within the /curtus clade, which means that Hormographiella candelabrata is the anamorph of Coprinellus curtus. With that information, all known anamorphs of the Psathyrellaceae family are identified and shown in Table 6.
Coprinellus sect. Hepthemeri Wächter & A. Melzer, sect. nov. MB 831456 (Fig. 54)
Description: Basidiomata tiny to small-sized, fimicolous. Lamellae deliquescent. Veil well developed, granular, consisting of globose, slightly thick-walled, brownish pigmented and usually encrusted cells with 10–50 μm in diam.; Moreover, lageniform, pigmented, basally strongly encrusted cells can be present. Spores medium to large-sized with an eccentric germ pore. Basidia 4-spored. Marginal cells of the lamellar edge clavate, sphaeropedunculate. Pleurocystidia absent. Relatively pointed pileocystidia present, additionally sclerocystidia can occur. Clamps absent.
Type species: Coprinellus hepthemerus (M. Lange & A.H. Sm.) Vilgalys, Hopple & Jacq. Johnson, Taxon 50(1):234, 2001. The spelling hepthemerus is correct because the meaning is hepta = seven, hemera = day.
Representatives:
Coprinellus hepthemerus (M. Lange & A.H. Sm.) Vilgalys, Hopple & Jacq. Johnson; Ref.v.: SZMC-NL-0589 (Nagy et al. 2011)
Coprinellus pusillulus (Svrček) Házi, L. Nagy, T. Papp & Vágvölgyi; Ref.v.: SZMC-NL-0589 (Nagy et al. 2011)
Remarks:
So far, only one species has been established beyond doubt, because whether C. pusillulus is an independent taxon remains questionable. The smaller spores are considered to be the separating feature; however, there are also transitions in the spore size, so that this separating feature is not reliable. See Enderle et al. (1986) and Melzer (2009a).
Like many other sections of the historical genus Coprinellus, the clade /hepthemerus is difficult to get in the right position within the phylogram. If an insufficient workflow is used, it appears at a completely different position in the tree and looks like a separate genus. The cause is that the ITS region can be aligned with sequences of other genera very well (Candolleomyces and Punjabia, both see below). The gappy regions within the ITS regions are impossible to align correctly without a sufficient workflow. To solve this problem, it is especially important here to use a multigene guide tree at the initial alignment steps which takes loci of higher phylogeny into account. Otherwise, the alignment software causes an avalanche-like alignment in the wrong direction and assumes the incorrect positioning to be true. To control the success, a HLPGT (see chapter “Plausibility check using a “HLPGT” (high-level phylogeny guide tree)”) should be used.
Coprinellus sect. Deminuti Wächter & A. Melzer, sect. nov. MB 831457 (Fig. 54)
Description: Basidiomata tiny to small, terrestrial. Lamellae deliquescent. Veil brown, granular, consisting of subglobose, thick-walled, encrusted elements as well as subcylindrical to irregularly shaped cells. Spores medium-sized with a central germ pore. Basidia 4-spored. Marginal cells of the lamellae edge clavate, sphaeropedunculate. Pleurocystidia, pileocystidia and clamps absent.
Type species: Coprinellus deminutus (Enderle) Valade, Index Fungorum 160:1, 2014.
Representative:
Coprinellus deminutus (Enderle) Valade; Ref.v.: SZMC-NL-0761 (Nagy et al. 2011)
Remarks:
So far only one species is known.
Narcissea Wächter & A. Melzer, gen. nov. MB 831471 (Fig. 57)
Etymology: Named after the French mycologist Narcisse Théophile Patouillard.
Description: Basidiomata small-sized, fimicolous or on fertilized soil, sometimes on plant remnants. Veil well developed, granular, consisting of often incrusted globose elements and chains of subcylindrical cells. Spores small to medium-sized, with a tri- to polygonal outline, laterally strongly flattened, germ pore central, often prolonged. Basidia mostly 4-spored. Marginal cells of the lamellar edge lageniform, utriform, interspersed with numerous sphaeropedunculate and clavate cells. Pleurocystidia utriform. Pileocystidia and clamps absent. Figure 56 illustrates the microcharacters.
Type species: Narcissea patouillardii (Quél.) Wächter & A. Melzer (Fig. 55).
Phylogram part of the genus Narcissea; position in tree see Fig. 42
Representatives:
Coprinopsis cordispora (T. Gibbs) Gminder; Ref.v.: LO41-01 (Larsson and Örstadius 2008)
Coprinopsis patouillardii (Quél.) G. Moreno; Ref.v.: SZMC-NL-1687 (Nagy, Urban et al. 2010)
Remarks:
The two sister clades /cordisporus certainly represent different taxa, because the divergence seems to be too high for being a single species. It is possible to accept Coprinus cardiasporus Bender as a valid taxon. Keirle et al. (2004) have studied the complex in detail; there are also two sister clades, one of them including C. cardiasporus; however, the type was not available. At the moment, it can only be stated that the genus certainly includes two described species but with high probability further species as well.
It is not appropriate to use the name Furfurelli Fr. for this section. The original diagnosis “Furfurelli, pileo furfuraceo micaceove, lamellis adnatis, vulgo apice stipitis dilatato in annulum” would not completely contradict, but Fries (1838) mentions species here, which according to modern knowledge belong to different genera. Pennington (1918) defines his section Furfurelli as follows “Pileus with micaceous particles or mealy granules” and mentioned as belonging Coprinus patouillardii and Coprinus radiatus (Bolton) Gray.
New combinations:
Narcissea cordispora (T. Gibbs) Wächter & A. Melzer, comb. nov. MB 831731
Basionym: Coprinus cordisporus T. Gibbs 1908, The Naturalist 614 (March):100, 1908. References: Keirle et al. (2004), Nagy (2007), Uljé and Noordeloos (1993), Vila and Rocabruna (1996), Vila and Rocabruna (2002). Mat. exam.: Germany: Saxony, Kyhna, 1.IX.2008, A. Melzer (AM1171); Choren near Döbeln,19.VI.2010, A. Melzer (AM1408).
Narcissea patouillardii (Quél) Wächter & A. Melzer, comb. nov. MB 831732
Basionym: Coprinus patouillardii Quél. apud Patouillard: Tabulae Analyticae Fungorum 1(3):107, 1884. References: Breitenbach and Kränzlin (1995), Ludwig (2007), Melzer (2010), Uljé and Noordeloos (1993). Mat. exam.: Austria: Scharnstein, IV./2010, G. Wührleitner (AM1751). Germany: Leipzig, 16.IV.2018, L. Kreuer (AM1943).
Tulosesus Wächter & A. Melzer, gen. nov. MB 831799 (Fig. 59)
Etymology: The name is an anagram of Setulosus (see remarks).
Description: Basidiomata tiny to medium-sized, terrestrial, lignicolous, subfimicolous or fimicolous. Lamellae deliquescent or withering. Veil absent or present, if present consisting of chains of diverticulate, subcylindrical cells, sometimes also mixed with subglobose, occasionally encrusted elements, if so then spores with a polygonal outline. Spores mostly large-sized, occasionally medium-sized, rarely small, germ pore predominantly eccentric, rarely central. Basidia mostly 4-spored. Marginal cells of the lamellar edge very often clavate and sphaeropedunculate, sometimes mixed with lageniform cystidia, rarely purely lageniform. Pleurocystidia present or absent. Pileocystidia always present, often (sub-) capitate, sclerocystidia often present. Clamps present or absent.
Type species: Tulosesus callinus (M. Lange & A.H. Sm.) Wächter & A. Melzer (see Fig. 58).
Representatives:
Coprinellus amphithallus (M. Lange & A.H. Sm.) Redhead, Vilgalys & Moncalvo; Ref.v.: L128 (Nagy et al. 2012)
Coprinellus angulatus (Peck) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-0906 (Nagy et al. 2011)
Coprinellus bisporiger (Buller ex P.D. Orton) Redhead, Vilgalys & Moncalvo; Ref.v.: Daams7198 (Nagy et al. 2011)
Coprinellus bisporus (J.E. Lange) Vilgalys, Hopple & Jacq. Johnson; Ref.v.: SZMC-NL-0152 (Hazi et al. 2011)
Coprinellus brevisetulosus (Arnolds) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-1445 (Hazi et al. 2011)
Coprinellus callinus (M. Lange & A.H. Sm.) Vilgalys, Hopple & Jacq. Johnson; Ref.v.: SZMC-NL-1931 (Nagy, Walther et al. 2011)
Coprinellus canistri (Uljé & Verbeken) Doveri & Sarrocco; Ref.v.: Walleyn 877 (Nagy et al. 2012)
Coprinellus christianopolitanus Örstadius & E. Larss.; Ref.v.: LO141-08/type (Örstadius et al. 2015)
Coprinellus cinereopallidus L. Nagy, Házi, Papp & Vágvölgyi; Ref.v.: SZMC-NL-0177/type (Nagy et al. 2012)
Coprinellus congregatus P. Karst.; Ref.v.: SZMC-NL-8588 (Hazi et al. 2011)
Coprinellus doverii (L. Nagy) Házi, L. Nagy, T. Papp & Vágvölgyi; Ref.v.: SZMC-NL-1035 (Nagy et al. 2012)
Coprinellus eurysporus (M. Lange & A.H. Sm.) Redhead, Vilgalys & Moncalvo; Ref.v.: Hoijer 95067 (Nagy et al. 2011)
Coprinellus fuscocystidiatus L. Nagy, Házi, Papp & Vágvölgyi; Ref.v.: SZMC-NL-2720/type (Nagy et al. 2011).
Coprinellus heterothrix (Kühner) Redhead, Vilgalys & Moncalvo; Ref.v.: Ulje 1063 (Nagy et al. 2012)
Coprinellus hiascens (Fr.) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-1350 (Hazi et al. 2011)
Coprinellus impatiens (Fr.) J.E. Lange; Ref.v.: SZMC-NL-1164 (Nagy et al. 2011)
Coprinellus marculentus (Britzelm.) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-1167 (Hazi et al. 2011)
Coprinellus mitrinodulisporus Doveri & Sarrocco; Ref.v.: HQ180171 (Doveri et al. 2010)
Coprinellus pallidus L. Nagy, Házi, Papp & Vágvölgyi; Ref.v.: SZMC-NL-1556/type (Nagy et al. 2012)
Coprinellus plagioporus (Romagn.) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-1365 (Nagy et al. 2012)
Coprinellus pseudoamphithallus (Uljé) Doveri & Sarrocco; Ref.v.: Ulje1288 (Nagy et al. 2012)
Coprinellus radicellus Házi, L. Nagy, Papp & Vágvölgyi; Ref.v.: SZMC-NL-3168/type (Házi et al. 2011)
Coprinellus sabulicola L. Nagy, Házi, Papp & Vágvölgyi; Ref.v.: SZMC-NL-1027 (Nagy et al. 2011)
Coprinellus sassii (M. Lange & A.H. Sm.) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-1495 (Nagy, Walther et al. 2011)
Coprinellus sclerocystidiosus (M. Lange & A.H. Sm.) Vilgalys, Hopple & Jacq. Johnson; Ref.v.: SZMC-NL-1022 (Nagy et al. 2011)
Coprinellus subdisseminatus (M. Lange) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-1482 (Nagy et al. 2012)
Coprinellus subimpatiens (M. Lange & A.H. Sm.) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-0162 (Nagy et al. 2011)
Coprinellus uljéi L. Nagy, Házi, Papp & Vágvölgyi; Ref.v.: SZMC-NL-2492 (Nagy et al. 2012)
Coprinellus velatopruinatus (Bender) Redhead, Vilgalys & Moncalvo; Ref.v.: Ulje 1264 (Nagy et al. 2011)
Remarks:
The /sabulicola and /christianopolitanus clade is particularly sensitive to the applied technique of phylogeny and the input data used. A small change in the parameters causes the clade to shift near the genus Psathyrella or even fall into it. However, the better the guide tree reflects the reality with every iteration step (see chapter MSA of the problematic ITS1 and ITS2 regions); these clades and Psathyrella get more and more separated from each other. The high support values (see Fig. 59) speak for the positioning shown. However, additional sequences from loci of higher phylogeny would be needed to consolidate this hypothesis.
Phylogram part of the genus Tulosesus; position in tree see Fig. 42
The clades /sp. (Hoijer 95067) to /sclerocystidiosus represent a considerable phylogenetic problem because there are only few sequences from loci which can determine the higher phylogeny. Again, small changes in the technique used (or loci used) cause a major change in positioning in the entire tree. Very high support values were reached for the calculated position (see Fig. 59) but the position must nevertheless be declared as relatively uncertain.
The subclade /congregatus has a higher divergence to the other species in the section than all other clades. In family-spanning phylogenetic studies, the clade falls between other genera with similar ITS regions (e.g. Typhrasa), if insufficient alignment methods are used.
Members of the genus Coprinellus s. str. have not infrequently also pileocystidia, but there are always (sub-) globose veil cells or chains of such elements present (see Fig. 60a–c). In the genus Tulosesus, this applies only to the species with rounded-angular spores (see Fig. 61f), as far as known consequently for Tulosesus doverii, S. marculentus and S. mitrinodulisporus. All other species have (sub-) cylindrical veil elements (Fig. 61e), or the veil is missing (Fig. 61a–d). This is important for the distinction of both genera.
a–d Pileocystidia and spores of some members of the genus Tulosesus. a Tulosesus heterosetulosus, AM1169; b T. ephemerus, AM1271; c T. plagioporus, AM1608; d T. fuscocystidiatus, AM1678; e, f Pileocystidia, veil elements and spores; e T. heterothrix, AM1681; f T. marculentus, AM1763; scale bar 5 μm (spores), 10 μm (other); Drawing: A. Melzer
C. christianopolitanus is the only species with deposits on the cystidia, which turn greenish in ammonia solution.
Certainly, the type also belongs in this genus, because the only differences to C. congregatus are the presence of clamps and minimally smaller spores. Because of the morphological features are undoubtedly also the following species (not included in the phylogram) members of this genus: C. allovelus (Uljé) Doveri & Sarrocco, C. aokii (Hongo) Vilgalys, Hopple & Jacq. Johnson; C. cinnamomeotinctus (P.D. Orton) D.J. Schaf.; C. fallax (M. Lange & A.H. Sm.) Redhead, Vilgalys & Moncalvo; C. minutisporus (Uljé) Doveri & Sarrocco; C. singularis (Uljé) Redhead, Vilgalys & Moncalvo; C. subpurpureus (A.H. Sm.) Redhead, Vilgalys & Moncalvo. Several sequences became available after the tree was already finished for the latter species (Vu et al. 2018), which confirm this position.
A suitable name would be Setulosus, with the basionym Coprinus subsect. Setulosi J.E. Lange, Dansk bot. Ark. 2(3):38, 1915. But that is a violation of art. 20.2 ICN. An anagram was therefore chosen as a homage. The names Impatientes Citerin, Docums Mycol 22(86):6, 1992, and Hiascentes Citérin, Docums Mycol 22(86):11, 1992, are not applicable for this genus because the former subsection includes only species without veil, the second those with veil.
New combinations:
Tulosesus allovelus (Uljé) Wächter & A. Melzer, comb. nov. MB 831800
Basionym: Coprinus allovelus Uljé, Persoonia 18(2):261, 2003. References: Uljé and Bas (1991, as Coprinus species 952 Uljé), Uljé and Noordeloos (2003).
Tulosesus amphithallus (M. Lange & A.H. Sm.) Wächter & A. Melzer, comb. nov. MB 831801
Basionym: Coprinus amphithallus M. Lange & A.H. Sm., Mycologia 45(5):774, 1953. References: Bender and Enderle (1988), Bender et al. (1984), Lange and Smith (1953), Ludwig (2007), Uljé (1984), Uljé and Bas (1991). Mat. exam.: Germany: Saxony, Delitzsch, 10.X.2006, A. Melzer (AM1744).
Tulosesus angulatus (Peck) Wächter & A. Melzer, comb. nov. MB 831802
Basionym: Coprinus angulatus Peck, Bulletin of the Buffalo Society of Natural Sciences 1:54, 1873. References: Breitenbach and Kränzlin (1995), Iglesias et al. (2014), Krieglsteiner and Gminder (2010), Lange and Smith (1953), Ludwig (2007), Uljé and Bas (1991), Smith (1948), Vila and Rocabruna (1996).
Tulosesus aokii (Hongo) Wächter & A. Melzer, comb. nov. MB 831803
Basionym: Coprinus aokii Hongo, J Jap Bot 41:167, 1966. Reference: Hongo (1966).
Tulosesus bisporiger (Buller ex P.D. Orton) Wächter & A. Melzer, comb. nov. MB 831804
Basionym: Coprinus bisporiger Buller, Trans Brit Mycol Soc 3:350, 1912 (inval.) ≡ Coprinus bisporiger Buller ex P.D. Orton, Notes R bot Gdn Edinb 35(1):147, 1976. References: Buller (1920), Giercyk et al. (2014), Ludwig (2007), Uljé and Bas (1991). Mat. exam.: Germany: Hessen, Gießen, 1.XI. 2015, W. Schößler (AM1777).
Tulosesus bisporus (J.E. Lange) Wächter & A. Melzer, comb. nov. MB 831805
Basionym: Coprinus bisporus J.E. Lange, Dansk Bot Arkiv 2(3):50, 1915. References: Breitenbach and Kränzlin (1995), Cacialli et al. (1999), Enderle and Bender (1990), Krieglsteiner and Gminder (2010), Lange and Smith (1953), Ludwig (2007), Prydiuk (2010), Uljé and Bas (1991), Watling (1967).
Tulosesus brevisetulosus (Arnolds) Wächter & A. Melzer, comb. nov. MB 831806
Basionym: Coprinus brevisetulosus Arnolds, Bibl mycol 90(3):309, 1982. References: Cacialli et al. (1999, as Coprinus bulleri Cacialli, Caroti & Doveri), Enderle and Bender (1990) Krieglsteiner and Gminder (2010, as Coprinus bulleri), Lange and Smith (1953 as Coprinus stellatus Buller), Ludwig (2007), Melzer (2009), Prydiuk (2010), Uljé and Bas (1991, as Coprinus stellatus). Mat. exam.: Germany: Saxony, Kyhna, 12.IX.2006, A. Melzer (no voucher); 25.IX.2008, A. Melzer (AM1182); 5.X.2008, A. Melzer (AM1240); 8.XII.2010, A. Melzer (no voucher); Choren near Döbeln, 23.V.2010, A. Melzer (no voucher).
Tulosesus callinus (M. Lange & A.H. Sm.) Wächter & A. Melzer, comb. nov. MB 831807
Basionym: Coprinus callinus M. Lange & A.H. Sm., Mycologia 45(5):770, 1953. References: Enderle and Bender (1990), Gierczyk et al. (2011), Lange and Smith (1953), Ludwig (2007), Melzer (2009), Ruiz Mateo and Garcia Murilo (2012), Uljé and Bas (1991). Mat. exam.: Germany: Saxony: Kyhna, 25.XI.2006, A. Melzer (AM915); 30.VIII.2010, A. Melzer (AM1419); 22.X.2011, A. Melzer (AM1482); 30.X.2011, A. Melzer (no voucher); Delitzsch, 9.IX.2008, A. Melzer (AM1163).
Tulosesus canistri (Uljé & Verbeken) Wächter & A. Melzer, comb. nov. MB 831808
Basionym: Coprinus canistri Uljé & Verbeken, Persoonia 18(1):143, 2002. References: Uljé and Verbeken (2002).
Tulosesus christianopolitanus (Örstadius & E. Larss.) Wächter & A. Melzer, comb. nov. MB 831809
Basionym: Coprinellus christianopolitanus Örstadius & E. Larss. Mycol Progr 14(25):14, 2015. Reference: Örstadius et al. (2015).
Tulosesus cinereopallidus (L. Nagy, Házi, Papp & Vágvölgyi) Wächter & A. Melzer, comb. nov. MB 831810.
Basionym: Coprinellus cinereopallidus L. Nagy, Házi, Papp & Vágvölgyi, Mycologia 104(1):257, 2011. Reference: Nagy et al. (2011).
Tulosesus cinnamomeotinctus (P.D. Orton) Wächter & A. Melzer, comb. nov. MB 831811
Basionym: Coprinus cinnamomeotinctus P.D. Orton, Trans Brit mycol Soc 91(4):547, 1988. References: P.D. Orton (1988), Schafer (2012a).
Tulosesus congregatus (P. Karst.) Wächter & A. Melzer, comb. nov. MB 831812
Basionym: Agaricus congregatus Bull., Herb Fr: tab. 94, 1786. References: Krieglsteiner and Gminder (2010), Lange and Smith (1953), Ludwig (2007), Melzer (2009c), Moreno and Faus (1984), Nagy (2007), Prydiuk (2010), Uljé and Bas (1991), Vila and Rocabruna (1996), Watling (1967). Mat. exam.: Germany: Saxony, Kyhna, 27.IV.2007, A. Melzer (AM950); 20.V.2009, A. Melzer (AM1253), 23.V.2009, A. Melzer (AM1254); 21.V.2010, A. Melzer (AM1407).
Tulosesus doverii (L. Nagy) Wächter & A. Melzer, comb. nov. MB 831813
Basionym: Coprinus doverii L. Nagy, Mycotaxon 98:148, 2007a, “2006”. References: Doveri (2010), Nagy (2007).
Tulosesus ephemerus (Bull.) Wächter & A. Melzer, comb. nov. MB 831927
Basionym: Agaricus ephemerus Bull., Hist champ France 394, 1792. References: Breitenbach and Kränzlin (1995), Krieglsteiner and Gminder (2010), Lange and Smith (1953), Locquin (1947), Prydiuk (2010), Vila and Rocabruna (1996). Mat. exam.: Germany: Thuringia, Hemleben, 15.XI.05, N. Heine (AM1271).
Tulosesus eurysporus (M. Lange & A.H. Sm.) Wächter & A. Melzer, comb. nov. MB 831928
Basionym: Coprinus eurysporus M. Lange & A.H. Sm., Mycologia 45:773, 1953. References: Lange and Smith (1953), Uljé and Bas (1991).
Tulosesus fallax (M. Lange & A.H. Sm.) Wächter & A. Melzer, comb. nov. MB 831929
Basionym: Coprinus fallax M. Lange & A.H. Sm., Mycologia 45:765, 1953. References: Lange and Smith (1953), Uljé and Bas (1991).
Tulosesus fuscocystidiatus (L. Nagy, Házi, Papp & Vágvölgyi) Wächter & A. Melzer, comb. nov. MB 831930
Basionym: Coprinellus fuscocystidiatus L. Nagy, Házi, Papp & Vágvölgyi, Mycologia 104(1):251, 2011. References: Nagy et al. (2011), Vesper and Melzer (2015). Mat. exam.: Germany: Thuringia, Stadtroda, 29.VI.2014, A. Vesper (AM1678).
Tulosesus heterosetulosus (Locq. ex Watling) Wächter & A. Melzer, comb. nov. MB 831931
Basionym: Coprinus heterosetulosus Locq., Bull Soc mycol Fr 63:78, 1947 (inval., without latin diagn.) ≡ Coprinus heterosetulosus Locq. ex Watling, Notes R bot Gdn Edinb 35(1):153, 1976. References: Breitenbach and Kränzlin (1995), Cacialli et al. (1999), Enderle et al. (1986), Gierczyk et al. (2011), Krieglsteiner and Gminder (2010), Lange and Smith (1953), Locquin (1947), Ludwig (2007), Prydiuk (2010), Uljé and Bas (1991), Watling (1967). Mat. exam.: Germany: Saxony, Kyhna, 1.XII.2006, A. Melzer (no voucher preserved); 20.III.2007, A. Melzer (no voucher preserved); 1.IX.2008, A. Melzer (AM1167); Spröda near Delitzsch, 10.VIII.2013, A. Melzer (no voucher preserved). Saxony-Anhalt, Querfurt, 31.X.2009, A. Melzer (no voucher preserved).
Tulosesus heterothrix (Kühner) Wächter & A. Melzer, comb. nov. MB 831932
Basionym: Coprinus heterothrix Kühner, Bull Soc Nat Oyonnax 10–11:3, 1957. References: Breitenbach and Kränzlin (1995), Enderle et al. (1986), Enderle (2004), Giercyk et al. (2014), Gröger (1985), Kaya et al. (2010), Ludwig (2007), Melzer (2010), Schafer (2012b), Uljé and Bas (1991). Mat. exam.: Germany: Saxony, Kyhna, 4.X.2010, A. Melzer (no voucher preserved); 12.X.2014, A. Melzer (AM1681); Oschatz, 14.IX.2012, T. Rödel (no voucher preserved). Mecklenburg-Vorpommern, Saßnitz, 14.X.2013, T. Richter (AM1648).
Tulosesus hiascens (Fr.) Wächter & A. Melzer, comb. nov. MB 831933
Basionym: Agaricus hiascens Fr., Syst Mycol 1:303, 1821. References: Enderle (2004), Enderle et al. (1986), Lange and Smith (1953), Ludwig (2007), Pegler and Legon (1994), Uljé and Bas (1991), Vila and Rocabruna (1996).
Tulosesus impatiens (Fr.) Wächter & A. Melzer, comb. nov. MB 831934
Basionym: Agaricus impatiens Fr., Syst Mycol 1:302, 1821. References: Breitenbach and Kränzlin (1995), Enderle et al. (1986), Krieglsteiner and Gminder (2010), Lange and Smith (1953), Ludwig (2007), Uljé and Bas (1991). Mat. exam.: Germany: Saxony, Delitzsch, 13.X.2000, A. Melzer (AM38).
Tulosesus marculentus (Britzelm.) Wächter & A. Melzer, comb. nov. MB 831935
Basionym: Coprinus marculentus Britzelm., Bot Centralbl 54(3):70, 1893. References: Breitenbach and Kränzlin (1995), Enderle et al. (1986), Gierczyk et al. (2011), Lange and Smith (1953, as C. hexagonosporus Joss.), Ludwig (2007), Nagy (2007), Smith (1948, as C. hexagonosporus), Uljé and Bas (1991, as C. hexagonosporus). Mat. exam.: Malta: Xewkija, Gozo, 17.IV.2013, D. Dandria (AM1763).
Tulosesus minutisporus (Uljé) Wächter & A. Melzer, comb. nov. MB 831936
Basionym: Coprinus minutisporus Uljé, Persoonia 18(2):260, 2003. References: Uljé and Bas (1991, as Coprinus species Uljé 926), Uljé and Noordeloos (2003).
Tulosesus mitrinodulisporus (Doveri & Sarrocco) Wächter & A. Melzer, comb. nov. MB 831937
Basionym: Coprinellus mitrinodulisporus Doveri & Sarrocco, Mycotaxon 114:353, 2010. Reference: Doveri et al. (2010).
Tulosesus pallidus (L. Nagy, Házi, Papp & Vágvölgyi) Wächter & A. Melzer, comb. nov. MB 831938
Basionym: Coprinellus pallidus L. Nagy, Házi, Papp & Vágvölgyi, Mycologia 104(1):261, 2011. Reference: Nagy et al. (2011).
Tulosesus pellucidus (P. Karst.) Wächter & A. Melzer, comb. nov. MB 831939
Basionym: Coprinus pellucidus P. Karst., Meddn Soc Fauna Fl Fenn 9:61, 1882. References: Cacialli et al. (1999), Gierczyk et al. (2011), Keirle et al. (2004), Krieglsteiner et al. (1982), Lange and Smith (1953), Ludwig (2007), Melzer (2009), Nagy (2007), Orton (1957), Prydiuk (2010), Uljé and Bas (1991), Vila and Rocabruna (1996). Mat. exam.: Germany: Saxony, Kyhna, 4.V.2007, A. Melzer (no voucher preserved); 16.I.2009, A. Melzer (AM1246).
Tulosesus plagioporus (Romagn.) Wächter & A. Melzer, comb. nov. MB 831940
Basionym: Coprinus plagioporus Romagnesi, Revue Mycol 6:126,1941. References: Enderle et al. (1986), Gierczyk et al. (2011), Iglesias and Vincente (2015), Keirle et al. (2004), Lange and Smith (1953), Ludwig (2007), Uljé and Bas (1991). Mat. exam.: Germany: Saxony, Kyhna, 15.IX.2013, A. Melzer (AM1608).
Tulosesus pseudoamphithallus (Uljé) Wächter & A. Melzer, comb. nov. MB 831941
Basionym: Coprinus pseudoamphithallus Uljé, Persoonia 18(2):263, 2003. Reference: Uljé and Noordeloos (2003).
Tulosesus radicellus (Házi, L. Nagy, Papp & Vágvölgyi) Wächter & A. Melzer, comb. nov. MB 831942
Basionym: Coprinellus radicellus Házi, L. Nagy, Papp & Vágvölgyi, Mycol Progr 10:366, 2011. References: Giercyk et al. (2014), Házi et al. (2011).
Tulosesus sabulicola (L. Nagy, Házi, Papp & Vágvölgyi) Wächter & A. Melzer, comb. nov. MB 831947
Basionym: Coprinellus sabulicola L. Nagy, Házi, Papp & Vágvölgyi, Mycologia 104(1):264, 2011. References: Melzer et al. (2016), Nagy et al. (2011). Mat. exam.: Germany: Saxony-Anhalt, Angern, 3.XI.2012, T. Richter (AM1689).
Tulosesus sassii (M. Lange & A.H. Sm.) Wächter & A. Melzer, comb. nov. MB 831948
Basionym: Coprinus sassii M. Lange & A.H. Sm., Mycologia 45(5):755, 1953 (nom. nov. for Coprinus ephemerus f. bisporus Sass, Amer J Bot 16:669, 1929). References: Doveri et al. (2005), Lange and Smith (1953), Ludwig (2007), Ruiz Mateo (2012), Uljé and Bas (1991). Mat. exam.: Sweden: Lapland, Gällivara-Porjusvägen, 28.VI.2016, M. Kamke (AM1879).
Tulosesus sclerocystidiosus (M. Lange & A.H. Sm.) Wächter & A. Melzer, comb. nov. MB 831949
Basionym: Coprinus sclerocystidiosus M. Lange & A.H. Sm., Dansk bot Ark 14(6):121, 1952. References: Enderle and Bender (1990), Gierczyk et al. (2011), Lange and Smith (1953), Uljé and Bas (1991).
Tulosesus singularis (Uljé) Wächter & A. Melzer, comb. nov. MB 831950
Basionym: Coprinus singularis Uljé, Persoonia 13(4):486, 1988. References: Uljé (1988), Uljé and Bas (1991).
Tulosesus subdisseminatus (M. Lange) Wächter & A. Melzer, comb. nov. MB 831951
Basionym: Coprinus subdisseminatus M. Lange, Dansk bot Ark 14(6):125, 1952. References: Krieglsteiner and Gminder (2010), Lange and Smith (1953), Ludwig (2007), Uljé and Bas (1991), Watling (1967).
Tulosesus subimpatiens (M. Lange & A.H. Sm.) Wächter & A. Melzer, comb. nov. MB 831952
Basionym: Coprinus subimpatiens M. Lange & A.H. Sm., Dansk bot Ark 14(6):56, 1952. References: Bender (1989), Krieglsteiner et al. (1982), Lange and Smith (1953), Ludwig (2007), Ortega and Esteve-Raventós (2003), Uljé and Bas (1991). Mat. exam.: Germany: Saxony, Kyhna, 24.VII.2004, A. Melzer (AM425); 9.VII.2008, A. Melzer (AM1142).
Tulosesus subpurpureus (A.H. Sm.) Wächter & A. Melzer, comb. nov. MB 831953
Basionym: Coprinus subpurpureus A.H. Sm., Mycologia 40(6):684, 1948. References: Gierczyk et al. (2011), Lange and Smith (1953), Ludwig (2007), Smith (1948), Uljé & Bas (1991).
Tulosesus uljei (L. Nagy, Házi, Papp & Vágvölgyi) Wächter & A. Melzer, comb. nov. MB 831954
Basionym: Coprinellus uljei (“uljéi”) L. Nagy, Házi, Papp & Vágvölgyi, Mycologia 104(1):267, 2011. Reference: Nagy et al. (2011).
Tulosesus velatopruinatus (Bender) Wächter & A. Melzer, comb. nov. MB 831955
Basionym: Coprinus velatopruinatus Bender, Beitr Kenntn Pilze Mitteleur V:80, 1989. References: Bender (1989), Ludwig (2007), Uljé and Bas (1991).
Britzelmayria Wächter & A. Melzer, gen. nov. MB 831472 (Fig. 64)
Etymology: Named after the German mycologist Max Britzelmayr.
Description: Basidiomata medium-sized, terrestrial, with tendance to caespitose or tightly gregarious growth, with a distinctly rooting stipe. Veil minimally developed, consisting of subcylindrical hyphae. Spores medium to large in size, laterally at most inconspicuously phaseoliform, dark, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge lageniform, with deposits turning greenish in ammonia solution, interspersed with numerous clavate cells. Pleurocystidia similar to the cheilocystidia. Pileipellis with pileocystidia or cystidium-like elements (Fig. 63). Clamps present.
Type species: Britzelmayria supernula (Britzelm.) Wächter & A. Melzer (Fig. 62).
Phylogram part of the genus Britzelmayria; position in tree see Fig. 42
Representatives:
Psathyrella multipedata (Peck) A.H. Sm.; Ref.v.: LO237-04 (Örstadius et al. 2015)
Psathyrella supernula (Britzelm.) Örstadius & Enderle; Ref.v.: LO250-04 (Örstadius et al. 2015)
Remarks:
Included is the section Multipedata Romagn., Bull Soc mycol Fr 98:11, 1982. The use of this name was discarded to avoid a tautonym. Moreover, Agaricus supernulus Britzelm. is the older name.
New combinations:
Britzelmayria supernula (Britzelm.) Wächter & A. Melzer, comb. nov. MB 831796
Basionym: Agaricus supernulus Britzelm., Ber naturhist Ver Augsburg 27:176, 1883. References (partly as Psathyrella narcotica Kits van Wav.): Christan et al. (2017), Deschuyteneer (2018), Einhellinger (1987), Enderle (1989), Kits van Waveren (1971), Kits van Waveren (1985), Krisai-Greilhuber (1992), Ludwig (2007), Örstadius and Enderle (2009). Mat. exam.: Belgium: Brabant, 13.XI.2016, D. Deschuyteneer (AM1859). Germany: Thuringia, Gera-Kaimberg, 06.XI. 2011, A. Vesper (AM1570).
Britzelmayria multipedata (Peck) Wächter & A. Melzer, comb. nov. MB 831797
Basionym: Psathyra multipedata Peck, Bull. Torrey bot. Club 32:80, 1905. References (all as Psathyrella multipedata): Breitenbach & Kränzlin (1995), Enderle (2000), Gröger (1984), Kits van Waveren (1985), Ludwig (2007) Muñoz and Caballero (2013), Smith (1972). Mat. exam.: Belgium: Perk, 6.I.2016, D. Deschuyteneer (AM1917). Germany: Saxony, Kyhna, 15.XI.2009, A. Melzer (AM1315); 12.IX.2010, A. Melzer (AM1422).
Psathyrella (Fr.) Quél.
Overview
It was possible to identify 18 sections within the genus Psathyrella with the currently available sequence data and morphological features. Figure 65 shows the 18 sections of genus Psathyrella as 360° radial consensus phylogram. The radial cladogram in Fig. 66 illustrates the numbers and the relationships of the taxa in the sections. Figure 67 shows an overview as a partial phylogram of the complete tree, collapsed to section level and serves the further orientation for the following detailed phylograms. The red brackets refer to these detailed phylograms.
Psathyrella sect. Pennatae Romagn. ex Romagn., Bull Soc mycol Fr 98:11, 1982 emend. Wächter & A. Melzer (Fig. 68)
Phylogram part of the section Pennatae; position in tree see Fig. 67
Description: Basidiomata small to medium-sized, terrestrial, lignicolous or rarely fimicolous. Veil mostly well developed. Stipe often with an annulus or an annular zone. Spores mostly medium-sized, rarely small or large, laterally very often phaseoliform, in one case with a rough surface, pale to dark, germ pore mostly visible, central. Basidia 4-spored. Marginal cells of the lamellar edge lageniform, subtriform, rarely utriform, sometimes with thickened walls, always interspersed with clavate and sphaeropedunculate cells. Pleurocystidia similar to the cheilocystidia. Clamps present.
Type species: Drosophila pennata (Fr.) Quél. sensu Ricken, Blätterp: 259, tab. 67 fig. 7 ≡ Psathyrella pennata (Fr.) A. Pearson & Dennis, Trans Br mycol Soc 31(3–4):184, 1948, designated by Romagnesi (1944:53).
Representatives:
Psathyrella atomatoides (Peck) A.H. Sm.; Ref.v.: LO249-82 (Örstadius et al. 2015)
Psathyrella conica T. Bau & J.Q. Yan; Ref.v.: HMJAU37846 (Yan and Bau 2018)
Psathyrella cortinarioides P.D. Orton; Ref.v.: LO77-00 (Örstadius et al. 2015)
Psathyrella dicrani (A.E. Jansen) Kits van Wav.; Ref.v.: LO270-04 (Larsson et Örstadius 2008)
Psathyrella fibrillosa (Pers.) Maire; Ref.v.: LO138-00 (Larsson and Örstadius 2008)
Psathyrella fimiseda Örstadius & E. Larsson; Ref.v.: LO56-96/type (Larsson and Örstadius 2008)
Psathyrella flexispora T. Wallace & P.D. Orton; Ref.v.: LO228-00 (Örstadius et al. 2015)
Psathyrella hirta Peck; Ref.v.: LO142-00 (Larsson and Örstadius 2008)
Psathyrella hololanigera (G.F. Atk.) A.H. Sm.; Ref.v.: Hausknecht071109 (Örstadius et al. 2015).
Psathyrella ichnusae Örstadius, Contu, E. Larss.; Ref.v.: Contu080106/type (Örstadius et al. 2015).
Psathyrella impexa (Romagn.) Bon; Ref.v.: LO162-03 (Örstadius et al. 2015)
Psathyrella jilinensis T. Bau & J.Q. Yan; Ref.v.: HMJAU37822 (Yan and Bau 2018)
Psathyrella kitsiana Örstadius; Ref.v.: LO217-85/type (Larsson and Örstadius 2008)
Psathyrella laricina A.H. Sm.; Ref.v.: Smith64604/type (Örstadius et al. 2015)
Psathyrella madida Örstadius & E. Larss.; Ref.v.: LO369-06 (Örstadius et al. 2015)
Psathyrella merdicola Örstadius & E. Larss.; Ref.v.: LO45-02/type (Larsson and Örstadius 2008)
Psathyrella orbicularis (Romagn.) Kits. v. Wav..; Ref.v.: LO149-11 (Örstadius et al. 2015)
Psathyrella parva A.H. Sm.; Ref.v.: LO23-08 (Örstadius et al. 2015)
Psathyrella pennata (Fr.) A. Pearson & Dennis; Ref.v.: BRNM705608 (Vasutová et al. 2008)
Psathyrella pseudocasca (Romagn.) Romagn. ex Kits van Wav.; Ref.v.: LO17-04 (Larsson and Örstadius 2008)
Psathyrella rostellata Örstadius; Ref.v.: LO228-85/type (Larsson et Örstadius 2008)
Psathyrella sabuletorum Örstadius & E. Larss.; Ref.v.: LO196-98/type (Örstadius et al. 2015)
Psathyrella scanica Örstadius & E. Larss.; Ref.v.: LO183-09/type (Örstadius et al. 2015)
Psathyrella scatophila Örstadius & E. Larss.; Ref.v.: LO64-95/type (Larsson et Örstadius 2008)
Psathyrella seymourensis A.H. Sm.; Ref.v.: LO42-87 (Örstadius et al. 2015)
Psathyrella siccophila Örstadius & E. Larss.; Ref.v.: LO417-06/type (Örstadius et al. 2015)
Psathyrella sphagnicola (Maire) J. Favre; Ref.v.: LO233-99 (Örstadius et al. 2015)
Psathyrella spintrigeroides P.D. Orton; Ref.v.: LO122-86 (Larsson and Örstadius 2008)
Psathyrella squamosa (P. Karst.) M.M. Moser ex A.H. Sm.; Ref.v.: LO104-95 (Larsson and Örstadius 2008)
Psathyrella suavissima Ayer; Ref.v.: LO4-87 (Örstadius et al. 2015)
Psathyrella umbrina Kits van Wav.; Ref.v.: SZMC-NL-1949 (Nagy, Urban et al. 2010)
Psathyrella vesterholtii Örstadius & E. Larss.; Ref.v.: JHP10.086/type (Örstadius et al. 2015)
Remarks:
The morphology in this section is highly variable and special features are not strictly limited to individual subclades. There are species with an annulus, e.g. P. vesterholtii, P. sphagnicola, and many with thickened, pigmented cystidial walls, e.g. P. pennata, P. sphagnicola. P. pseudocasca has rough spores; P. seymourensis has lentiform spores. Many species have pale spores with a hardly visible germ pore, e.g. P. siccophila and P. kitsiana. This diversity makes a concise diagnosis impossible, so that a further differentiation does not appear to make sense at present. The emendation of the section Pennatae by Kits van Waveren (1985) was primarily done to integrate all species with more or less lageniform pleurocystidia into this section. A renewed extension is necessary because species with utriform cystidia are also included.
P. dondlii has not yet been validly described (Melzer in prep.).
Psathyrella sect. Cystopsathyra (Singer) Kits van Wav., Persoonia Suppl. 2:280, 1985 (Fig. 69)
Phylogram part of the sections Cystopsathyra and Noli-tangere; position in tree see Fig. 67
Description: Basidiomata small to medium-sized, terrestrial, fimicolous, in one case parasitic. Veil strongly developed, granular, predominantly consisting of subglobose to globose elements. Spores medium-sized, pale to dark, germ pore usually visible, central. Basidia 4-spored. Marginal cells of the lamellar edge lageniform to utriform, partially interspersed with clavate and sphaeropedunculate cells. Pleurocystidia similar to the cheilocystidia. Pileocystidia rarely present. Clamps present.
Type species: Psathyrella kellermanii (Peck) Singer, Mycologia 51(3):392, 1959, designated by Singer (1962 “1961”:68).
Representatives:
Psathyrella albofloccosa Arenal, Villareal & Esteve-Raventós; Ref.v.: Sivertsen65-89 (Örstadius et al. 2015)
Psathyrella globosivelata Gröger; Ref.v.: Schumacher035 (Örstadius et al. 2015)
Psathyrella kellermanii (Peck) Singer; Ref.v.: de Meulder11242 (Örstadius et al. 2015)
Psathyrella lyckebodensis Örstadius & E. Larss.; Ref.v.: LO301-11/type (Örstadius et al. 2015)
Psathyrella sphaerocystis P.D. Orton; Ref.v.: LO126-99 (Larsson and Örstadius 2008)
Psathyrella tenuicula (P. Karst.) Örstadius & Huhtinen; Ref.v.: LO37-04 (Larsson and Örstadius 2008)
Remarks:
This section comprises species with pileocystidia, previously only P. tenuicula, and species without pileocystidia. For the former (as P. berolinensis Ew. Gerhardt), the section Setulopsathyra Arnolds & C. Perini was proposed (Arnolds and Perini 2006). However, only a subsection would be justified. The pileocystidia are not the main feature, instead the structure of the veil. P. tenuicula seems to be a complex of two taxa.
The voucher de Meulder11242 described by De Haan (1993) as P. cf. kellermanii deviates from the descriptions by Peck (1906) and Singer (1959) due to the more delicate habit and spores with a very large germ pore. In addition, this collection grew on rotten mushroom remainders. So it is not quite beyond doubt whether this record is identical to the original P. kellermanii. The same applies to the collection AM1705.
Undoubtedly, P. utriformcystis S.J. Seok & Y.S. Kim belongs to the section Cystopsathyra (see Seok et al. 2010).
Psathyrella sect. Noli-tangere Wächter & A. Melzer, sect. nov. MB 831458 (Fig. 69)
Description: Basidiomata small to medium-sized, terrestrial, lignicolous, subfimicolous. Veil sparse. Spores predominantly medium-sized, rarely small or large, laterally often phaseoliform, medium dark to dark, germ pore central. Basidia mostly 4-spored. Marginal cells of the lamellar edge mostly utriform, mixed with moderately numerous clavate and sphaeropedunculate cells that can sometimes have a thickened wall. Pleurocystidia primarily utriform, rarely lageniform or fusiform. Clamps present.
Type species: Psathyrella noli-tangere (Fr.) A. Pearson & Dennis, Trans Br mycol Soc 31(3–4):184, 1948.
Representatives:
Psathyrella fagetophila Örstadius & Enderle; Ref.v.: LO210-85/type (Örstadius et al. 2015)
Psathyrella fennoscandica Örstadius & E. Larss.; Ref.v.: LO484-05/type (Örstadius et al. 2015)
Psathyrella fulvescens (Romagn.) M.M. Moser ex A.H. Sm.; Ref.v.: WU13965 (Vasutová et al. 2008)
Psathyrella noli-tangere (Fr.) A. Pearson & Dennis; Ref.v.: LO83-03 (Larsson and Örstadius 2008)
Psathyrella perpusilla Kits van Wav.; Ref.v.: LO213-96 (Larsson and Örstadius 2008)
Psathyrella pseudocorrugis (Romagn.) Gallant ex Bon; Ref.v.: LO226-06 (Örstadius et al. 2015)
Psathyrella romagnesii Kits van Wav.; Ref.v.: LO267-04 (Larsson and Örstadius 2008)
Psathyrella rubiginosa A.H. Sm.; Ref.v.: LO107-96 (Örstadius et al. 2015)
Psathyrella seminuda A.H. Sm.; Ref.v.: Smith34091/type (Örstadius et al. 2015)
Psathyrella senex (Peck) A.H. Sm.; Ref.v.: LO115-02 (Larsson and Örstadius 2008)
Psathyrella warrenensis A.H. Sm.; Ref.v.: Smith70162/type (Örstadius et al. 2015)
Remarks:
The vouchers LO267-04, LO85-98 and LO213-96 have small deviations in the LSU sequences, but almost identical ITS sequences. It is very likely that these are the same species and P. perpusilla is merely a 2-spored form of P. romagnesii.
P. pseudocorrugis is considered here sensu Romagnesi (1952, 1982) as well as Örstadius and Knudsen (2008), not sensu Kits van Waveren (1985).
Psathyrella sect. Hydrophilae Romagn. ex Singer, Sydowia 15:68, 1962 “1961” (Fig. 70)
Phylogram part of the section Hydrophilae; position in tree see Fig. 67
Description: Basidiomata small to medium-sized, all lignicolous. Veil sparse to strongly developed. Spores small in size, laterally mostly phaseoliform, predominantly pale to medium-coloured, germ pores often indistinct or absent. Basidia 4-spored. Marginal cells of the lamellar edge lageniform, utriform, very often mucronate, undermixed with moderately numerous clavate and sphaeropedunculate cells. Pleurocystidia similar to the cheilocystidia. Clamps present.
Type species: Drosophila hydrophila (Bull.) Quél., Enchir fung:116, 1886 ≡ Psathyrella piluliformis (Bull.) P.D. Orton, Notes R bot Gdn Edinb 29:116, 1969, designated by Romagnesi (1944:52).
Representatives:
Psathyrella echinata (Cleland) Grgurinovic; Ref.v.: ZT12073 (Örstadius et al. 2015)
Psathyrella maculata (Parker) A.H. Sm.; Ref.v.: CBS206.33 (Nagy et al. 2011)
Psathyrella mucrocystis A.H. Sm.; Ref.v.: LO103-98 (Larsson and Örstadius 2008)
Psathyrella oboensis Desjardin & B.A. Perry; Ref.v.: SFSU DED 8234/type (Desjardin and Perry 2016)
Psathyrella obscurotristis Enderle & M. Wilhelm ex Enderle & M. Wilhelm; Ref.v.: Wilhelm489/type (Örstadius et al. 2015)
Psathyrella pertinax (Fr.) Örstadius; Ref.v.: LO259-91 (Larsson and Örstadius 2008)
Psathyrella piluliformis (Bull.) P.D. Orton; Ref.v.: LO162-02 (Larsson and Örstadius 2008)
Remarks:
The characteristics of P. echinata differ from those of the other species. In particular, the spores are darker and not phaseoliform. In addition, the cystidia have thickened walls and partly crystals, similar to P. olympiana A.H. Sm. It is positioned at the base of the clade.
P. laevissima (Romagn.) Singer very probably belongs to this section. References: Romagnesi (1975), Kits van Waveren (1982, 1985), Muñoz and Sánchez (2018), Musumeci (2006), Örstadius & Knudsen (2008). Mat. exam.: Belgium: Hofstade, 23.XII.2015, D. Deschuyteneer (AM1858). Germany: Nordrhein-Westfalen, Mönchengladbach, 25.X.2012, H. Bender (AM1585). Baden-Württemberg, Leimen, 18.I.2014, A. Oppolzer & P. Schäfer (AM1657). Rheinland-Pfalz, Kandel, 06.XII.2015, R. Ziebarth (AM1785); 09.VIII.2017, R. Ziebarth (AM1908); 26.IX.2017, R. Ziebarth (AM1909). Baden-Württemberg, Bad Mergentheim, 14.I.2018, R. Markones (AM1911). La Réunion: Forét de Bélouve, 16.III.2007, T. Rödel (AM1846); 18.III.2007, T. Rödel (AM1848).
Muñoz and Sánchez (2018) suppose P. laevissima could be conspecific with P. oboensis. The latter name would then be a younger synonym.
Psathyrella sect. Pygmaeae Romagn., Bull Soc mycol Fr 98:10, 1982 emend. Wächter & A. Melzer (Fig. 71)
Phylogram part of the sections Pygmaeae, Saponaceae, Stridvalliorum, Arenosae and Confusae; position in tree see Fig. 67
Description: Basidiomata small to medium-sized, terrestrial or lignicolous. Veil at most sparsely developed. Spores small to medium-sized, frontally ellipsoid, laterally phaseoliform, pale to medium dark coloured, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge mostly utriform, sometimes with thickened walls and crystalline deposits. Pleurocystidia similar to the cheilocystidia. Clamps present.
Type species: Psathyrella pygmaea (Bull. ex Schum.) Singer, Lilloa 22:467, 1951, designated by Romagnesi (1982:10).
Representatives:
Psathyrella olympiana A.H. Sm.; Ref.v.: SZMC-NL-2935 (Nagy et al. 2011)
Psathyrella pygmaea (Bull. ex Schum.) Singer; Ref.v.: LO97-04 (Larsson and Örstadius 2008)
Psathyrella rybergii Örstadius & E. Larss.; Ref.v.: LO373-06/type (Örstadius et al. 2015)
Remarks:
Romagnesi (1982) mentions P. pygmaea as the only species in his section Pygmaeae and wrote “Lignicolae inter Coprinos disseminatus”; an emendation was therefore necessary.
Whether HMJAU 37810 and H6038514 are really P. amaura (Berk. & Broome) Pegler cannot be answered. This species was originally described from Sri Lanka (Berkeley and Broome 1871).
Psathyrella sect. Saponaceae Wächter & A. Melzer, sect. nov. MB 831459 (Fig. 71)
Description: Basidiomata small to large-sized, terrestrial, lignicolous or fimicolous. Veil sparse. Spores medium in size, frontally ellipsoid, ovoid, rarely angular-ovoid, laterally often phaseoliform, dark, germ pore central or slightly to distinctly eccentric. Basidia 4-spored. Marginal cells of the lamellar edge predominantly utriform, sometimes with thickened walls and mucoid deposits. Pleurocystidia similar to the cheilocystidia. Clamps present.
Type species: Psathyrella saponacea F.H. Møller, Fungi of the Faeröes 1:179, 1945.
Representatives:
Psathyrella abieticola A.H. Sm.; Ref.v.: Smith58673/type (Örstadius et al. 2015)
Psathyrella conferta Eyssart. & Chiaffi; Ref.v.: GE02.007/type (Örstadius et al. 2015)
Psathyrella panaeoloides (Maire) Svrček ex Arnolds; Ref.v.: LO293-04 (Larsson and Örstadius 2008)
Psathyrella saponacea F.H. Møller; Ref.v.: LO204-96 (Larsson and Örstadius 2008)
Psathyrella tephrophylla (Romagn.) M.M. Moser ex Bon; Ref.v.: SZMC-NL-0630 (Nagy, Urban et al. 2010)
Remarks:
Most of the species in this section have more or less distinct deposits on the cystidia. P. saponacea and sometimes P. tephrophylla display the rare phenomenon of an eccentric germ pore. P. panaeoloides probably contains several taxa. The same applies to P. tephrophylla; P. abieticola is considered independent for this reason, especially because sequences of the type are present. P. fusca (Schumach.) A. Pearson, a name that has often come into use for P. tephrophylla (Örstadius 2007), is illegitimate, because Agaricus fuscus Schumach. 1803 is a younger homonym of Agaricus fuscus Schaeff. 1774.
Psathyrella sect. Stridvalliorum Wächter & A. Melzer, sect. nov. MB 831460 (Fig. 71)
Description: Basidiomata medium-sized, terrestrial. Veil moderately developed. Spores small-sized, pale, germ pore absent. Basidia 4-spored. Marginal cells of the lamellar edge utriform, clavate. Pleurocystidia utriform, clavate, sometimes with slightly thickened walls. Clamps present.
Type species: Psathyrella stridvallii Örstadius & E. Larss., Mycol Progr 14(25):27, 2015.
Representative:
Psathyrella stridvallii Örstadius & E. Larss., Ref.v.: LO104-98/type (Örstadius et al. 2015)
Remarks:
So far only one species is known.
Psathyrella sect. Arenosae Wächter & A. Melzer, sect. nov. MB 831461 (Fig. 71)
Description: Basidiomata small-sized, terrestrial. Veil well developed. Spores medium-sized, laterally partly phaseoliform, nearly opaque, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge utriform, lageniform, also clavate. Pleurocystidia utriform or lageniform. Clamps present.
Type species: Psathyrella arenosa Örstadius & E. Larss., Mycol Progr 14(25):17, 2015.
Representative:
Psathyrella arenosa Örstadius & E. Larss.; Ref.v.: LO220-96/type (Örstadius et al. 2015)
Remarks:
Psathyrella salina Broussal, G. Mir, J. Carbó & Pérez-De-Greg. also belongs to this section; see the phylogenetic results in Broussal et al. (2018).
Psathyrella sect. Confusae Wächter & A. Melzer, sect. nov. MB 831462 (Fig. 71)
Etymology: Derived from confusa = confusing, because of the differing characteristics.
Description: Basidiomata small to large-sized, terrestrial, lignicolous, in one case parasitic. Veil absent to strongly developed. Spores medium-sized, dark, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge mostly utriform, clavate cells practically absent. Pleurocystidia similar to the cheilocystidia. Clamps present.
Type species: Psathyrella gordonii (Berk. & Broome) A. Pears. & Dennis, Trans Br mycol Soc 31(3–4):184, 1948.
Representatives:
Psathyrella epimyces (Peck) A.H. Sm.; Ref.v.: WU19965 (Örstadius et al. 2015)
Psathyrella gordonii (Berk. & Broome) A. Pears. & Dennis; Ref.v.: LO220-95 (Örstadius et al. 2015)
Psathyrella violaceopallens Contu; Ref.v.: LO96-11 (Örstadius et al. 2015)
Remarks:
The characteristics of the three known species of this section are extremely different, especially the size of the basidiomata, the development of the veil and the spore measurements. Moreover, P. epimyces is a parasite on large Coprinus- and Coprinopsis-species; P. violaceopallens has violet pileus colours. A common feature could be the pileipellis; Örstadius et al. (2015) note “the pileipellis is similar to a cutis”.
Psathyrella sect. Obtusatae (Fr.) Singer, The Agaricales in modern taxonomy, 2th edn.:509,1962 “1961” (Fig. 72)
Phylogram part of the section Obtusatae; position in tree see Fig. 67
Description: Basidiomata small to medium-sized, terrestrial. Veil sparse. Spores medium-sized, mostly very dark, rarely medium dark, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge predominantly clavate and sphaeropedunculate, sometimes mucronate or with thickened walls, also lageniform, utriform or fusiform. Pleurocystidia lageniform, utriform, fusiform. Clamps present.
Type species: Agaricus obtusatus Pers.: Fr., Syst mycol 1:293, 1821 ≡ Psathyrella obtusata (Pers.) A.H. Sm., Contr Univ Mich Herb 5:55, 1941, designated by Singer (1962:509).
Representatives:
Psathyrella obtusata (Pers.) A.H. Sm.; Ref.v.: LO88-01 (Larsson and Örstadius 2008)
Psathyrella nitens A.H. Sm.; Ref.v.: AHS30388 (Frank et al. 2010)
Psathyrella dunensis Kits van Wav.; Ref.v.: WU19387 (Vasutová et al. 2008)
Psathyrella psammophila A.H. Sm.; Ref.v.: Smith67836/type (Örstadius et al. 2015)
Remarks:
P. psammophila is very likely to be a synonym of P. obtusata.
Psathyrella sect. Spadiceogriseae Kits van Wav., Persoonia, Suppl. 2:280, 1985 (Fig. 73)
Phylogram part of the section Spadiceogriseae; position in tree see Fig. 67
Description: Basidiomata medium-sized to large, terrestrial, lignicolous, in one case on rhizomes of grasses. Veil sparse to strongly developed. Spores predominantly medium-sized, rarely large, almost always phaseoliform, mostly medium dark, rarely pale or opaque, germ pore central, rarely invisible. Basidia 4-spored. Marginal cells of the lamellar edge dominating clavate and sphaeropedunculate shaped, less often utriform, sublageniform. Pleurocystidia utriform. Clamps present.
Type species: Psathyrella spadiceogrisea (Schaeff.) Maire, Mém Soc Sci Nat Maroc 45:113, 1937, designated by Kits van Waveren (1985:280)
Representatives:
Psathyrella ammophila (Durieu & Lév.) P.D. Orton; Ref.v.: LO169-01 (Örstadius et al. 2015)
Psathyrella carminei Örstadius & E. Larss.; Ref.v.: LO5-09/type (Örstadius et al. 2015)
Psathyrella casca (Fr.) Konr. & Maubl.; Ref.v.: AM1814/ GLM-F111048/type (Melzer 2018)
Psathyrella cascoides A. Melzer, Karich & Wächter; Ref.v.: AM1893/GLM-F111050/type (Melzer 2018)
Psathyrella clivensis (Berk. & Broome) P.D. Orton; Ref.v.: SZMC-NL-1952 (Nagy et al. 2009)
Psathyrella fatua (Fr.) Konr. & Maubl.; Ref.v.: LO231-08/type (Örstadius et al. 2015)
Psathyrella hellebosensis Deschuyteneer & A. Melzer; Ref.v.: AM1816/LZP-7615/type (Deschuyteneer and Melzer 2017)
Psathyrella mammifera (Romagn.) Courtec.; Ref.v.: HMJAU37882 (Yan and Bau 2018)
Psathyrella marquana A. Melzer, Wächter & Kellner; Ref.v.: AM1693/GLM-F111049/type (Melzer 2018)
Psathyrella phegophila Romagn.; Ref.v.: SZMC-NL-3527 (Nagy et al. 2011)
Psathyrella spadiceogrisea (Schaeff.) Maire; Ref.v.: AM1894/GLM-F111047/type (Melzer 2018)
Psathyrella striatoannulata Heykoop, G. Moreno & M. Mata; Ref.v.: INB:4162132/type (Crous et al. 2017)
Psathyrella subspadiceogrisea T. Bau & J.Q. Yan; Ref.v.: HMJAU35992 (Yan and Bau 2017)
Psathyrella thujina A.H. Sm.; Ref.v.: Smith66720/type (Örstadius et al. 2015)
Remarks:
This section is well characterized by its main feature, the dominance of clavate and sphaeropedunculate marginal cells in the lamellar edge and is essentially consistent with the species placed here by Kits van Waveren (1985). P. ammophila with very large dark spores and a special ecology, as well as Psathyrella clivensis with bright spores without a germ pore, are the notable exceptions in the morphology. The status of P. groegeri G. Hirsch is still unclear, see Melzer (2016, 2018). Whether P. mammifera is the taxon in the original sense must remain unanswered; however, this species belongs undoubtedly in this section.
Psathyrella sect. Jacobssoniorum Wächter & A. Melzer, sect. nov. MB 831463 (Fig. 74)
Phylogram part of the sections Jacobssoniorum, Microrhizae, Pseudostropharia and Lutenses; position in tree see Fig. 67
Description: Basidiomata small to medium-sized, terrestrial, stipe with a pseudorrhiza. Veil sparse. Spores large-sized, dark, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge lageniform, fusiform, sometimes also clavate. Pleurocystidia lageniform, fusiform, sometimes with deposits turning greenish in ammonia solution. Clamps present.
Type species: Psathyrella jacobssonii Örstadius, Windahlia 24:15, 2001.
Representatives:
Psathyrella jacobssonii Örstadius; Ref.v.: LO256-92/type (Örstadius et al. 2015)
Psathyrella sublatispora Örstadius, S.- Å Hanson & E. Larss.; Ref.v.: LO190-97/type (Örstadius et al. 2015)
Remarks:
The long connection branch of section Spadiceogriseae indicates that section Jacobssoniorum is more closely related to section Microrhizae (see below) than to section Spadiceogriseae. In addition, the morphological features are different between section Jacobssoniorum and section Spadiceogriseae.
Psathyrella sect. Microrhizae Romagn. ex Singer, Sydowia 15:68, 1962 (Fig. 74)
Description: Basidiomata medium-sized, terrestrial, lignicolous, mostly with a pseudorrhiza. Veil sparse. Spores medium to large-sized, dark, laterally never phaseoliform, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge lageniform, fusiform, rarely clavate. Pleurocystidia lageniform, subutriform, fusiform. Pileocystidia rarely present. Clamps present.
Type species: Drosophila microrhiza (Lasch) Quél. Enchir fung:118, 1886, erroneous as Drosophila microrhiza (Fr. ex Lasch) Romagn. ≡ Psathyrella microrhiza (Lasch) Konr. & Maubl., Encyclop Mycol 14:123, 1949, designated by Romagnesi (1944:54).
Representatives:
Psathyrella microrhiza (Lasch) Konr. & Maubl.; Ref.v.: LO185-02 (Larsson and Örstadius 2008)
Psathyrella uskensis A.H. Sm.; Ref.v.: AHS73377 (Frank et al. 2010)
Psathyrella boreifasciculata Kytöv & Liimat.; Ref.v.: IK08-1565/type (von Bonsdorff et al. 2014)
Psathyrella alluviana A.H. Sm.; Ref.v.: AHS30217 (Frank et al. 2010)
Remarks:
Örstadius et al. (2015) contains an unfortunate misprint. The voucher LO136-08 (GenBank access.-numbers KC992868.1, KC992868.1, KJ664850.1, KJ732764.1) is not the type of P. microrhiza. It is very likely P. alluviana, which according to Smith (1972) has pileocystidia, a rare appearance in the genus. The same clade also contains the examined voucher Belgium: Brabant Flamand, Perk, 30.XI.2017, D. Deschuyteneer (AM1883, GenBank number MK053806.1), with pileocystidia.
The historical spelling “microrhiza” should be retained. Lasch named the species “microrhizus” (modified Greek) and Fries (1832) sanctioned this.
Psathyrella sect. Pseudostropharia A.H. Sm. emend. Wächter & A. Melzer (Fig. 74)
Description: Basidiomata medium-sized to large, lignicolous or terrestrial. Veil moderately to strongly developed, sometimes forming an annulus on the stipe. Spores medium to large-sized, pale or dark, germ pore small to indistinct. Basidia 4-spored. Marginal cells of the lamellar edge lageniform, utriform, rarely clavate. Pleurocystidia similar to the cheilocystidia. Clamps present.
Type species: Psathyrella caput-medusae (Fr.) Konr. & Maubl., Encyclop Mycol 14:127, 1949, designated by Smith (1972:61).
Representatives:
Psathyrella caput-medusae (Fr.) Konr. & Maubl.; Ref.v.: LO36-94 (Örstadius et al. 2015)
Psathyrella cotonea (Quél.) Konr. & Maubl.; Ref.v.: LO136-00 (Örstadius et al. 2015)
Psathyrella magnispora Heykoop & G. Moreno; Ref.v.: AH24929/type (Örstadius et al. 2015)
Remarks:
Smith (1972) defined the subgenus Pseudostropharia and the homonymous section: “Stipes annulatus; annulus membraneus vel floccosus et crassus, persistens vel evanescens”. The diagnosis does not mention any micro-features; therefore, the micro-features had to be extended to species without annulus.
From a phylogenetic point of view it is beyond doubt that P. caput-medusae divides into two taxa, the considerable distance speaks for even two good species.
P. duchesnayensis A.H. Sm. is certainly a younger synonym of P. cotonea.
Psathyrella sect. Lutenses (Kits van Wav.) Wächter & A. Melzer, stat. nov. MB 832431 (Fig. 74)
Basionym: Subsection Lutenses Kits van Wav., Persoonia Suppl. 2:280, 1985
Description: Basidiomata small to large, terrestrial. Veil sparse. Spores medium-sized, dark, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge utriform, lageniform, also clavate, with deposits turning greenish in ammonia solution. Pleurocystidia similar to the cheilocystidia. Clamps present.
Type species: Psathyrella lutensis (Romagn.) M.M. Moser ex Bon, Docums Mycol 12(46):53, 1982, designated by Kits van Waveren (1985:280).
Representatives:
Psathyrella lutensis (Romagn.) M.M. Moser ex Bon; Ref.v.: LO98-03 (Larsson and Örstadius 2008)
Psathyrella ramicola A.H. Sm.; Ref.v.: PBM871 (Frank et al. 2010)
Remarks:
Whether P. ramicola is actually a synonym of P. lutensis cannot be answered clearly, because the voucher PBM871 is not the type.
Psathyrella sect. Psathyrella (Fig. 75)
Phylogram part of the section Psathyrella; position in tree see Fig. 67
Description: Basidiomata small-sized to large, terrestrial or lignicolous. Pileus sometimes with pink tones, especially when drying. Veil practically absent or minimally developed. Stipe often with a pseudorrhiza. Spores large-sized, dark, laterally never phaseoliform, germ pore in most species central, rarely eccentric. Basidia mostly 4-spored. Marginal cells of the lamellar edge lageniform, utriform, also often clavate or irregular. Pleurocystidia lageniform, utriform. Clamps present.
Representatives:
Psathyrella amarescens Arnolds; Ref.v.: Arnolds02-78/type (Örstadius et al. 2015)
Psathyrella aquatica J.L. Frank, Coffan & Southworth; Ref.v.: SOC1097/type (Frank et al. 2010)
Psathyrella bipellis (Quél.) A.H. Sm.; Ref.v.: LO207-96 (Larsson and Örstadius 2008)
Psathyrella brooksi A.H. Sm.; Ref.v.: MICH11888 (Frank et al. 2010)
Psathyrella corrugis (Pers.) Konr. & Maubl.; Ref.v.: LO171-01 (Larsson and Örstadius 2008)
Psathyrella fontinalis A.H. Sm.; Ref.v.: AHS25652 (Frank et al. 2010)
Psathyrella longicauda P. Karst.; Ref.v.: LO254-91 (Örstadius et al. 2015)
Psathyrella pseudogracilis (Romagn.) M.M. Moser; Ref.v.: LO287-06 (Örstadius et al. 2015)
Psathyrella subincarnata A.H. Sm.; Ref.v.: LO190-97 (Frank et al. 2010)
Psathyrella superiorensis A.H. Sm.; Ref.v.: AHS32107/type (Frank et al. 2010)
Remarks:
This section contains by definition the type of the genus, designated by Earle (1909:393): Psathyrella gracilis (Fr.) Quél., Mém Soc Émul Montbéliard Sér. 2, 5:122, 1872 ≡ Psathyrella corrugis (Pers.) Konr. & Maubl., Encyclop Mycol 14:123, 1949.
Psathyrella bipellis is separated into two clades. The difference between the two clades is approx. 0.02 exp. changes/site. The divergence of the ITS and LSU regions is small and barely allows a separation. The β-tubulin and the ef-1α region show a higher divergence. Whether this is also reflected in morphological features remains to be examined. The species is quite plastic, because the cystidia have either an obtuse or an acute apex. In addition, the spores can have a central or an eccentric germ pore. P. brooksi and P. subincarnata are possibly younger synonyms, but this can only be decided after examining the types. Whether P. aquatica and P. fontinalis are identical needs to be investigated.
Psathyrella amarescens is certainly a form of P. corrugis, which also shows the spotty pigmented lamellar edge (see Friebes and Melzer 2009).
The separation of the clade /pseudobifrons from the clade /longicauda is based on new data (after the finalization of the tree) given in Vu et al. (2018). Several collections are assigned to P. pseudobifrons Romagn. (inval.). In addition, the investigation of the voucher Germany: Saxony, Kyhna, 20.X.2013, A. Melzer (AM1650) showed that it is placed in the same position. Morphological differences are present. For a final clarification, further work is necessary.
Kits van Waveren (1976) emended the section Psathyrella to unite the sections Graciles Romagn. ex. Romagn., Bull Soc mycol Fr 98:11, 1982 and Microrrhizae Romagn. ex Singer, Sydowia 15:68, 1962 “1961”; the emendation must be discarded, because both sections are phylogenetically well separated.
Psathyrella sect. Atomatae Romagn. ex Singer, Sydowia 15:68, 1962 “1961” (Fig. 76)
Phylogram part of the sections Atomatae and Sinefibularum; position in tree see Fig. 67
Description: Basidiomata very small to medium-sized, terrestrial, subfimicolous or fimicolous. Lamellar edge, and drying pileus often with pink tones. Veil sparse. Spores large in size, laterally never phaseolioform, dark, rarely medium dark, germ pore central. Basidia 4- or 2-spored, occasionally 1-spored. Marginal cells of the lamellar edge predominantly lageniform, subutriform, very frequently also clavate. Pleurocystidia similar to the cheilocystidia. Clamps present.
Type species: Drosophila prona (Fr.) Quél., 1886 sensu Ricken ≡ Psathyrella prona (Fr.) Gillet, Hyménomycètes:618, 1878, designated by Romagnesi (1944:53).
Representatives:
Psathyrella calcarea (Romagn.) M.M. Moser; Ref.v.: LO211-03 (Larsson and Örstadius 2008)
Psathyrella calvini A.H. Sm.; Ref.v.: AHS34788 (Frank et al. 2010)
Psathyrella liliputana Örstadius & E. Larss.; Ref.v.: LO130-09/type (Örstadius et al. 2015)
Psathyrella mycenoides T. Bau; Ref.v.: HMJAU37993 (Yan and Bau (2018)
Psathyrella orbitarium (Romagn.) M.M. Moser; Ref.v.: LO257-90 (Larsson and Örstadius 2008)
Psathyrella potteri A.H. Sm.; Ref.v.: LO271-01 (Larsson and Örstadius 2008)
Psathyrella prona (Fr.) Gillet; Ref.v.: LO237-00/type (Örstadius et al. 2015)
Psathyrella stercoraria (Kühn. & Joss.) M.M. Moser ex Kits van Wav.; Ref.v.: LO460-05/type (Larsson and Örstadius 2008)
Psathyrella tenera Peck; Ref.v.: LO382-89 (Örstadius et al. 2015)
Remarks:
Striking morphological exceptions are P. liliputana and P. mycenoides with relatively small and bright spores; from a phylogenetic point of view, however, there is no doubt that they belong to this section.
Psathyrella sect. Sinefibularum Wächter & A. Melzer, sect. nov. MB 831464 (Fig. 76)
Etymology: Derived from sine = without, fibula = clamp; no clamps present.
Description: Basidiomata small to medium-sized, terrestrial, lignicolous or fimicolous. Veil sparse to rich. Spores medium to large-sized, laterally never phaseolioform, mostly dark, rarely medium dark, germ pore central to slightly eccentric. Basidia 4-spored. Marginal cells of the lamellar edge utriform, lageniform, also often clavate. Pleurocystidia predominantly utriform, very rarely lageniform. Clamps absent.
Type species: Psathyrella vinosofulva P.D. Orton, Trans Br mycol Soc 43(2):378, 1960.
Representatives:
Psathyrella complutensis Heykoop & G. Moreno; Ref.v.: AH23895 (Crous et al. (2015).
Psathyrella effibulata Örstadius & E. Ludwig; Ref.v.: LO37-96/type (Larsson and Örstadius (2008)
Psathyrella purpureobadia Arnolds; Ref.v.: Arnolds99-56A/type (Larsson and Örstadius (2008)
Psathyrella romellii Örstadius; Ref.v.: LO240-01/type (Örstadius et al. (2015)
Psathyrella vinosofulva P.D. Orton; Ref.v.: LO2-88 (Örstadius et al. (2015)
Remarks:
Psathyrella complutensis supposedly should not have pleurocystidia (Crous et al. 2015). The close relationship with P. effibulata suggests that these are potentially present but extremely rare. Neither Enderle (1994, 1998) nor Melzer (2008) found pleurocystidia in P. effibulata, and Muñoz and Caballero (2013:30) explicitly wrote “Pleurocistidios muy raros, visibles sólo tras varias preparaciones y no en todos los ejemplares, …”. P. citerinii Eyssart., unless identical to P. effibulata, also belongs in this section. Örstadius et al. (2015) found that P. riparia A.H. Sm. is a more recent synonym of P. vinosofulva, which can be confirmed by the authors of the present study. More difficult to assess is the problem whether P. purpureobadia is a separate species because of its growth on dung, as proposed by Örstaidus et al. (2015).
Candolleomyces Wächter & A. Melzer, gen. nov. MB 832256 (Fig. 79)
Etymology: Named after the type.
Description: Basidiomata small to large, terrestrial, lignicolous, rarely fimicolous. Veil most likely always present but often very fugacious, as far as known fibrillose, scaly or granulose, consisting of chains of subcylindrical, partially slightly thick-walled and brownish pigmented cells; sphaerocysts may be characteristic as the second component of the veil. (see Fig. 78). Stipe occasionally with an annulus. Spores mostly medium-sized, laterally often phaseoliform, pale to medium dark, germ pore central, but often invisible. Basidia 4-spored. Marginal cells of the lamellar edge utriform, subutriform, subcylindrical, never predominantly lageniform, as well as clavate or sphaeropedunculate, rarely exclusively showing the latter forms. Pleurocystidia absent. Clamps at least in the majority of species present.
Type species: Candolleomyces candolleanus Wächter & A. Melzer (see Fig. 77).
Phylogram part of the genus Candolleomyces; position in tree see Fig. 42
Representatives:
Psathyrella badhyzensis Kalamees; Ref.v.: TAA79478/type (Örstadius et al. 2015)
Psathyrella badiophylla (Romagn.) Bon; Ref.v.: SZMC-NL-2347 (Nagy et al. 2011)
Psathyrella cacao Desjardin & B. A. Perry; Ref.v.: SFSU DED 8339/type (Desjardin and Perry 2016)
Psathyrella candolleana (Fr.) Maire; Ref.v.: LAS73030/type (Örstadius et al. 2015)
Psathyrella efflorescens (Sacc.) Pegler; Ref.v.: Pegler2133 (Örstadius et al. 2015)
Psathyrella leucotephra (Berk. & Broome) P.D. Orton; Ref.v.: LO138-01 (Örstadius et al. 2015)
Psathyrella luteopallida A.H. Sm.; Ref.v.: Sharp20863/type (Örstadius et al. 2015)
Psathyrella singeri A.H. Sm.; Ref.v.: HMJUA37867 (Yan and Bau 2018)
Psathyrella subsingeri T. Bau & J.Q. Yan; Ref.v.: HMJAU37913 (Yan and Bau 2018)
Psathyrella sulcatotuberculosa (J. Favre) Einhell.; Ref.v.: LO55-12 (Battistin et al. 2014)
Psathyrella trinitatensis R.E.D. Baker & W.T. Dale; Ref.v.: TL9035 (Örstadius et al. 2015)
Psathyrella typhae (Kalchbr.) A. Pearson & Dennis; Ref.v.: LO21-04 (Larsson and Örstadius 2008)
Remarks:
See also radial phylogram Fig. 35.
The numerous subclades or clusters with vouchers designated as P. candolleana illustrate the already known fact that it is a collective species. More information about this could give the sequencing of Romagnesis types of P. elegans (Romagn.) Bon, P. proxima (Romagn.) Bon and P. scotospora (Romagn.) Bon, which are currently considered synonyms of P. candolleana, but are probably independent species. The type of P. candolleana is in the subclade /candolleana ss. str.; whether these would split up is questionable and they are surely morphologically hardly comprehensible. Also interesting is the fact that several well-demarcated subclades contain no designated vouchers. This seems to be an indication that the closer circle of P. candolleana on the one hand is more species-rich than previously thought. On the other hand, hardly unique features are present, which allow an identification by conventional methods. Whether P. badhyzensis and P. trinitatensis are independent species is questionable.
For the following species, the determination sensu orig. is genetically insufficiently confirmed, because the types were not tested, but the morphology clearly refers to the genus: P. albipes (Murill) A.H. Sm., P. paecilosperma Pacioni, P. pseudocandolleana A.H. Sm. and P. tuberculata (Pat.) A.H. Sm. Phylogenetically unequivocal is the position of P. halophila Esteve-Raventós & Enderle because of the appropriate investigation by Broussal et al. (2018). The same applies to P. secotioides G. Moreno, Heykoop, Esqueda & Olariaga due to the results in Moreno et al. (2015), and P. aberdarensis A. Melzer, Kimani & R. Ullrich, see Melzer et al. (2019, “2018”).
The following species should be checked: P. acutisquamosa Dennis, P. aequatoriae Singer, P. albocapitata Dennis, P. araguana Dennis, P. argillospora Singer, P. armeniaca Pegler, P. atroumbonata Pegler, P. avilana Dennis, P. coprinoceps (Berk. & M.A. Curtis) Dennis, P. erinensis Dennis, P. glandispora Pegler, P. glaucescens Dennis, P. lacuum Huijsman, P. lignatilis Singer, P. longicystidiata Heykoop & Moreno, P. marthae Singer, P. microsporoides Heykoop & G. Moreno, P. naivashaiensis Pegler, P. pallidispora Dennis, P. pervelatoides S.J. Seok & Y.S. Kim, P. pruinosa Rawla, P. pusilla Pegler, P. roystoniae (Earle) A.H. Smith, P. trigonospora Dennis, P. varicosa A. Pearson.
Certainly, there are many more species, but they are often not very characteristic and therefore difficult to distinguish from others. A limitation to reasonably known species is advised for this reason.
Within section Candolleana, there several sequences of Cercospora spec. are included. This phenomenon occurs exclusively in section Candolleana. A sequencing error can be excluded for explanation, especially since the authors and the collection regions of the Cercospora spec. sequences vary. This would mean that two unrelated genera have an almost identical ITS region, which is extremely unlikely. This phenomenon cannot be clarified at present.
New combinations:
Candolleomyces aberdarensis (A. Melzer, Kimani & R. Ullrich) Wächter & A. Melzer, comb. nov. MB 832425
Basionym: Psathyrella aberdarensis A. Melzer, Kimani & R. Ullrich, Österr Z Pilzk 27:27, 2019. Reference: Melzer et al. (2019, “2018”).
Candolleomyces albipes (Murill) Wächter & A. Melzer, comb. nov. MB 832260
Basionym: Astylospora (“Atylospora”) albipes Murrill, Mycologia 10(1):22, 1918. References: Smith (1972), Wilhelm (2017, as Psathyrella “alba”).
Candolleomyces badhyzensis (Kalamees) Wächter & A. Melzer, comb. nov. MB 832261
Basionym: Psathyrella badhyzensis Kalamees, Folia cryptog Estonica 15:7, 1981. Reference: Kalamees (1981).
Candolleomyces badiophyllus (Romagn.) Wächter & A. Melzer, comb. nov. MB 832262
Basionym: Drosophila badiophylla Romagn., Bull mens Soc linn Lyon 21:155, 1952. References: Bon and van Haluwyn (1983), Enderle and Christan (1992), Enderle (1994), Hausknecht and Krisai-Greilhuber (2012), Kasik et al. (2004), Kits van Waveren (1985), Krisai-Greilhuber (1992). Mat. exam.: Germany: Saxony, Delitzsch, 25.IX.2008, A. Melzer (AM1175). Saxony-Anhalt, Querfurt, 10.X.2006 A. Melzer (AM814); Querfurt, 15.VII.2007 A. Melzer (AM974).
Candolleomyces bivelatus (Contu) Wächter & A. Melzer, comb. nov. MB 832263
Basionym: Psathyrella bivelata Contu, Bull Soc mycol Fr 107(3):86, 1991. References: Contu (1991), Mifsud (2017). Mat. exam.: Malta: Buskett, 16.XII.2009, C. Sammut (AM1503); 16.XII.2009, C. Sammut (AM1555); 31.XII.2011, C. Sammut (AM1557); 11.IV.2012, C. Sammut (AM1572); 24.IV.2012, C. Sammut (AM1573); 18.X.2012, C. Sammut (AM1759); 18.X.2012, C. Sammut (AM1761); 05.XI.2015, C. Sammut (AM1764); 18.X.2012, C. Sammut (AM1798); 16.IV.2014, C. Sammut (AM1799); 6.XI.2014, C. Sammut (AM1801); 18.X.2014, C. Sammut (AM1802); Wied il-Qleghja, 28.XI.2012., C. Sammut (AM1760).
Candolleomyces cacao (Desjardin & B.A. Perry) Wächter & A. Melzer, comb. nov. MB 832264
Basionym: Psathyrella cacao Desjardin & B.A. Perry, Mycosphere 7(3):378, 2016. Reference: Desjardin and Perry (2016).
Candolleomyces candolleanus (Fr.) Wächter & A. Melzer, comb. nov. MB 832265
Basionym: Agaricus candolleanus Fr., Observ mycol 2:182 (1818). References: Breitenbach and Kränzlin (1995), Consiglio (2000), El-Assfouri et al. (2009), Ludwig (2007), Mifsud (2017), Muñoz and Caballero (2012), Smith (1941). Mat. exam.: Germany: Saxony, Kyhna, 18.X.2001, A. Melzer (AM110); 17.VII.2005, A. Melzer (AM528); 14.VIII.2005, A. Melzer (AM556); 30.VII.2007, A. Melzer (AM972); Wellaune, 20.VI.2004, A. Melzer (AM391); Delitzsch, 26.VI.2007, A. Melzer (AM958); 31.VII.2007, A. Melzer (AM965); Brinnis, 28.IX.2013, A. Melzer (AM1623). Saxony-Anhalt, Querfurt, 19.V.2002, A. Melzer (AM148);15.VIII.2007, A. Melzer (AM971); Landsberg, 14.VI.2002, A. Melzer (AM160); 29.VI.2007, A. Melzer (AM957). Hessen, Gießen, 1.VI.2011, W. Schößler (AM1465); 1.VII.2013, W. Schößler (AM1465). Malta: Buskett, 16.12.09, C. Sammut (AM1554); 08.10.11, C. Sammut (AM1498).
Candolleomyces efflorescens (Sacc.) Wächter & A. Melzer, comb. nov. MB 832266
Basionym: Psathyra efflorescens Sacc., Syll Fung 5:1067, 1887. References: Berkeley et Broome (1871), Kits van Waveren (1995), Pegler (1977), Saccardo (1887).
Candolleomyces fimicola (Atri, Munruchi Kaur & Amandeep Kaur) Wächter & A. Melzer, comb. nov. MB 832268
Basionym: Psathyrella fimicola Atri, Munruchi Kaur & Amandeep Kaur, J New Biol Rep 2(3):276, 2013. References: Amandeep et al. (2015), Kaur et al. (2013). Mat. exam.: India: Punjab, 18.VI.2011, A. Kaur (PUN4317, holotype).
Candolleomyces floccosus (Earle) Wächter & A. Melzer, comb. nov. MB 832269
Basionym: Stropharia floccosa Earle, Inf an Estac Cent agr Cuba 1:241, 1906. References: Morgan (1908), Pegler (1987), Smith (1972).
Candolleomyces graminus (Kalamees) Wächter & A. Melzer, comb. nov. MB 832270
Basionym: Psathyrella gramina Kalamees, Folia Cryptogam Estonica 27:7, 1989. Reference: Kalamees (1989).
Candolleomyces halophilus (Esteve-Raventós & Enderle) Wächter & A. Melzer, comb. nov. MB 832271
Basionym: Psathyrella halophila Esteve-Raventós & Enderle, Z Mykol 58(2):206, 1992. References: Carbó and Pérez-de-Gregorio (1999), Corriol (2014), Esteve-Raventós and Enderle (1992). Mat. exam.: Portugal: District Faro, Algarve, Valle do Garrao, 31.XII.2017, L. Krieglsteiner (LK2).
Candolleomyces leucotephrus (Berk. & Broome) Wächter & A. Melzer, comb. nov. MB 832272
Basionym: Agaricus leucotephrus Berk. & Broome, Ann Mag nat Hist, Ser. 4 6:468, 1870. References: Arnolds (2003), Breitenbach and Kränzlin (1995), Consiglio (2005), El-Assfouri et al. (2009), Enderle and Hübner (2005), Fasciotto (2009), Kits van Waveren (1985), Ludwig (2007), Orton (1960). Mat. exam.: Germany: Baden-Württemberg, Mühlen-Ottenhau, 12.IX.2008, J. Marqua (AM1236).
Candolleomyces luteopallidus (A.H. Sm.) Wächter & A. Melzer, comb. nov. MB 832273
Basionym: Psathyrella luteopallida A.H. Sm., Mem N Y bot Gdn 24:101, 1972. Reference: Smith (1972).
Candolleomyces paecilospermus (Pacioni) Wächter & A. Melzer, comb. nov. MB 832274
Basionym: Psathyrella paecilosperma Pacioni, Micol Veg Medit 13(2):149, 1999. Reference: Pacioni (1999).
Candolleomyces pseudocandolleanus (A.H. Sm.) Wächter & A. Melzer, comb. nov. MB 832276
Basionym: Psathyrella pseudocandolleana A.H. Sm., Mem N Y bot Gdn 24:81, 1972. Reference: Smith (1972).
Candolleomyces rupchandii (A.H. Sm.) Wächter & A. Melzer, comb. nov. MB 832278
Basionym: Psathyrella rupchandii A.H. Sm. Reference: Smith (1972).
Candolleomyces secotioides (G. Moreno, Heykoop, Esqueda & Olariaga) Wächter & A. Melzer, comb. nov. MB 832280
Basionym: Psathyrella secotioides G. Moreno, Heykoop, Esqueda & Olariaga, Mycol Progr 14(6/34):3, 2015. Reference: Moreno et al. (2015).
Candolleomyces singeri (A.H. Sm.) Wächter & A. Melzer, comb. nov. MB 832281
Basionym: Psathyrella singeri A.H. Sm., Mem N Y bot Gdn 24:83, 1972. Reference: Smith (1972). Mat. exam.: Ethiopia: Awurado, 5.XII.2014, A. Gminder; Komba, 10.XII.2014, A. Gminder (both herbar Gminder).
Candolleomyces subsingeri (T. Bau & J.Q. Yan) Wächter & A. Melzer, comb. nov. MB 832282
Basionym: Psathyrella subsingeri T. Bau & J.Q. Yan, Mycokeys 33:94, 2018. References: Ferisin and Melzer (2019, “2018”), Yan and Bau (2018). Mat. exam.: Slovenia, Nova Gorica, 28.VII.2018, G. Ferisin (AM1934, AM1936); 4.VIII.2018, G. Ferisin (AM1940); 5.VIII.2018, G. Ferisin (AM1939); 11.VIII.2018, G. Ferisin (AN1935).
Candolleomyces sulcatotuberculosus (J. Favre) Wächter & A. Melzer, comb. & stat. nov. MB 832287
Basionym: Psathyrella typhae var. sulcatotuberculosa J. Favre, Beitr Kryptfl Schweiz 10(3):215, 1948. References: Battistin et al. (2014), Einhellinger (1976), Ferisin and Melzer (2019, “2018”). Mat. exam.: Germany: Baden-Württemberg, Heidelberg, 1.VII.2012, M. Rave (AM1575). Slovenia: Nova Gorica, 5.VIII.2018, G. Ferisin (AM1937); 16.VIII.2018, G. Ferisin (AM1933, AM1938).
Candolleomyces trinitatensis (R.E.D. Baker & W.T. Dale) Wächter & A. Melzer, comb. nov. MB 832283
Basionym: Psathyrella trinitatensis R.E.D. Baker and W.T. Dale, Mycol Pap 33:93, 1951. References: Baker and Dale (1951), Pegler (1983).
Candolleomyces tuberculatus (Pat.) Wächter & A. Melzer, comb. nov. MB 832284
Basionym: Hypholoma tuberculatum Pat., Bull Soc mycol Fr 15:196, 1899. References: Morgan (1908, as Stropharia tuberculata (Pat.) Morgan), Pegler (1983), Smith (1972).
Candolleomyces typhae (Kalchbr.) Wächter & A. Melzer, comb. nov. MB 832285
Basionym: Agaricus typhae Kalchbr., Mathem Természettud Közlem 2:160, 1863. References: Aronsen (1993), Breitenbach and Kränzlin (1995), Christan et al. (2017), Consiglio (2000), Enderle (1989), Kits van Waveren (1985), Kotlaba (1952), Kreisel (1961), Ludwig (2007). Mat. exam.: Germany: Mecklenburg-Vorpommern, Blankenburg, 11.VII.2009, T. Richter (AM1268); Roduchelstorf, 28.VI.2009, T. Richter (AM1269). Nordrhein-Westfalen, Düsseldorf, 1.VI.2006, K. Büchler (AM791).
Candolleomyces caespitosus (Murill) Wächter & A. Melzer, comb. nov. MB 832286
Basionym: Stropharia caespitosa Murrill, Mycologia 10(2):71, 1918. References: Baker and Dale (1951), Pegler (1983), Pegler (1987), Smith (1972).
Genus Hausknechtia Wächter & A. Melzer, gen. nov. MB 831465 (Fig. 84)
Etymology: Named after the Austrian mycologist Anton Hausknecht.
Description: Basidiomata small, terrestrial on sandy soil. Lamellae deliquescent. Young pileus strikingly sulcate, the margin splitting radially. Pileipellis a hymeniderm. Veil distinct but fugacious, consisting of subcylindrical, branched hyphae. Spores medium-sized, subcylindrical, pale, germ pore absent. Basidia 4-spored. Pleuro- and cheilocystidia absent. Clamps present.
Type species: Hausknechtia floriformis (Hauskn.) Wächter & A. Melzer (Fig. 80).
Representative:
Galerella floriformis Hauskn.; Ref.v.: WU22833 (Tóth et al. 2013)
New combination:
Hausknechtia floriformis (Hauskn.) Wächter & A. Melzer, comb. nov. MB 831956
Basionym: Galerella floriformis Hauskn., Österr Z Pilzk 12:34, 2003. References: Hausknecht and Contu (2003).
Olotia Wächter & A. Melzer, gen. nov. MB 831466 (Fig. 84)
Etymology: Named after the city Olot (Spain), the type locality of the type species.
Description: Basidiomata small, lignicolous. Veil sparse. Spores medium-sized, ellipsoid to slightly ovoid in frontal view, dark, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge predominantly lageniform or clavate. Pleurocystidia mostly spatula-shaped and strongly pediculated, walls often slightly thickened and brownish pigmented. Clamps present. Figure 81 illustrates the microcharacters.
Type species: Olotia codinae (Deschuyteneer, A. Melzer, Pérez-De-Gregorio) Wächter & A. Melzer.
Representative:
Psathyrella codinae Deschuyteneer, A. Melzer, Pérez-De-Gregorio; Ref.v.: GLM-F112430/type (Deschuyteneer et al. 2018).
New combination:
Olotia codinae (Deschuyteneer, A. Melzer, Pérez-De-Gregorio) Wächter & A. Melzer, comb. nov. MB 832424
Basionym: Psathyrella codinae Deschuyteneer, A. Melzer, Pérez-De-Gregorio, Bulletin de l’Association des Mycologues francophones de Belgique 11:6, 2018. Reference: Deschuyteneer & al. (2018). Mat. exam.: Spain, Catalonia, Olot, 06.V.2017, leg. Miquel À. Pérez-De-Gregorio (holotype, GLM-F112430).
Remarks:
So far, only a single species is known. A colour photograph is shown in Deschuyteneer et al. (2018) on page 4.
Punjabia Wächter & A. Melzer, gen. nov. MB 831468 (Fig. 84)
Etymology: Named after the Pakistani province of Punjab with the type locality.
Description: Basidiomata medium-sized, terrestrial. Pileus strongly plicate, with greenish tones, “light yellow green, greyish-greenish-yellow” (Hussain et al. 2018). Veil sparse, composed of subglobose and globose cells with slightly thickened walls. Spores medium-sized, broadly ellipsoid to slightly ovoid in frontal view, dark, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge subcylindrical, utriform, with some crystals at the apex. Pleurocystidia absent. Pileocystidia present. Clamps present.
Type species: Punjabia pakistanica (Usman & Khalid) Wächter & A. Melzer. See Fig. 82.
Representative:
Coprinellus pakistanicus Usman & Khalid; Ref.v.: MEL 2382843A in accordance with LAH35322 (Hussain et al. 2018)
New combination:
Punjabia pakistanica (Usman & Khalid) Wächter & A. Melzer, comb. nov. MB 831957
Basionym: Coprinellus pakistanicus Usman & Khalid, Mycokeys 39:53, 2018. Reference: Hussain et al. (2018).
Remarks:
Distinctly greenish hues are very rare in the family, so far only known in Candolleomyces tuberculatus (Pat.) Wächter & A. Melzer, Psathyrella glaucescens Dennis (both with a greenish pileus), and Coprinopsis piepenbroekorum (Uljé & Bas) Redhead, Vilgalys & Moncalvo (greenish veil).
In Hussain et al. (2018), the drawing (Fig. 6d) shows subcylindrical and utriform cheilocystidia, although lageniform cystidia are mentioned in the description. Perhaps there is an error in the study of Hussain et al. (2018); the authors in the present paper gave priority to the drawing (Fig. 6d).
Cystoagaricus Singer emend. Örstadius & E. Larss. (Fig. 84).
Sequences exist for the following recognized species: Cystoagaricus hirtosquamulosus (Peck) Örstadius & E. Larss., C. olivaceogriseus (A.H. Sm.) Örstadius & E. Larss., C. squarrosiceps (Singer) Örstadius & E. Larss. and C. strobilomyces (Murrill) Singer. Psathyrella lepidotoides A.H. Sm. and P. populina Britzelm. undoubtedly belong to Cystoagaricus. On one hand, these are certainly different species, but on the other hand they are not newer synonyms of Hypholoma sylvestre Gillet. The name Cystoagaricus silvestris Örstadius & E. Larss. is not applicable. A detailed study of Cystoagaricus is in progress (Muñoz in prep.).
Typhrasa Örstadius & E. Larss. (Fig. 84)
Currently two species are known: Typhrasa gossypina (Bull.) Örstadius & E. Larss. and T. nanispora Örstadius, Hauskn. & E. Larss. Whether Psathyrella delineata (Peck) A.H. Sm. represents an independent species is not clear yet.
Kauffmania Örstadius & E. Larss. (Fig. 84)
The genus currently only includes Kauffmania larga (Kauffman) Örstadius & E. Larss (Fig. 83).
Phylogram part of the genera Hausknechtia, Olotia, Punjabia, Cystoagaricus, Typhrasa and Kauffmania; position in tree see Fig. 42
Phylogram part of the section Coprinopsis; position in tree see Fig. 87
Phylogram part of the section Cinereae; position in tree see Fig. 87
Phylogram part of the sections Narcoticae and Mitraesporae; position in tree see Fig. 87
Phylogram part of the sections Alopeciae and Xenobiae; position in tree see Fig. 87
Phylogram part of the sections Geesteranorum and Atramentariae; position in tree see Fig. 87
Phylogram part of the sections Phlyctidosporae, Krieglsteinerorum, and Erythrocephalae; position in tree see Fig. 87
Phylogram part of the section Lanatulae; position in tree see Fig. 87
ITS alignment of the “/lagopus A” down to “/pachyderma” clade. Coloured sites (in AliView colour code) represent the difference from majority rule consensus. Grey areas are sites which match the consensus. White sites are gaps. The yellow area highlights the sites which were used for the sequence key. Presentation from AliView. The scale represents the site numbering
Genetic key to separate the nine clades: 1 = C. lagopus A; 2 = C. lagopus B (note the 2 different rows); 3 = C. lagopus C; 4 = C. lagopus D; 5 = C. lagopus E; 6 = C. lagopus F; 7 = C. lagopus G; 8 = C. brunneofibrillosa; 9 = C. pachyderma. The yellow framed area is suspect. The scale shows the site numbering of the ITS1 loci. Presentation created with AliView; colours: Nucleotides in AliView colour code
Phylogram part of the section Radiatae; position in tree see Fig. 87
Phylogram part of the sections Filamentiferae, Picaceae and Melanthinae; position in tree see Fig. 87
Phylogram part of the sections Subniveae, Niveae, Canocipes and Quartoconatae; position in tree see Fig. 87
Phylogram part of the genera Lacrymaria and Homophron; position in tree see Fig. 42
Phylogram part of the genus Parasola; position in tree see Fig. 42
Coprinopsis P. Karst.
Overview
Twenty sections of genus Coprinopsis were possible to identify with the currently available sequences data and morphological features. The 360° radial consensus phylogram in Fig. 85 clearly shows their divergence and further that the sections Quartoconatae to Subniveae form a distinct basal group within the genus Coprinopsis.
The radial cladogram in Fig. 86 illustrates the numbers and the relationships of the taxa in the proposed sections.
Figure 87 shows an overview phylogram collapsed to section level and serves above all for further orientation; the red brackets refer to the detailed phylograms.
As all others before, Fig. 85, Fig. 86 and Fig. 87 are partial cuts from the complete main phylogram (see Fig. 42).
Coprinopsis sect. Coprinopsis (Fig. 88)
Description: Basidiomata small to medium-sized, fimicolous, terrestrial, herbiolous or lignicolous. Lamellae deliquescent. Veil strongly developed, consisting of chains of diverticulate, thin or thick-walled colourless to brownish pigmented more or less coralloid cells. Spores medium to large-sized, in frontal view ellipsoid, ovoid, subtriangular or subglobose, laterally sometimes distinctly flattened. Basidia 4-spored. Marginal cells of the lamellar edge utriform, clavate, fusiform. Pleurocystidia utriform, subcylindrical, clavate, fusiform, sometimes mucronate. Clamps present in most cases.
Representatives:
Coprinopsis alcobae (A. Ortega) Valade; Ref.v.: SZMC-NL-0767 (Nagy et al. 2012)
Coprinopsis argentea (P.D. Orton) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-1678 (Nagy et al. 2012)
Coprinopsis episcopalis (P.D. Orton) Redhead, Vilgalys & Moncalvo; Ref.v.: F-062,769 (Gonzalez del Val et al. 2003)
Coprinopsis gonophylla (Quél.) Redhead, Vilgalys & Moncalvo; Ref.v.: ST-R-9 (Li et al. 2016)
Coprinopsis kubickae (Pilát & Svrček) Redhead, Vilgalys & Moncalvo; Ref.v.: CID1342 (Shipunov et al. 2008)
Coprinopsis phaeopunctata (Esteve-Rav. & A. Ortega) Valade; Ref.v.: AH 18881/type (Nagy et al. 2012)
Coprinopsis pseudofriesii (Pilát & Svrček) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-2631 (Nagy et al. 2012)
Coprinopsis sclerotiorum (Horvers & de Cock) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-0564 (Nagy et al. 2012)
Coprinopsis spilospora (Romagn.) Redhead, Vilgalys & Moncalvo; Ref.v.: LO 73-97 (Nagy et al. 2012)
Coprinopsis urticicola (Berk. & Broome) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-0170 (Nagy et al. 2012)
Coprinopsis vermiculifera (Joss. ex Dennis) Redhead, Vilgalys & Moncalvo; Ref.v.: CBS132.46 (Nagy et al. 2011)
Coprinus subdomesticus Murill; Ref.v.: Murrill459/type (Nagy et al. 2012)
Remarks:
The section includes the type of the genus designated by Earle (1909:384): Coprinopsis friesii (Quél.) P. Karst., Acta Soc Fauna Flora fenn 2(1):27, 1881. For this reason, the hitherto used name Alachuani Singer is to be rejected.
There was no usable sequence of C. friesii (only voucher SZMC-NL-0565 without ITS). Therefore, the voucher Germany: Saxony, Kyhna, 24.VI.2007, A. Melzer (AM954) was examined and the sequence deposited at GenBank as MK072829.1. The phylogenetic position is near M200T-4-EM2, but with a clear distance, so that these are not identical species. After completion of the phylogram, the sequence MH422562.1 of Coprinopsis kubickae (voucher CNF 1/6614, Tkalcec et al. unpubl.) was available. A comparison showed the identity with CID1342, and therefore this species unquestionably belongs to the section Coprinopsis.
The following species are most likely to be included here; however, the legitimacy of the status of an independent species should be checked using molecular biological methods:
Coprinopsis austrofriesii (Redhead & Pegler) Redhead, Vilgalys & Moncalvo. References: Dennis (1961, as Coprinus friesii), Redhead and Traquair (1981)
Coprinopsis burkii (A.H. Sm.) Redhead, Vilgalys & Moncalvo. References: Smith and Hesler (1946)
Coprinopsis herinkii (Pilát & Svrček) Redhead, Vilgalys & Moncalvo. References: Pilát and Svrček (1967), Uljé and Noordeloos (1997)
Coprinopsis phaeospora (P. Karst.) P. Karst. References: Aronsen (1993), Gierczyk et al. (2011), Ludwig (2007), Pilát and Svrček (1967), Reid (1958, as Coprinus saichiae D.A. Reid), Uljé and Noordeloos (1997), Vila and Rocabruna (1996), Watling (1967, as Coprinus saichiae)
Coprinopsis subtigrinella (Dennis) Redhead, Vilgalys & Moncalvo. References: Dennis (1961);
Coprinopsis tigrinella (Boud.) Redhead, Vilgalys & Moncalvo. References: Aronsen (1993), Gierczyk et al. (2011), Krieglsteiner and Gminder (2010), Krieglsteiner et al. (1982), Mifsud (2017), Nagy (2007), Uljé and Noordeloos (1997)
Coprinopsis xantholepis (P.D. Orton) Redhead, Vilgalys & Moncalvo. References: Krisai-Greilhuber (1992), Orton (1972), Uljé and Noordeloos (1997), Ruiz Mateo (2013)
Compared to other sections of Coprinopsis, the sequences in this section show much higher divergency, namely with longest path length = 0.24 expected changes/site rather than 0.03–0.12 expected changes/site on average. This explains why apparently closely adjacent taxa have distinctly different morphological features. Therefore, the scale bar of the phylogram must be observed. See collapsed triangle phylogram of section Coprinopsis Fig. 87 for this as well.
New combination:
Coprinopsis subdomestica (Murill) Wächter & A. Melzer, comb. nov. MB 831728
Basionym: Coprinus subdomesticus Murill, Proceedings of the Florida Academy of Sciences 7 (2/3):126, 1945. References: Murill (1945).
Coprinopsis sect. Cinereae Wächter & A. Melzer, sect. nov. MB 831473 (Fig. 89)
Description: Basidiomata medium to large-sized, gregarious to caespitose, fimicolous or lignicolous, never terrestrial, stipe often with a pseudorrhiza. Lamellae deliquescent. Veil distinct, consisting of chains of more or less hyaline, subcylindrical, not diverticulate cells. Spores large-sized, ellipsoid to slightly ovoid in frontal view, smooth or warty, with a truncate central germ pore often surrounded by a ridge. Basidia 4-spored. Marginal cells of the lamellar edge globose, ellipsoid, sometimes mixed with utriform cheilocystidia. Pleurocystidia, subglobose, ellipsoid, subcylindrical, utriform or lageniform. Clamps present.
Type species: Coprinopsis cinerea (Schaeff.) Redhead, Vilgalys & Moncalvo, Taxon 50(1):227, 2001.
Representatives:
Coprinopsis afrocinerea Mešić, Tkalčec, Čerkez, I. Kušan & Matočec; Ref.v.: CNF 1/5838 (Crous et al. 2018)
Coprinopsis annulopora (Enderle) P. Specht & H. Schubert; Ref.v.: Enderle 30.71987 (Nagy et al. 2013)
Coprinopsis calospora (Bas & Uljé) Redhead, Vilgalys & Moncalvo; Ref.v.: Bas 8795a/type (Nagy et al. 2013)
Coprinopsis cinerea (Schaeff.) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-1266 (Nagy et al. 2013)
Coprinopsis neocinerea nom. prov.; Ref.v.: CBM-FB39575 (Nguyen et al. unpubl.)
Remarks:
In addition to Coprinopsis “neocinerea”, the section most likely contains more undescribed species. Morphological details are not available; the collections are from India, Puerto Rica and New Mexico and were examined in the context of specific ecological questions, which do not take into account the morphology. The ecology is partly remarkable; see Porras-Alfaro et al. (2008) and Cantrell et al. (2013).
The name Tomentosi Fr. is not applicable beyond doubt. Fries (1838:245) characterized Coprinus Tribus Pelliculosi * 4. Tomentosi Fr. as follows: “Tomentosi, pileo squamulis floccosis discretis villove laxo secedentibus primo velato. Annulus 0! Subfimicolae”. The type, unique by the choice of the name, is Coprinus tomentosus (Bull.) Fr. However, Bulliard (1783: Plate 138) wrote about Agaricus tomentosus Bull. “... dans le bois, les jardins. Il ne vient qu’en bonne terre, sur du terreau ou sur de vieills couches”, so the designated location is not entirely fimicolous. Plate 138 is more likely to be interpreted as Coprinopsis lagopus (Fr.) Redhead, Vilgalys & Moncalvo. This note is also given by Redhead et al. (2001). Pennington (1918:214) describes his section Tomentosi with the words “Universal veil a loose villose web which becomes torn into distinct floccose scales”. This concurs with Fries (1838) and leaves no doubt that Pennington refers to Fries (1838). The first of the following species is Coprinus fimetarius Fr. (≡ Coprinopsis cinerea), but section Tomentosi is not explicitly typified. In addition, there are doubts about Pennington’s identification, because he mentioned a yellowish ozonium, but no pseudorrhiza. Also not applicable is Lentispora Fayod, Ann Sci Nat Bot 9:379, 1889. The description alone is problematic: “Spores lenticulaires, rarement aplaties légèrement sur les côtés”. Lentispora Fayod, Ann Sci Nat Bot 9:379, 1889 is also not applicable. The description of this genus alone is problematic: “Spores lenticulaires, rarement aplaties légèrement sur les côtés”. The type of this genus, Coprinus tomentosus was designated by Earle (1909:435), so it remains an unclear species (see above).
For these reasons, the authors of the present study decided to create a new section with a separate type for the clade shown in Fig. 89.
Note the anamorph of Coprinopsis cinerea is Hormographiella aspergillata (see Table 6 and Fig. 89).
Coprinopsis sect. Narcoticae (Uljé & Noordel.) D.J. Schaf., Field Mycology 11(2):51, 2010 (Fig. 90)
Description: Basidiomata tiny to medium-sized, terrestrial, lignicolous or fimicolous. Lamellae deliquescent. Some species with an unpleasant odour. Veil strongly developed, predominantly consisting of globose, mostly hyaline, thin-walled or slightly thick-walled, densely warty cells, connected by thin, often also warty and diverticulate hyphae; the globose elements are rarely directly connected. Spores small to large-sized, ellipsoid in frontal view, always with a more or less distinct perisporium and a central germ pore. Basidia 4-, 2-, rarely 3-spored. Marginal cells of the lamellar edge consisting of subglobose cells and utriform, fusiform or lageniform cheilocystidia. Pleurocystidia always present, shaped like the cheilocystidia, usually slightly larger. Clamps present or absent.
Type species: Coprinus narcoticus (Batsch) Fr., Epicr Syst Mycol:250, 1838 ≡ Coprinopsis narcotica (Batsch) Redhead, Vilgalys & Moncalvo, Taxon 50(1):229, 2001, designated by Uljé and Noordeloos (1993:262).
Representatives:
Coprinopsis foetidella (P.D. Orton) A. Ruiz, G. Muñoz; Ref.v.: SZMC-NL-3187 (Nagy et al. 2012)
Coprinopsis laanii (Kits van Wav.) Redhead, Vilgalys & Moncalvo; Ref.v.: CBS476.70 (Nagy et al. 2011)
Coprinopsis narcotica (Batsch) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-2342 (Nagy, Urban et al. 2010)
Coprinopsis sclerotiger (Watling) Redhead, Vilgalys & Moncalvo; Ref.v.: CBS596.80 (Nagy et al. 2011)
Coprinopsis semitalis (P.D. Orton) Redhead, Vilgalys & Moncalvo; Ref.v.: CBS291.77/type (Nagy et al. 2011)
Coprinopsis stercorea (Fr.) Redhead, Vilgalys & Moncalvo; Ref.v.: SFSU DEH2074A (Keirle et al. 2004)
Remarks:
The clade /trispora could be named by comparison with the voucher Germany: Bavaria, Waldershof, 22.VII.2018, M. Reul (MR180722). For this reason, Coprinopsis trispora (Kemp & Watling) Redhead, Vilgalys & Moncalvo also belongs in this section. The sequences of this voucher are deposited at GenBank as: ITS: MN227299.1; LSU: MN227300.1.
The following species certainly belong here, too:
Coprinopsis cinereofloccosa (P.D. Orton) Redhead, Vilgalys & Moncalvo. References: Breitenbach and Kränzlin (1995), Giercyk et al. (2014), Kits van Waveren (1968), Ludwig (2007) Orton (1972), Ruiz Mateo and Cerdán (2016)
Coprinopsis martinii (P.D. Orton) Redhead, Vilgalys & Moncalvo. References: Breitenbach and Kränzlin (1995), Kits van Waveren (1968), Krieglsteiner and Gminder (2010), Krieglsteiner et al. (1982), Ludwig (2007), Ruiz Mateo and Cerdán (2016), Watling (1967)
Coprinopsis radicans (Romagn.) Redhead, Vilgalys & Moncalvo. References: Kits van Waveren (1968), La Chiusa and Mauri (1996), Ludwig (2007), Romagnesi (1951)
Coprinopsis saccharomyces (P.D. Orton) P. Roux & Guy Garcia. References: Breitenbach and Kränzlin (1994, 1995), Ludwig (2007), Orton (1960)
Coprinopsis tuberosa (Quél.) Doveri, Granito & Lunghini. References: Breitenbach and Kränzlin (1995), Cacialli et al. (1999), Iglesias et al. (2015), Krieglsteiner et al. (1982), Krieglsteiner and Gminder (2010), Ludwig (2007), Melzer (2009b), Vila and Rocabruna (1996)
This section also includes C. poliomalla (Romagn.) Doveri, Granito & Lunghini. However, the voucher SZMC-NL-2336 is not useful for a positioning, because the ITS (FM163182.1) is exactly the same as that of the voucher SZMC-NL-2336 of C. bellula (Uljé) P. Roux & Eyssartier (FM163176.1). This is not possible; there is probably a mistake or a typographical error. For the set FM163182.1, FM160727.1, FN396275.1, FM897244.1 (Nagy et al. 2011), only the LSU sequence is correct. To clarify this question, voucher Germany: Nordrhein-Westfalen, Brüggen, 19.XII.2015, H. Bender (HB20151219A) was examined. The ITS sequence allows an assignment in the neighbourhood of C. foetidella. The morphology differs from the other members of the section: the veil does not consist of densely warty subglobose cells, but of strongly encrusted cells; furthermore, the spores have no perisporium. It is possible that the clade /foetidella forms a separate section. For a concrete assessment, further species must be found. The sequences of the voucher HB20151219A are deposited at GenBank as MK072612.1 (ITS) and MK072613.1 (LSU).
Whether C. clastophylla (Maniotis) Redhead, Vilgalys & Moncalvo is an independent species or an aberrant form of C. sclerotiger has to be left unclear. According to the description of Maniotis (1964), the species does not belong in the section Narcotici, because the typical veil is missing and the spores are without a perisporium.
Coprinopsis sect. Mitraesporae Wächter & A. Melzer, sect. nov. MB 831474 (Fig. 90)
Description: Basidiomata rather large-sized, lignicolous, with a preference for tree caves. Lamellae deliquescent. Veil strongly developed, consisting of chains of subcylindrical, hyaline or brownish pigmented cells. Spores medium-sized, fusiform to rhomboid in frontal view, laterally somewhat flattened, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge utriform and clavate. Pleurocystidia subglobose, ellipsoid, subcylindrical, utriform. Clamps present.
Type species: Coprinopsis mitraespora (Bohus) L. Nagy, Vágvölgyi & Papp, Mycologia 105(1):120, 2013.
Representative:
Coprinopsis mitraespora (Bohus) L. Nagy, Vágvölgyi & Papp; Ref.v.: WU14574 (Nagy et al. 2013)
Remarks:
This section forms the basal group of the section Narcoticae, with strongly divergent morphology. The habit is completely different, the spores have no perisporium and a different shape and the veil is differently structured.
The priority of Coprinus mitraesporus Bohus before Coprinus spelaiophilus Bas & Ulje (≡ Coprinopsis spelaiophila (Bas & Uljé) Redhead, Vilgalys & Moncalvo) is conclusively proven in Nagy et al. (2013).
Coprinopsis sect. Alopeciae Wächter & A. Melzer, sect. nov. MB 831475 (Fig. 91)
Description: Basidiomata small to rather large-sized, terrestrial or lignicolous. Lamellae deliquescent. Veil moderately to vigorously developed, fugacious, consisting of chains of subcylindrical to subglobose, hyaline cells. Spores large-sized, warty or smooth, apically prolonged or conspicuously conical to limoniform, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge utriform, subcylindrical, clavate. Pleurocystidia absent or present, subcylindrical, utriform. Clamps present.
Type species: Coprinopsis alopecia (Lasch) La Chiusa & Boffelli, Index Fungorum 333:1, 2017.
Representatives:
Coprinopsis alopecia (Lasch) La Chiusa & Boffelli; Ref.v.: SZMC-NL-1510 (Nagy et al. 2013)
Coprinopsis fusispora L. Nagy, Vágvölgyi and Papp; Ref.v.: SZMC-NL-1227/type (Nagy et al. 2013)
Coprinopsis rugosomagnispora Gierczyk, Pietras, Piatek, Gryc, Czerniawski & Rodriguez-Flakus; Ref.v.: KRAM F-58717/type (Gierczyk et al. 2017)
Remarks:
Morphological reasons include the vouchers SZMC-NL-2141 (FN396149.1, FN396190.1, FN396291) and SZMC-NL-1054 (JX118739, JX118813.1) in the section Alopeci. SZMC-NL-2141 was originally named Coprinopsis cinerea, but then recognized as Coprinopsis fusispora (Nagy et al. 2013). However, the phylogenetic position speaks against it. About SZMC-NL-1054 is noted in Nagy et al. (2013) “The species designated as C. sp. 3 could not be separated from C. fusispora morphologically, whereas the phylogeny provides strong support for its status as a separate species”.
The conspecificity of Coprinopsis alopecia (≡ Coprinus alopecius Lasch) and Coprinopsis insignis (Peck) Redhead, Vilgalys & Moncalvo (≡ Coprinus insignis Peck) is not clarified beyond doubt; for the time being, the older name was preferred here.
Coprinopsis sect. Xenobiae Wächter & A. Melzer, sect. nov. MB 831476 (Fig. 91)
Description: Basidiomata small to medium-sized, terrestrial or fimicolous. Lamellae deliquescent. Veil primarily composed of chains of branched, slightly diverticulate, hyaline to brownish, sometimes encrusted cells. Spores medium to large-sized. Basidia 4-spored. Marginal cells of the lamellar edge utriform, clavate, subcylindrical. Pleurocystidia utriform, ellipsoid. Clamps present.
Type species: Coprinopsis xenobia (P.D. Orton) Redhead, Vilgalys & Moncalvo, Taxon 50(1):232, 2001.
Representatives:
Coprinopsis fluvialis (Lanconelli & Uljé) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-0840 (Nagy et al. 2012)
Coprinopsis ochraceolanata (Bas) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-0192 (Nagy et al. 2013)
Coprinopsis xenobia (P.D. Orton) Redhead, Vilgalys & Moncalvo; Ref.v.: BR 302 (Ruiz Mateo et al. 2013)
Remarks:
C. ochraceolanata takes an intermediate position. The morphological features do not match either the section Xenobiae or the section Alopeciae; for example, the rhizomorphs or the pseudorrhiza as well as the heavily encrusted veil are particularities. However, the establishment of a separate section is currently being dispensed with. The examination of other species of these sections should provide a better overview.
Coprinopsis sect. Geesteranorum Wächter & A. Melzer, sect. nov. MB 831477 (Fig. 92)
Description: Basidiomata very small to medium-sized, terrestrial. Lamellae deliquescent. Veil strongly developed but very fugacious, consisting of chains of subcylindrical, hyaline to pale brownish cells. Spores small to medium-sized, ellipsoid in frontal view with a conical base, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge utrifom. Pleurocystidia utriform. Clamps present.
Type species: Coprinopsis geesterani (Uljé) Redhead, Vilgalys & Moncalvo, Taxon 50(1):228, 2001.
Representative:
Coprinopsis geesterani (Uljé) Redhead, Vilgalys & Moncalvo; Ref.v.: Ulje 1078/type (Nagy et al. 2013)
Remarks:
So far only one species is known.
Coprinopsis sect. Atramentariae (Fr.) D.J. Schaf., Field Mycology 11(2):51, 2010 (Fig. 92)
Description: Basidiomata medium to large-sized, terrestrial or lignicolous with a volva-like annular zone near the stipe base. Lamellae deliquescent. Veil strongly developed, consisting of chains of subcylindrical, hyaline or pale brownish pigmented, usually thin-walled, occasionally branched and slightly diverticulate, sometimes encrusted cells. Spores medium-sized, base often conspicuously conical, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge mainly utriform or subcylindrical, sometimes clavate. Pleurocystidia subcylindrical, utriform. Clamps present.
Type species: Agaricus atramentarius Bull., Herb Fr 6:tab. 164, 1786, designated by Fries (1838:243), uniquely determined by the choice of the name.
≡ Coprinopsis atramentaria (Bull.) Redhead, Vilgalys & Moncalvo, Taxon 50(1):226, 2001.
Representatives:
Coprinopsis acuminata (Romagn.) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-3028 (Nagy et al. 2012)
Coprinopsis atramentaria (Bull.) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-3399 (Nagy et al. 2011)
Coprinopsis romagnesiana (Singer) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-2148 (Nagy et al. 2013)
Remarks:
The neighbouring sections Atramentariae and Geesteranorum hardly show any characteristic transitions, let alone similarities. Only a tendentially conical spore base is to be noted.
Coprinopsis sect. Phlyctidosporae Wächter & A. Melzer, sect. nov. MB 831478 (Fig. 93)
Description: Basidiomata small to very large-sized, lignicolous, fimicolous or growing on very fertilized soil. Veil moderately developed, consisting of chains of thin-walled, colourless, branched, slightly diverticulate cells. Spores medium to large-sized, ellipsoid, ovoid or limoniform in frontal view, laterally hardly flattened, rough to warty, germ pore central, sometimes prolonged. Basidia mostly 4-spored. Marginal cells of the lamellar edge predominantly utriform, globose or clavate. Pleurocystidia voluminous, utriform, subcylindrical, clavate. Clamps present.
Type species: Coprinopsis phlyctidospora (Romagn.) Redhead, Vilgalys & Moncalvo, Taxon 50(1):230, 2001.
Representatives:
Coprinopsis asiaticiphlyctidospora Fukiharu & Horigome; Ref.v.: CBM-FB38668/type (Fukiharu et al. 2013)
Coprinopsis austrophlyctidospora Fukiharu; Ref.v.: 201NZ (CHU3002)/type (Suzuki et al. 2002)
Coprinopsis echinospora (Buller ex Buller) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-4249 (Nagy et al. 2012)
Coprinopsis neophlyctidospora Raut, Fukiharu & A. Suzuki; Ref.v.: CBM-FB-37998/type (Raut et al. 2011)
Coprinopsis novorugosobispora Fukiharu & Yamakoshi; Ref.v.: AB978534.1 (Raut et al. 2015)
Coprinopsis phlyctidospora (Romagn.) Redhead, Vilgalys & Moncalvo; Ref.v.: CBM-FB21061 (Suzuki et al. 2002)
Coprinopsis rugosobispora (J. Geesink & Imler ex Walleyn) A. Melzer & Schößler; Ref.v.: BR-44338-09 (Raut et al. 2015)
Remarks:
C. novorugosobispora and C. rugosobispora may be only 2-spored forms of C. phlyctidospora; at least they are not two separate species. For a final decision, further investigations are necessary.
The clade /echinospora could be interpreted as a separate section. But since morphology does not show striking differences, this is discarded for the time being.
Coprinopsis sect. Krieglsteinerorum Wächter & A. Melzer, sect. nov. MB 831479 (Fig. 93)
Description: Basidiomata medium-sized, terrestrial. Lamellae deliquescent. Veil sparse, consisting of chains of hyaline cells. Stipe with a pseudorrhiza. Spores large-sized, smooth, ellipsoid, germ pore central. Marginal cells of the lamellar edge utriform, lageniform, sometimes subcapitate, undermixed with clavate and sphaeropedunculate cells. Pleurocystidia utriform, sublageniform, subcylindrical. Clamps present.
Type species: Coprinopsis krieglsteineri (Bender) Redhead, Vilgalys & Moncalvo, Taxon 50(1):229, 2001.
Representative:
Coprinopsis krieglsteineri (Bender) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-2345 (Nagy et al. 2013)
Remarks:
From a phylogenetic point of view, it is not possible to tell whether the section Krieglsteinerorum and the neighbour section Erythrocephalae are actually independent sections. Both are located at the base of the clade Phlyctidospori, which means that they may have different morphological characteristics as usual for basal clades. Because these differences are relatively significant (especially the spore surface), a separation is currently made. On the other hand, C. krieglsteineri and C. erythrocephala are the only representatives of their sections that have striking similarities; both have a pseudorrhiza, spores and cystidia are quite similar. The main difference only concerns the veil. It is conceivable that the abovementioned species could be assigned to a common section if further information is available, in particular by finding and examining other species from this group.
Coprinopsis af. krieglsteineri might well be a new taxon, as it differs from C. kriegelsteineri in the non-rooting basidiomata and variations in the pleurocystidia, but it shares habit and pileus colour with that species (Nagy et al. 2013).
Coprinopsis sect. Erythrocephalae Wächter & A. Melzer, sect. nov. MB 831489 (Fig. 93)
Description: Basidiomata medium-sized, terrestrial or lignicolous. Lamellae deliquescent. Veil reddish, moderately developed, fugacious, consisting of chains of subcylindrical, hyaline to brownish, partially strongly encrusted cells. Stipe with a pseudorrhiza. Spores large-sized, smooth, ellipsoid to ovoid, base conical, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge utriform, clavate. Pleurocystidia utriform. Clamps present.
Type species: Coprinopsis erythrocephala (Lév.) Redhead, Vilgalys & Moncalvo, Taxon 50(1):228, 2001.
Representative:
Coprinopsis erythrocephala (Lév.) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-1701 (Nagy et al. 2013)
Remarks:
So far only one species is known. See also the notes at Krieglsteinerorum.
Coprinopsis sect. Lanatulae (Fr.) D.J. Schaf., Field Mycology 11(2): 51, 2010 (Fig. 94)
Description: Basidiomata small to rather large-sized, terrestrial or fimicolous. Lamellae deliquescent. Veil strongly developed, consisting of chains of subcylindrical to subglobose, hyaline or brownish pigmented cells. Spores medium to large-sized, ellipsoid in frontal view, regularly without a perisporium, germ pore central. Basidia mostly 4-spored. Marginal cells of the lamellar edge globose, ellipsoid, clavate, sometimes mixed with utriform cheilocystidia. Pleurocystidia present in most species. Clamps present.
Type species: Coprinus lagopus (Fr.) Fr. ≡ Coprinopsis lagopus (Fr.) Redhead, Vilgalys & Moncalvo, Taxon 50(1):229, 2001, designated by Singer (1975:494).
Representatives:
Coprinopsis babosiae L. Nagy, Vágvölgyi & Papp; Ref.v.: SZMC-NL-4139/type (Nagy et al. 2011, 2013)
Coprinopsis bicornis (Uljé & Horvers) Redhead, Vilgalys & Moncalvo; Ref.v.: Ulje 1216/type (Nagy et al. 2013)
Coprinopsis brunneofibrillosa (Dennis) Redhead, Vilgalys & Moncalvo; Ref.v.: Dennis 1120 (Nagy et al. 2013)
Coprinopsis brunneistragulata (Bogart ex Bogart) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-FVBD 3821/type (Nagy et al. 2013)
Coprinopsis jonesii (Peck) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-0154 (Nagy et al. 2013)
Coprinopsis lagopus (Fr.) Redhead, Vilgalys & Moncalvo; Ref.v.: Kemp 1431 (Nagy et al. 2013)
Coprinopsis pachyderma (Bogart ex Bogart) Redhead, Vilgalys & Moncalvo; Ref.v.: FVDB 3237/type (Nagy et al. 2013)
Coprinopsis pseudoradiata (Kühner & Joss. ex Watling) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-0956 (Nagy et al. 2013)
Coprinopsis scobicola (P.D. Orton) Redhead, Vilgalys & Moncalvo; Ref.v.: Orton 964/type (Nagy et al. 2012)
Remarks:
There are sequences of the type of C. brunneofibrillosa available (voucher Dennis 1120). According to Nagy et al. (2013), the sequencing was not successful, so that the voucher Nicholson 426 was used. However, a review revealed that the LSU region of Dennis 1120 is faulty and unusable; ITS and β-tubulin, on the other hand, are fine. ITS1 lacks a subset, which proved to be unproblematic. In any case, the two vouchers mentioned above represent different species. Nicholson 426 is most likely C. cinerea.
C. bicornis and C. scobicola are in the /pseudoradiata clade, which is undoubtedly correct, because of the inclusion of the type. The same was stated by Nagy et al. (2013): “Although the phylogenetic analyses do not support the recognition of these species as separate (except C. babosiae), on the basis of clear-cut morphological differences, we raise the possibility that more variable loci would provide unambiguous support for them”. This statement may well apply; but it should also be considered that it could be 2-spored forms of other species. To be similarly assessed is C. brunneistragulata within the /jonesii clade. The peculiarity is the presence of a perisporium, while the other characteristics are not fundamentally different. C. babosiae should not have pleurocystidia in contrast to the other species. Incidentally, this species is provisionally named C. subgeesterani in the phylogram of Nagy et al. (2013).
The species group around C. lagopus is the most difficult to assess. While C. jonesii can be addressed reasonably precisely, several subclades remain, where members are difficult to identify in a conventional way (see Nagy et al. 2013). The status of C. lagopus var. vacillans (Uljé) P. Roux & Guy Garcia, contained in several subclades, must be regarded as unclear; maybe they are only hunger forms of different species and resulting in erroneous determinations. For the characteristics of C. lagopus var. vacillans, see Uljé et al. (2000).
Within the loci analysed, the members of the lagopus group show a distinct divergence in a small segment of the ITS1 region only. There are some small differences in the ITS2 and the LSU region as well, but these are negligible. This might explain the surely difficult morphological separation. The phylogenetic divergence from about site 120 to site 235 of the ITS1 region is distinct. At this point, some deletions or insertions also took place. For a better phylogenetic resolution of the lagopus aggregate, it is necessary to use indel matrices. Figure 95 shows the genetic differences across the entire ITS1 to ITS2 region for the complete clades “/lagopus A” down to “/pachyderma” with highlighted non-consensus sites and indels in white areas. Other parts of the analysed loci are negligible for this purpose. Figure 96 shows a genetic key, generated from these input ITS sequences, which can be used to separate these 9 clades without computing a complete phylogram.
Coprinopsis sect. Radiatae Wächter & A. Melzer, sect. nov. MB 831490 (Fig. 97)
Description: Basidiomata tiny to medium-sized, fimicolous, terrestrial or herbiolous. Lamellae deliquescent. Veil strongly developed, consisting of chains of subcylindrical, hyaline cells, sometimes additionally consisting of chains of thin, diverticulate cells or ventricose elements. Spores predominantly large-sized, germ pore central or nearly central, mostly without a perisporium. Marginal cells of the lamellar edge clavate, sometimes mixed with utriform cheilocystidia. Pleurocystidia present. Clamps present in most species.
Type species: Coprinopsis radiata (Bolton) Redhead, Vilgalys & Moncalvo, Taxon 50(1):230, 2001.
Representatives:
Coprinopsis candidolanata (Doveri & Uljé) Keirle, Hemmes & Desjardin; Ref.v.: CAND1 (Nagy et al. 2013)
Coprinopsis neolagopus (Hongo & Sagara) Redhead, Vilgalys & Moncalvo; Ref.v.: NBRC100013 (Barua et al. 2012)
Coprinopsis nevellei Guy Garcia & Vellinga; Ref.v.: GG08090401/type (Garcia and Vellinga 2010)
Coprinopsis radiata (Bolton) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-1548 (Nagy et al. 2013)
Coprinopsis tectispora (Bogart ex Bogart) Redhead, Vilgalys & Moncalvo; Ref.v.: FVDB 6016/type (Nagy et al. 2013)
Coprinopsis villosa L. Nagy, Desjardin, Vágvölgyi & Papp; Ref.v.: SZMC-NL-1758 (Nagy et al. 2013)
Remarks:
The clade /“uljei” contains C. uljei Bender & Guardian, which will be validly described in the near future (Bender in prep.). This species was identified by examination of the voucher Germany: Nordrhein-Westfalen, Mönchengladbach, 31.VII.2003, H. Bender (HB20030827A). The ITS (deposited at GenBank as MK069601.1) shows 100% coverage with JX624300.1; whether the other vouchers in the clade are absolutely identical cannot be answered clearly.
C. macrocephala (Berk.) Redhead, Vilgalys & Moncalvo certainly belongs here, but the distinction of C. radiata is difficult. References: Amandeep et al. (2014), Krieglsteiner et al. (1982), Ludwig (2007), Uljé and Noordeloos (1999).
C. tectispora deviates noticeably by the presence of a perisporium, but is included in the /radiata clade. A reduction to subspecific level under C. radiata could be considered, but is not the subject here. Compare C. brunneistragulata in the section Lanatuli too. C. macrocephalus var. perisporalis, a variety with a perisporium was described by Ludwig (2007), and it appears that such exceptional variants do exist. C. nevellei is certainly a synonym of C. radiata, even if the holotype grew on a stem of Polygonatum, not on dung. Unlike the other species, C. candidolanata is located in a sister clade. Special features are missing clamps and a trimorphic veil. The creation of a separate section is currently ignored, because the constancy of the morphological properties should be checked.
Coprinopsis sect. Filamentiferae Wächter & A. Melzer, sect. nov. MB 831491 (Fig. 98)
Description: Basidiomata small-sized, fimicolous. Lamellae deliquescent. Veil strongly developed, but fugacious, consisting of chains of branched, diverticulate, partially encrusted cells, and subglobose elements. Spores small to medium-sized, subcylindrical to submitriform with a truncated base, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge mostly clavate, less often utrifom. Pleurocystidia utriform, subcylindrical, clavate. Clamps present.
Type species: Coprinopsis filamentifera (Kühner) Redhead, Vilgalys & Moncalvo, Taxon 50(1):228, 2001.
Representative:
Coprinopsis filamentifera (Kühner) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-0380 (Nagy et al. 2012)
Remarks:
So far only one species is known. The proximity to the section Picaceae is reflected in similar cystidia and main veil elements; significant differences are spore size and shape. In addition, the species is relatively small and grows on dung. Coprinopsis “subfilamentifer” is a provisional name (Nagy 2011). Whether it is an independent species is unclear.
Coprinopsis sect. Picaceae (Penn. in Kauffman) Wächter & A. Melzer, comb. nov. MB 832467 (Fig. 98)
Basionym: Coprinus sect. Picacei Penn. in Kauffman, The Agaricaceae of Michigan:213, 1918
Description: Basidiomata rather large-sized, terrestrial or lignicolous. Veil initially covering the entire pileus, later tearing into patches, consisting of chains of somewhat diverticulate, thin-walled, hyaline cells. Spores medium to large-sized, ellipsoid to slightly ovoid in frontal view, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge versiform, often mixed with clavate cells. Pleurocystidia utriform, lageniform, subcylindrical, fusiform, ellipsoid, globose. Clamps present.
Type species: Coprinus picaceus (Bull.) Gray ≡ Coprinopsis picacea (Bull.) Redhead, Vilgalys & Moncalvo, Taxon 50(1):230, 2001, designated by Citérin (1992:23).
Representatives:
Coprinopsis picacea (Bull.) Redhead, Vilgalys & Moncalvo; Ref.v.: Champ-44 (Perez-Izquierdo et al. 2017)
Coprinopsis stangliana (Enderle, Bender & Gröger) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-2153 (Nagy, Urban et al. 2010)
Coprinopsis strossmayeri (Schulzer) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-0774 A (Nagy et al. 2012)
Remarks:
Whether C. variegata (Peck) Redhead, Vilgalys & Moncalvo actually belongs here or in section Atramentarii must remain unclear. The pronounced volva fits for the latter, the veil for the former.
The name takes over the unranked group Picacei Fries, Epicr Syst Mycol:244, 1838.
Coprinopsis sect. Melanthinae Wächter & A. Melzer, sect. nov. MB 831492 (Fig. 98)
Etymology: Named after the type.
Description: Basidiomata large, lignicolous. Pileus not radially sulcate, lamellae not deliquescent. Veil strongly developed, consisting of chains of subcylindrical, often encrusted cells. Spores medium to large-sized, ellipsoid to ovoid in frontal view, strikingly pale and thin-walled, germ pore absent or very indistinct. Basidia 4-spored, always clavate, never polymorphic. Marginal cells of the lamellar edge predominantly utriform. Pleurocystidia absent. Clamps present.
Type species: Coprinopsis melanthina (Fr.) Örstadius & E. Larss., Mycol Progr 14(25):37, 2015.
Representatives:
Coprinopsis cineraria (Har. Takah.) Örstadius & E. Larss.; Ref.v.: CBM-FB-24142/type (Örstadius et al. 2015)
Coprinopsis melanthina (Fr.) Örstadius & E. Larss.; Ref.v.: WU19918 (Örstadius et al. 2015)
Coprinopsis uliginicola (McKnight & A.H. Sm.) Örstadius & E. Larss.; Ref.v.: Smith34903/type (Örstadius et al. 2015)
Remarks:
This section is morphologically and ecologically very uniform and well recognizable. The clade /lignicola contains Coprinopsis lignicola nom. prov. (Rockefeller unpubl.).
Coprinopsis sect. Subniveae Wächter & A. Melzer, sect. nov. MB 831493 (Fig. 99)
Etymology: Derived from sub = under, nivea = snow-white; in the vicinity of C. nivea.
Description: Basidiomata small to medium-sized, terrestrial, lignicolous or fimicolous. Lamellae not or hardly deliquescent. Veil strongly developed, consisting of globose to subglobose and subcylindrical, hyaline or brownish pigmented, encrusted or diverticulate elements, sometimes with slightly thickened walls. Spores small to large-sized, ellipsoid in front view, germ pore central. Basidia 4-spored. Marginal cells of the lamellar edge utriform, clavate. Pleurocystidia predominantly absent. Clamps present in most species.
Type species: Coprinopsis cortinata (J.E. Lange) Gminder, Die Großpilze Baden-Württembergs (Stuttgart) 5:650, 2010.
Representatives:
Coprinopsis bellula (Uljé) P. Roux & Eyssartier; Ref.v.: SZMC-NL-2341 (Nagy et al. 2009)
Coprinopsis cerkezii Tkalčec, Mešić, I. Kušan & Matočec; Ref.v.: CNF 1/7253/type (Tibpromma et al. 2017)
Coprinopsis coniophora (Romagn.) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-3414 (Nagy et al. 2011)
Coprinopsis cortinata (J.E. Lange) Gminder; Ref.v.: SZMC-NL-1621 (Nagy et al. 2011)
Coprinopsis utrifera (Joss. ex Watling) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-0591 (Nagy et al. 2011)
Remarks:
C. bellula is the only species with large spores, most likely because of the 2-spored basidia. Possibly, it is simply a form of C. cortinata with a reduced number of sterigmata; the other features are very similar. In particular, the absence of true cystidia is remarkable.
Coprinopsis sect. Niveae (Citérin) D.J. Schaf., Field Mycology 11(2):51, 2010 (Fig. 99)
Description: Basidiomata small to medium-sized, fimicolous or lignicolous. Lamellae hardly deliquescent, rather withering. Veil strongly developed, consisting of globose to subglobose, encrusted elements and chains of subcylindrical, often diverticulate cells. Spores medium to large-sized, ellipsoid, mitriform or rounded-angular in frontal view, in side view distinctly flattened, apically prolonged, germ pore central to slightly eccentrical. Basidia 4-spored. Marginal cells of the lamellar edge utriform, clavate. Pleurocystidia present or absent. Clamps present.
Type species; Coprinus niveus (Pers.) Fr., Epicr Syst Mycol:246, 1838 ≡ Coprinopsis nivea (Pers.) Redhead, Vilgalys & Moncalvo, Taxon 50(1):229, 2001, designated by Citérin (1992:17).
Representatives:
Coprinopsis afronivea Desjardin & B.A. Perry; Ref.v.: SFSU BAP 619/type (Desjardin and Perry 2016)
Coprinopsis igarashi Fukiharu & K. Shimizu; Ref.v.: CBM-FB39186 (Fukiharu et al. 2015)
Coprinopsis nivea (Pers.) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-0847 (Nagy et al. 2012)
Coprinopsis pseudonivea (Bender & Uljé) Redhead, Vilgalys & Moncalvo; Ref.v.: SZMC-NL-2340 (Nagy et al. 2009)
Coprinopsis yokdonensis nom. prov.; Ref.v.: CBM-FB41367 (Nguyen et al. unpubl.)
Remarks:
Section Nivei shares several morphological features with section Subnivei; however, the spores tend to be larger and differently shaped. It is possible that the overall picture changes after finding more species, as there could be a transition between the two sections. C. “yokdonensis” is a provisional name for a Vietnamese species on elephant dung.
Coprinopsis sect. Canocipes Wächter & A. Melzer, sect. nov. MB 831494 (Fig. 99)
Description: Basidiomata medium to large. Pileus not radially sulcate, lamellae not deliquescent. Veil strongly developed, consisting of chains of subcylindrical, occasionally branched, hyaline, sometimes slightly encrusted cells. Spores medium to large-sized, ellipsoid to slightly ovoid. Basidia mostly 4-spored, clavate or sphaeropedunculate, never ululiform. Marginal cells of the lamellar edge utriform, lageniform or remarkably versiform, mixed with only a few globose, subglobose or ellipsoidal cells. Pleurocystidia absent or extremely rare. Clamps present.
Type species: Coprinopsis canoceps (Kauffman) Örstadius & E. Larss., Mycol Progr 14(25):37, 2015.
Representatives:
Coprinopsis aesontiensis A. Melzer, G. Ferisin & F. Dovana; Ref.v.: LZ P-7614/type (Melzer et al. 2017)
Coprinopsis canoceps (Kauffman) Örstadius & E. Larss.; Ref.v.: LO148-95 (Örstadius et al. 2015)
Coprinopsis pannucioides (J.E. Lange) Örstadius & E. Larss.; Ref.v.: LO143-03 (Larsson and Örstadius 2008)
Coprinopsis submicrospora (Heykoop & G. Moreno) Örstadius & E. Larss.; Ref.v.: AH27055/type (Örstadius et al. 2015)
Coprinopsis udicola Örstadius, A. Melzer & E. Larss.; Ref.v.: AM1240/type (Örstadius et al. 2015)
Remarks:
The members of this section are habitually more reminiscent of Psathyrella than of coprinoid species.
Coprinopsis lotinae (Picón) Picón very probably also belongs here. References: Iglesias et al. (2011), Iglesias et al. (2015), Picón (2003, as Coprinus lotinae Picón), Ruiz Mateo et al. (2011, as Psathyrella marcescibilis var. virginea J.E. Lange ex Surault, Tassi & Coué), Sammut and Melzer (2010). Mat. exam.: Malta: Buskett, 26.X.2010, C. Sammut (AM1504).
Coprinopsis sect. Quartoconatae Wächter & A. Melzer, sect. nov. MB 832307 (Fig. 99)
Etymology: Derived from quartus = four, conatus = attempt; it is the fourth attempt to establish a section.
Description: Basidiomata medium to large. Pileus not radially sulcate, lamelleae not deliquescent. Veil present at the margin of the pileus, consisting of chains of subcylindrical, occasionally branched, hyaline, sometimes slightly encrusted cells. Spores small to large-sized, ellipsoid to slightly ovoid. Basidia 4-spored, clavate or sphaeropedunculate, never ululiform. Marginal cells of the lamellae edge predominantly utriform, occasionally subcapitate, mixed with only a few globose, subglobose or ellipsoidal cells. Pleurocystidia absent. Clamps present.
Type species: Coprinopsis marcescibilis Örstadius & E. Larss., Mycol Res 112(10):1180, 2008.
Representatives:
Coprinopsis marcescibilis (Britzelm.) Örstadius & E. Larss.; Ref.v.: LO31-03 (Larsson and Örstadius 2008)
Coprinopsis musae Örstadius & E. Larss.; Ref.v.: JV06-179/type (Örstadius et al. 2015)
Coprinopsis pseudomarcescibilis Heykoop, G. Moreno & P. Alvarado; Ref.v.: AH:33711/type (Crous et al. 2017)
Remarks:
Previous (illegitimate) attempts to establish a section Fragilissimae have been the following: Romagnesi (1944), Singer (1951, “1949”), Romagnesi (1982). Romagnesi (1944) designated as type Drosophila marcescibilis (Britzelm.) Romagn. and mentioned “Sporis 10-15 μm longis, opacis”. However, C. musae has smaller and much brighter spores. Therefore, the original concept of section Fragilissimae ss. Romagnesi (1944) could not be adopted.
Lacrymaria Pat. (Fig. 100)
The species often appear difficult to distinguish from each other. Without a doubt are L. hypertropicalis (Guzmán, Bandala & Montoya) Cortez, L. lacrymabunda (Bull.) Pat. and L. subcinnamomea (A.H. Sm.) Watling, probably L. pyrrhotricha (Holmsk.) Konrad & Maubl., too. For the latter species, the spelling is pyrrho (Greek: fire red). A judgement whether L. glareosa (J. Favre) Watling and L. rigidipes (Peck) Watling are independent species must be avoided at present. Moreover, the phylogram shows the possibility that previously undescribed species exist, or that described species have not yet been sequenced and therefore there is no comparison sequence for them. It should also be noted that it is necessary to examine additional gene regions in order to obtain a better resolution of the phylogram. Sequences of the haploid nuclear genome are probably required.
Homophron (Britzelm.) Örstadius & E. Larss. (Fig. 100)
Representatives of the genus are currently only H. camptopodum (Sacc.) Örstadius & E. Larss., H. cernuum (Vahl) Örstadius & E. Larss. and H. spadiceum (P. Kumm.) Örstadius & E. Larss., very likely Psathyrella crenulata A.H. Sm., too, while Psathyrella avellaneifolia A.H. Sm. is still questionable.
Parasola Redhead, Vilgalys & Hopple emend. Wächter & A. Melzer (Fig. 101)
The original diagnosis of Parasola Redhead, Vilgalys & Hopple (Redhead et al. 2001) is “Basidiomata ephemera, terrestia. Pileus plicatus, membranaceus, glaber vel setosus, eglandulatus. Velum nullum. Lamellae deliquescentes in senectute. Pleurocystidia praesentia. Stipites centrales, friabiles. Basidiosporae atrae”. Here the authors make a mistake, because even Parasola misera (P. Karst.) Redhead, Vilgalys & Hopple has no pleurocystidia. Furthermore, the recombination of Psathyrella conopilea (Fr.) A. Pearson & Dennis to Parasola (Örstadius and Larsson 2008) requires an emendation because Parasola conopilea (Fr.) Örstadius & E. Larss. neither is ephemeral nor has a plicated pileus; moreover, pleurocystidia are absent. The diagnosis must therefore be changed in parts, as follows: “basidiomata maxima ephemera, ... Pileus maxima plicatus, ... Pleurocystidia praesentia vel absentia …”.
Parasola sect. Parasola
Description: Basidiomata small to medium-sized, sometimes fimicolous, withering. Pileus radially sulcate, without a veil, sometimes with brown hairs. Lamellae withering. Spores frontally ovoid, subglobose, hexagonal, rounded subtriangular to subpentangular, in side view mostly lentiform. Basidia regularly 4-spored. Marginal cells of the lamellar edge utriform, sublageniform, less often lageniform or purely clavate to spheropedunculate. Pleurocystidia mostly present, utriform or subcylindrical. Clamps present.
Representatives:
Parasola auricoma (Pat.) Redhead, Vilgalys & Hopple; Ref.v.: SZMC-NL-0268 (Nagy et al. 2009)
Parasola crataegi Schmidt-Stohn; Ref.v.: SZMC-NL-4175/type (Szarkándi et al. 2017)
Parasola glabra Hussain, Afshan, Ahmad & Khalid; Ref.v.: LAH-SAP-23 (Hussain, Ahmad et al. 2018)
Parasola hercules (Uljé & Bas) Redhead, Vilgalys & Hopple; Ref.v.: L146 (Nagy et al. 2012)
Parasola kuehneri (Uljé & Bas) Redhead, Vilgalys & Hopple; Ref.v.: Ulje 1241/type (Nagy et al. 2012)
Parasola lactea (A.H. Sm.) Redhead, Vilgalys & Hopple; Ref.v.: SZMC-NL-0466 (Nagy et al. 2009)
Parasola lilatincta (Bender & Uljé) Redhead, Vilgalys & Hopple; Ref.v.: SZMC-NL-0683 (Nagy et al. 2009)
Parasola malakandensis S. Hussain, N.S. Afshan & H. Ahmad; Ref.v.: LAH-SHP-17/type (Hussain et al. 2017)
Parasola megasperma (P.D. Orton) Redhead, Vilgalys & Hopple; Ref.v.: SZMC-NL-1924 (Nagy et al. 2009)
Parasola misera (P. Karst.) Redhead, Vilgalys & Hopple; Ref.v.: SZMC-NL-0280/type (Nagy et al. 2009)
Parasola neoplicatilis Fukiharu, P.T. Nguyen & Shimizu; Ref.v.: CBM-FB-40433 (Fujiharu and Nguyen unpubl.)
Parasola ochracea L. Nagy, Szarkándi & Dima; Ref.v.: SZMC-NL-3621 (Szarkándi et al. 2017)
Parasola plicatilis (Curtis) Redhead, Vilgalys & Hopple; Ref.v.: SZMC-NL-0075a/epitype (Nagy et al. 2009)
Parasola plicatilis-similis L. Nagy, Szarkándi & Dima; Ref.v.: SZMC-NL-0287/type (Szarkándi et al. 2017)
Parasola schroeteri (P. Karst.) Redhead, Vilgalys & Hopple; Ref.v.: Brier 10.5.1999 (Nagy et al. 2009)
Parasola setulosa (Berk. & Broome) Redhead, Vilgalys & Hopple; Ref.v.: L32 (Nagy et al. 2012)
Remarks:
The type of the genus is included: Parasola plicatilis (Curtis) Redhead, Vilgalys & Hopple, designated by Redhead et al., Taxon 50(1):235, 2001.
Like the whole genus, this section is relatively well researched, as the works of Hussain et al. (2016, 2017, 2018), Nagy et al. (2009, 2010, 2012) and Szarkándi et al. (2017) show. However, there is still a considerable amount of confusion. There are many obvious identification errors or mistakes; for example, P. schroeteri can be found in many different positions. Moreover, the phylogram clearly shows that there are still some undescribed species.
P. cuniculorum D.J. Schaf., Field Mycology 15(3):83, 2014 also belongs in this section, but it could be just a 2-spored form of P. misera. P. pseudolactea Sadiqullah, Hussain & Khalid, proposed in Hussain, Ahmad et al. (2018), is possibly another species, but the sequences are too short for a definitive decision.
The interpretation of P. ochracea appears to be extremely problematic. A molecular biological comparison (not yet included in the phylogram) with the collection Germany: Kandelberg, on cow dung, 11.IX.1987, H. Bender (HB19870911A), was made to clarify the situation. The result showed a complete agreement with the vouchers SZMC-NL-3167, SZMC-NL-3167, SZMC-NL-3621 and NL-3623, all currently named P. ochracea. In this species, pleurocystidia should be absent (Szarkándi et al. 2017), while pleurocystidia were found in HB19870911A. There is a suspicion that P. ochracea is an already known species that also grows on dung, in fact Parasola nudiceps (P.D. Orton) Redhead, Vilgalys & Hopple, Taxon 50(1):236, 2001 (≡ Coprinus nudiceps P.D. Orton, Notes R bot Gdn Edinb 32:142, 1972). If this is true, another consequence would be that P. nudiceps is an independent species and not a synonym of P. schroeteri. More concrete investigations are in progress (Bender in prep.). A complete description of the German record is given by Bender and Enderle (1988); the sequence is deposited at GenBank under the accession number MK063783.1.
Parasola sect. Conopileae Wächter & A. Melzer, sect. nov. MB 831495
Description: Basidiomata medium to large, not fimicolous, persevering. Pileus not radially sulcate, without a veil and long brown hairs. Lamellae neither deliquescent nor withering. Spores large-sized, ellipsoid, germ pore distinctly eccentric to almost central. Basidia 4-spored. Marginal cells of the lamellar edge lageniform, subutriform, utriform, mixed with clavate cells. Pleurocystidia absent. Clamps present.
Type species: Parasola conopilea (Fr.) Örstadius & E. Larss., Mycol Res 112(10):1180, 2008.
The following software, databases, models, and similar tools were used in this study:
-
AliView 1.20: Larsson (2014)
-
Bayesian Information Criterion (BIC): Schwarz (1978)
-
CIPRES Science Gateway V 3.3 (Cyberinfrastructure for Phylogenetic Research): Miller et al. (2010)
-
ClustalX 2.0: Larkin et al. (2007)
-
Corrected Akaike Information Criterion (AICc): Akaike (1974), Hurvich and Tsai (1989), Sugiura (1978)
-
Dendroscope 3.5.9: Huson and Scornavacca (2012)
-
Elongation Factor 1-α Protein Model – SMTL ID 2b7c: Pittman et al. (2006)
-
F81 Model: Felsenstein (1981)
-
Gblocks: Jose Castresana (2002)
-
GTR-Model: Tavaré (1986)
-
HMMER 3.1b2 (February 2015): http://hmmer.org/ – Copyright (C) 2015 Howard Hughes Medical Institute. Freely distributed under the GNU General Public License (GPLv3)
-
ITSx 1.1b: Bengtsson-Palme et al. (2013)
-
JModelTest – Version 2.1.10 v20160303: Darriba et al. (2012)
-
Mafft 7.372 (used over mafft.cbrc.jp) and 7.305b over Cipres: Katoh and Frith (2012), Katoh and Standley (2013), Katoh and Toh (2007), Katoh and Toh (2008), Katoh and Toh (2008a), Katoh and Toh (2010), Katoh et al. (2002), Katoh et al. (2005), Katoh et al. (2009), Katoh et al. (2016), Katoh et al. (2017), Kuraku et al. (2013), Nakamura et al. (2018), Yamada et al. (2016)
-
MCIC (modified complex indel coding): Müller (2006)
-
MEGA 6.06: Tamura et al. (2013)
-
MrBayes 3.2.6 64-Bit parallel version: Huelsenbeck and Ronquist (2001), Ronquist and Huelsenbeck (2003); parallel-version of MrBayes: Altekar et al. (2004)
-
Gamma-distribution used in MrBayes: Yang (1993), Yang (1994)
-
MCMCMC Metropolis-coupled Markov Chains with Monte Carlo Simulation used in MrBayes: Geyer (1991), Hastings (1970), Metropolis et al. (1953)
-
nst=2 models used in MrBayes: Hasegawa et al. (1984), Hasegawa et al. (1985), Kimura (1980)
-
nst=6 models used in MrBayes: Tavare (1986)
-
Simple Jukes-Cantor-like model for restriction sites used in MrBayes: Felsenstein (1992)
-
NCBI GenBank: National Center for Biotechnology Information, U.S. National Library of Medicine 8600 Rockville Pike, Bethesda MD, 20894 USA – https://www.ncbi.nlm.nih.gov/
-
Noisy 1.5.12: Dress et al. (2008)
-
PartitionFinder 2.1.1: Lanfear et al. (2012), Lanfear et al. (2016)
-
PhyML 3.0 used with PartitionFinder: Guindon et al. (2010)
-
PlutoF: Abarenkov et al. (2010)
-
Prank 140603: Löytynoja (2014), Löytynoja and Goldman (2005), Löytynoja and Goldman (2008a); Prank -F Option: Löytynoja and Goldman (2008b)
-
Probalign 1.4: Roshan and Livesay (2006)
-
RAxML Version 8.2.10: Stamatakis (2014)
-
Robinson-Foulds Distance (RF): Robinson and Foulds (1981)
-
SeqState 1.4.1: Müller (2005)
-
SIC (Simple Indel Coding): Simmons and Ochoterena (2000)
-
SWISS-MODEL: SWISS-MODEL Workspace / GMQE: Waterhouse et al. (2018); SWISS-MODEL Repository: Bienert et al. (2017); Swiss-PdbViewer / DeepView project mode: Guex et al. (2009); QMEAN: Benkert et al. (2011); Quaternary Structure Prediction / QSQE: Bertoni et al. (2017)
-
Tracer 1.6.0: MCMC Trace Analysis Tool – Andrew Rambaut, Marc A. Suchard, Walter Xie and Alexei J. Drummond (2003-2013)
-
TreeBASE: Piel et al. (2009), Vos et al. (2012)
-
Treegraph 2.14.0-771 beta: Stöver and Müller (2010)
-
Tubulin Beta Chain – SMTL ID 5fnv: Yang et al. (2016)
-
Two Parameter Model & Acquisition Bias Correction: Lewis (2001)
-
Unite: Kõljalg et al. (2013)
References
Abarenkov K, Tedersoo L, Nilsson RH, Vellak K, Saar I, Veldre V, Parmasto E, Prous M, Aan A, Ots M, Kurina O, Ostonen I, Jõgeva J, Halapuu S, Põldmaa K, Toots M, Truu J, Larsson K-H, Kõljalg U (2010) PlutoF – a web based workbench for ecological and taxonomic research, with an online implementation for fungal ITS sequences. Evol Bioinform Online 6:189–196. https://doi.org/10.4137/EBO.S6271
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Automat Contr 19(9):716–723. https://doi.org/10.1109/TAC.1974.1100705
Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F (2004) Parallel Metropolis-coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20:407–415. https://doi.org/10.1093/bioinformatics/btg427
Amandeep K, Atri NS, Munruchi K (2014) Taxonomic study on coprophilous species of Coprinopsis (Psathyrellaceae, Agaricales) from Punjab, India. Mycosphere 5(1):1–25
Amandeep K, Atri NS, Munruchi K (2015) Psathyrella (Psathyrellaceae, Agaricales) species collected on dung from Punjab, India. Curr Res Environ Appl Mycol J Fungal Biol 5(2):128–137. https://doi.org/10.5943/cream/5/2/6
Arnolds E (2003) Rare and interesting species of Psathyrella. Fungi non delineati 26:1–76
Arnolds E, Perini C (2006) Psathyrella berolinensis, a remarkabla fungus on dung of wild boar. Micol Veget Medit 21(1):35–40
Aronsen A (1993) Agarics from wetland in south-east Norway. Agarica 21:22–64
Baca SM, Toussaint EFA, Miller KB, Short AEZ (2016) Molecular phylogeny of the aquatic beetle family Noteridae (Coleoptera: Adephaga) with an emphasis on data partitioning strategies. Mol Phylogenet Evol 107:282–292. https://doi.org/10.1016/j.ympev.2016.10.016
Baker RED, Dale WT (1951) Fungi of Trinidad and Tobago. Mycol Pap 33:1–123
Barua BS, Suzuki A, Pham HN-C, Inatomi S (2012) Adaption of ammonia fungi to urea enrichment environment. Journal of Agricultural Technology 8(1):173–189
Battistin E, Chiarello O, Vizzini A, Örstadius L, Larsson E (2014) Morphological characterisation and phylogenetic placement of the very rare species Psathyrella sulcatotuberculosa. Sydowia 66(2):171–181
Bender H (1989) Coprinus subimpatiens und einige seiner nächsten Verwandten. Beitr Kenntn Pilze Mitteleur 5:75–82
Bender H, Enderle M (1988) Studien zur Gattung Coprinus (Pers.: Fr.) S.F. Gray in der BR Deutschland. IV. Z Mykol 54(1):45–68
Bender H, Enderle M, Krieglsteiner GJ (1984) Studien zur Gattung Coprinus (Pers.: Fr.) S.F. Gray in der BR Deutschland. II. Z Mykol 50(1):17–40
Bengtsson-Palme J, Veldre V, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, Bertrand Y, De Wit P, Sanchez M, Ebersberger I, Sanli K, de Souza F, Kristiansson E, Abarenkov K, Eriksson KM, Nilsson RH (2013) Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for use in environmental sequencing. Methods Ecol Evol 4(10):914–919. https://doi.org/10.1111/2041-210X.12073
Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350. https://doi.org/10.1093/bioinformatics/btq662
Berkeley MJ, Broome CE (1871) The fungi of Ceylon (Hymenomycetes, from Agaricus to Cantharellus). Bot J Linn Soc 11:494–567
Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T (2017) Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci Rep 7(1):10480. https://doi.org/10.1038/s41598-017-09654-8
Bienert S, Waterhouse A, de Beer TAP, Tauriello G, Studer G, Bordoli L, Schwede T (2017) The SWISS-MODEL Repository – new features and functionality. Nucleic Acids Res 45:D313–D319. https://doi.org/10.1093/nar/gkw1132
Bon M, van Haluwyn C (1983) Macromycetes des terrils de Charbonnages du nord de la France. 4 ème note. Docums Mycol 13(49):43–55
Breitenbach J, Kränzlin F (1994) Über einen kritischen Rübling, zwei seltene Tintlinge sowie ein kurioses Tintlings-Wachstum in der Schweiz. Z Mykol 60(1):25–33
Breitenbach J, Kränzlin F (1995) Pilze der Schweiz 4. Mykologia, Luzern
Brewer MJ, Butler A, Cooksley SL (2016) The relative performance of AIC, AICC and BIC in the presence of unobserved heterogeneity. Methods Ecol Evol 7(6):679–692. https://doi.org/10.1111/2041-210X.12541
Broussal M, Carbo J, Mir G, Pérez-de-Gregorio MÀ (2018) Psathyrella salina, nouvelle espèce des milieux halophiles méditerranéens. Bull FAMM, N.S 53:17–30
Brown JM, Lemmon AR (2007) The importance of data partitioning and the utility of Bayes factors in Bayesian phylogenetics. Syst Biol 56:643–655. https://doi.org/10.1080/10635150701546249
Buller AHR (1920, "1919") The production and liberation of spores in the genus Coprinus. Trans Br mycol Soc 3(5):348-350
Bulliard JBF (1783) Herbier de la France 3. Chez l'auteur, Didot, Debure, Belin, Paris
Cacialli G, Caroti V, Doveri F (1999) Contributio ad Cognitionem Coprinorum. Monografie di Pagine di Micologia 1:1–256
Cantrell SA, Tkavc R, Gunde-Cimerman N, Zalar P, Acevedo M, Baez-Felix C (2013) Fungal communities of young and mature hypersaline microbial mats. Mycologia 105(4):827–836. https://doi.org/10.3852/12-288
Carbó J, Pérez-de-Gregorio MÀ (1999) Cuartro especies de hongos interesantes citadas por primera vez en la península ibérica. Revista Soc Catalana Micol 22:7–90
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Christan J, Hussong A, Dondl M (2017) Beiträge zur Familie Psathyrellaceae: Psathyrella spintrigeroides, Psathyrella supernula, Psathyrella typhae. Mycol Bav 18:35–58
Citérin M (1992) Clé analytique du genre Coprinus Pers. Doc Mycol 22(86):1–28
Citérin M. (1994) Clé analytique du genre Coprinus Pers. (suite). Révision des sections Farinosi, Lanatuli, et Picacei. Doc Mycol 24(95):1–13
Consiglio F (2000) Contributo alla conoszenza dei Macromiceti dell´Emilia-Romagna. XX. Genre Psathyrella. Boll Gr micol G Bres (n.s.) 43(1):31–44
Consiglio F (2005) Contributo alla conoscenza dei Macromiceti dell’Emilia-Romagna. XXIII. Famiglia Coprinaceae – Parte terza. Boll Gr micol G Bres (n.s.) 48(2):7–22
Contu M (1991) Psathyrella bivelata spec. nov., une nouvelle espèce de la section Cystopsathyra. Bull Soc mycol Fr 107(3):85–89
Corriol GG (2014) Psathyrella litoralis sp. nov., una especie halófila de les marismas retrodunaes del sur de Córcega. Errotari 11:17–25
Crous PW, Wingfield MJ, Burgess TI et al (2017) Fungal Planet description sheets 558-624. Persoonia 38:240–384. https://doi.org/10.3767/003158517X698941
Crous PW, Wingfield MJ, Burgess TI et al (2018) Fungal Planet description sheets: 716-184. Persoonia 40:240–393
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9(8):772. https://doi.org/10.1093/molbev/mst197
De Haan A (1993) Twee Psathyrella’s uit de sectie Cystopsathyra: Psathyrella kellermanii (Peck) Sing. en Psathyrella globosivelata Gröger. AMK Mededelingen 93(3):69–71
Dennis RWG (1961, “1960”) Fungi venezuelani: IV, Agaricales. Kew Bull 15(1):67–156
Deschuyteneer D (2018) Psathyrella supernula (Britzelm.) Örstadius & Enderle, une espèce peu commune récoltée en Belgique. Bulletin de la Fédération des Associations Mycologiques de l'Ouest 7:3–9
Deschuyteneer D, Melzer A (2017) Psathyrella hellebosensis, a new species from Belgium. Bulletin de l’Association des Mycologues francophones de Belgique 10:3–10
Deschuyteneer D, Melzer A, Pérez-De-Gregorio MÀ (2018) Psathyrella codinae, a new species from Spain. Bulletin de l’Association des Mycologues francophones de Belgique 11:4–8
Desjardin DE, Perry BA (2016) Dark-spored species of Agaricineae from Republic of Sao Tome and Principe, West Africa. Mycosphere 7(3):359–391
Doveri F (2010) Occurrence of coprophilous Agaricales in Italy, new records, and comparisons with their European and extraeuropean distribution. Mycosphere 1(2):103–140
Doveri F, Granito VM, Lunghini D (2005) Nuovi ritrovamenti di Coprinus s.l. fimicoli in Italia. Riv Micol 48(4):319–340
Doveri F, Sarrocco S, Pecchia S, Forti M, Vannacci G (2010) Coprinellus mitrinodulisporus, a new species from chamois dung. Mycotaxon 114:351–360
Dress AWM, Flamm C, Fritzsch G, Gruenewald S, Kruspe M, Prohaska SJ, Stadler PF (2008) Noisy: Identification of Homoplastic Characters in Multiple Sequence Alignments. Alg Mol Biol 3:7. https://doi.org/10.1186/1748-7188-3-7
Earle FS (1909) The genera of the North American gill fungi. Bull New York Bot Gard 5:373–451
Einhellinger A (1976) Die Pilze in primären und sekundären Pflanzengesellschaften oberbayerischer Moore. Teil 1. Ber Bayer Bot Ges 47:75–149
Einhellinger A (1987) Erster sicherer mitteleuropäischer Nachweis von Psathyrella narcotica Kits van Waveren außerhalb der Niederlande. Beitr Kenntn Pilze Mitteleur 3:235–240
El-Assfouri A, Ouazzani Touhami A, Benkirana R, Douira A (2009) Etude de quelques espèces du genre Psathyrella (Fr.) Quél., nouvellement découvertes au Maroc. Bull de i´Institut Scientifique Rabat 31(1):7–11
Enderle M (1989) Bemerkenswerte Agaricales (Psathyrella)-Funde VIII. Beitr Kenntn Pilze Mitteleur 5:55–74
Enderle M (1994) Studien in der Gattung Psathyrella III. Beitr Kenntn Pilze Mitteleur 9:57–78
Enderle M (1998) Studien in der Gattung Psathyrella VII. Z Mykol 64(2):217–231
Enderle M (2000) Studien in der Gattung Psathyrella VIII. Z Mykol 66(1):3–26
Enderle M (2004) Die Pilzflora des Ulmer Raumes. Südd, Verlagsgesellschaft, Ulm
Enderle M, Bender H (1990) Studien zur Gattung Coprinus (Pers.: Fr.) S.F. Gray in der BR Deutschland V. Z Mykol 56(1):19–46
Enderle M, Christan J (1992) Studien in der Gattung Psathyrella I. Z Mykol 58(1):67–84
Enderle M, Hübner H-J (2005) Studien in der Gattung Psathyrella IX. Beitr. Kenntn Pilze Mitteleur 14:53–65
Enderle M, Krieglsteiner GJ, Bender H (1986) Studien zur Gattung Coprinus (Pers.: Fr.) S.F. Gray in der BR Deutschland. Z Mykol 52(1):101–132
Esteve-Raventós F, Enderle M (1992) Psathyrella halophila spec. nov., eine neue Art aus der Sektion Spintrigerae (Fr.) Konrad & Maublanc vom Meerestrand der Insel Mallorca (Spanien). Z Mykol 58(2):205–210
Fasciotto J-L (2009) Espèces rares ou intéressantes, étuidiées en 2007. Bull mycol bot Dauphiné-Savoie 194:5–16
Felsenstein J (1981) Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Felsenstein J (1992) Phylogenies from restriction sites: A maximum-likelihood approach. Evolution 46:159–173. https://doi.org/10.1111/j.1558-5646.1992.tb01991.x
Ferisin G, Melzer A (2019, “2018”) Three interesting species of Psathyrella from Slovenia. Micol Veget Medit 33(2):121–133
Frank JL, Coffan RA, Southworth D (2010) Aquatic gilled mushrooms: Psathyrella fruiting in the Rogue River in southern Oregon. Mycologia 102(1):93–107
Friebes G, Melzer A (2009) Psathyrella amarescens in Österreich. Österr Z Pilzk 18:53–57
Fries EM (1838) Epicrisis Systematis mycologici. Typographia Academica, Uppsala
Fukiharu T, Shimizu K, Utsunomiya H, Raut JK, Goto R, Okamoto T, Kato M, Horigome R, Furuki T, Kinjo N (2013) Coprinopsis asiaticiphlyctidospora sp. nov., an agaric ammonia fungus from Amami and Okinawa, southern Japan. Mycoscience 55(5):355–360. https://doi.org/10.1016/j.myc.2013.12.002
Fukiharu T, Shimizu K, Nakajima A, Miyamoto T, Raut JK, Kinjo N (2015) Coprinopsis igarashii sp. nov., a coprophilous agaric fungus from Hokkaido, northen Japan. Mycoscience 56:413–418. https://doi.org/10.1016/j.myc.2014.12.005
Garcia G, Vellinga EC (2010) Une nouvella espèce de coprin sur tiges de Polygonatum multiflorum: Coprinopsis nevillei sp. nov. Bull Féd Assoc Mycol Méditerr 37:37–58
Geyer CJ (1991) Markov chain Monte Carlo maximum likelihood. In: Keramidas EM (ed) Computing Science and Statistics: Proceedings of the 23rd Symposium on the Interface. Fairfax Station, Interface Foundation, pp 230–257
Gierczyk B, Kujawa A, Pachlewski T, Szczepkowski A, Wójtowski M (2011) Rare species of the genus Coprinus Pers. s. lato. Acta Mycol 46(1):27–73
Gierczyk V, Rodriquez-Flakus P, Pietras M, Gryc M, Czerniawski W, Piatek M (2017) Coprinopsis rugosomagnispora: a distinct new coprinoid species from Poland (Central Europe). Plant Syst Evol 303:915–925. https://doi.org/10.1007/s00606-017-1418-7
Gonzalez del Val A, Platas G, Arenal F, Orihuela JC, Garcia M, Hernandez P, Royo I, De Pedro N, Silver LL, Young K, Vicente MF, Pelaez F (2003) Novel illudins from Coprinopsis episcopalis (syn. Coprinus episcopalis), and the distribution of illudin-like compounds among filamentous fungi. Mycol Res 107(10):1201–1209. https://doi.org/10.1017/S0953756203008487
Gröger F (1984) Bemerkenswerte Psathyrella-Funde aus Thüringen. Boletus 1984(1):1–16
Gröger F (1985) Ein Fund von Coprinus heterothrix in der DDR. Agarica 6(12):67–72
Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 30:S162–S173. https://doi.org/10.1002/elps.200900140
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59(3):307–321. https://doi.org/10.1093/sysbio/syq010
Hasegawa M, Yano T, Kishino H (1984) A new molecular clock of mitochondrial DNA and the evolution of Hominoids. Proc Japan Acad 60B:95–98
Hasegawa M, Kishino H, Yano T (1985) Dating the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174. https://doi.org/10.1007/BF02101694
Hastings WK (1970) Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57:97–109. https://doi.org/10.2307/2334940
Hausknecht A, Contu M (2003) The genus Galerella. A world-wide survey. Österr Z Pilzk 12:31–40
Hausknecht A, Krisai-Greilhuber I (2012) Die Pilzflora der Lössgebiete im westlichen Weinviertel (Niederösterreich). Österr Z Pilzk 21:83–116
Hazi J, Nagy LG, Vágvölgyi C, Papp T (2011) Coprinellus radicellus, a new species with northern distribution. Mycol Progress 10:363–371. https://doi.org/10.1007/s11557-010-0709-y
Held BW, Blanchette RA (2017) Deception Island, Antarctica, harbors a diverse assemblage of wood decay fungi. Fungal Biol 121(2):145–157
Hongo T (1966) Notes on Japanese larger fungi (18). J Jap Bot 41:165–172
Hopple JS Jr, Vilgalys R (1999) Phylogenetic relationships in the mushroom genus Coprinus and dark spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: divergent domains, outgroups, and monophyly. Mol Phylogenet Evol 13:1–19
Horak E (1968) Synopsis generum Agaricalum (Die Gattungstypen der Agaricales). Beitr KryptFl Schweiz 13:1–741
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. https://doi.org/10.1093/bioinformatics/17.8.754
Huhtinen S, Vauras J (1992) Mythicomyces corneipes, a rare agaric, in Fennoscandia. Karstenia 32:7–12
Hurvich C, Tsai C (1989) Regression and time series model selection in small samples. Biometrika 76:297–307. https://doi.org/10.1093/biomet/76.2.297
Huson DH, Scornavacca C (2012) Dendroscope 3: An interactive viewer for rooted phylogenetic trees and networks. Syst Biol 61(6):1061–1067. https://doi.org/10.1093/sysbio/sys062
Hussain S, Afshan N-u-S, Ahmad H, Khalid AN, Niazi AR (2017) Parasola malakandensis sp. nov. (Psathyrellaceae; Basidiomycota) from Malakand, Pakistan. Mycoscience 58(2):69–76. https://doi.org/10.1016/j.myc.2016.09.002
Hussain S, Ahmad H, Ullah S, Afshan N, Pfister DH, Sher H, Ali H, Khalid AN (2018a) The genus Parasola in Pakistan with the description of two new species. MycoKeys 30:41–60
Hussain S, Usman M, Afshan NS, Ahmad H, Khan J, Khalid AN (2018b) The genus Coprinellus (Basidiomycota; Agaricales) in Pakistan with the description of four new species. Mycokeys 39:41–61
Iglesias P, Vincente JF (2015) Aportación al catálogo de macromicetos de los Parques Naturales del Gorbea-Urkiola y zona norte de la peninsula Ibérica. Errotari 12:80–207
Iglesias P, Vincente JF, Oyerzabal M (2011) Aportaciones al conocimento micológico de la isla de La Palma III. Errotari 8:159–198
Iglesias P, Vincente JF, Oyerzabal M (2014) Aportaciones al catálogo micológico de la isla de Madeira (Portugal). Errotari 11:99–165
Kalamees K (1981) Agaric fungi of Badhyz Nature Reserve. Folia Cryptog Estonica 15:5–8
Kalamees K (1989) On the Agaricales flora of the Zaamin National Park II. Folia Cryptog Estonica 27:1–24
Kasik G, Dogan HH, Öztürk C, Aktas S (2004) New Records in Coprinaceae and Bolbitaceae from Mut (Mersin) District. Turk J Bot 28:449–455
Katoh K, Frith MC (2012) Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics 28:3144–3146. https://doi.org/10.1093/bioinformatics/bts578
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Katoh K, Standley DM (2016) A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32:1933–1942. https://doi.org/10.1093/bioinformatics/btw108
Katoh K, Toh H (2007) Errata – PartTree: an algorithm to build an approximate tree from a large number of unaligned sequences. Bioinformatics 23:372–374. https://doi.org/10.1093/bioinformatics/btl592
Katoh K, Toh H (2008) Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics 9:212. https://doi.org/10.1186/1471-2105-9-212
Katoh K, Toh H (2008a) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298. https://doi.org/10.1093/bib/bbn013
Katoh K, Toh H (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26:1899–1900. https://doi.org/10.1093/bioinformatics/btq224
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. https://doi.org/10.1093/nar/gkf436
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. https://doi.org/10.1093/nar/gki198
Katoh K, Asimenos G, Toh H (2009) Multiple Alignment of DNA Sequences with MAFFT. Methods Mol Biol 537:39–64. https://doi.org/10.1007/978-1-59745-251-9_3
Katoh K, Rozewicki J, Yamada KD (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform n. pag. doi. https://doi.org/10.1093/bib/bbx108
Kaur H, Kaur M, Atri NS, Kaur A (2013) The Genus Psathyrella (Fr.) Quél. from India: New Records. Journal on New Biological Reports 2(1):55–63
Kaya A, Uzun Y, Keles A, Demirel K (2010) Three coprinoid macrofungi taxa, new to Turkey. Turk J Bot 34:351–353
Keirle M, Hemmes DE, Desjardin DE (2004) Agaricales of the Hawaiian Islands. 8. Agaricaceae: Coprinus and Podaxis; Psathyrellaceae: Coprinopsis, Coprinellus and Parasola. Fungal Divers 15:33–124
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Kits van Waveren E (1968) The ‘Stercorarius group’ of the genus Coprinus. Persoonia 5(2):131–176
Kits van Waveren E (1971) Notes on the genus Psathyrella – II. Three new species of Psathyrella. Persoonia 6(3):295–312
Kits van Waveren E (1985) The Dutch, French and British species of Psathyrella. Persoonia Suppl 2:1–300
Kits van Waveren E (1995) The Berkeley & Broome species of Psathyrella in the Kew Herbarium. Kew Bull 50(2):307–325
Ko KS, Lim YW, Kim YH, Jung HS (2001) Phylogeographic divergences of nuclear ITS sequences in Coprinus species sensu lato. Mycol Res 105 (12):1519-1526. doi: 10.1017}S0953756201005184
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiß M, Larsson K-H (2013) Towards a unified paradigm for sequence-based identification of Fungi. Mol Ecol 22(21):5272–5277. https://doi.org/10.1111/mec.12481
Kotlaba F (1952) Křehutička orobincová – Psathyrella typhae (Kalchbr.) Kühner in Favre v Československu. Česká Mykol 6:169–175
Kreisel (1961) Pilze der Moore und Ufer Norddeutschlands II. Psathyrella typhae, Galerina mycenoides und G. clavata. Westfälische Pilzbriefe 3(1):1–6
Krieglsteiner GJ, Gminder A (2010) Die Großpilze Baden-Württembergs. Band 5: Ständerpilze: Blätterpilze III. Eugen Ulmer KG, Stuttgart
Krieglsteiner GJ, Bender H, Enderle M (1982) Studien zur Gattung Coprinus (Pers. ex Fr.) S.F. Gray in der Bundesrepublik Deutschland. I. Z Mykol 48(1):65–88
Krisai-Greilhuber I (1992) Die Makromyceten im Raum von Wien, Ökologie und Floristik. Libri Botanici 6. IHW, Eching
Kühner R, Romagnesi H (1953) Flore Analytique des Champignons Supérieurs. Masson et cie, Paris
Kuraku S, Zmasek CM, Nishimura O, Katoh K (2013) aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res 41:W22–W28. https://doi.org/10.1093/nar/gkt389
La Chiusa L, Mauri F (1996) Due interessanti Coprini della Alpi Apuane. Riv Micol 3(1996):225–232
Lanfear R, Calcott B, Ho SY, Guindon S (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29(6):1695–1701. https://doi.org/10.1093/molbev/mss020
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2016) PartitionFinder 2: new methods for selecting partitioned models of evolution formolecular and morphological phylogenetic analyses. Mol Biol Evol 34(3):772–773. https://doi.org/10.1093/molbev/msw260
Lange JE (1915) Studies in the Agarics of Denmark. II. Amanita, Lepiota, Coprinus. Dansk bot Ark 3(2):1–50
Lange M, Smith AH (1953) The Coprinus ephemerus Group. Mycologia 45:747–780
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Larsson A (2014) AliView: a fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30(22):3276–3278. https://doi.org/10.1093/bioinformatics/btu531
Larsson E, Örstadius L (2008) Fourteen coprophilous species of Psathyrella identified in the Nordic countries using morphology and nuclear rDNA sequence data. Mycol Res 112:1165–1185. https://doi.org/10.1016/j.mycres.2008.04.003
Lewis PO (2001) A Likelihood Approach to Estimating Phylogeny from Discrete Morphological Character Data. Syst Biol 50(6):913–925. https://doi.org/10.1080/106351501753462876
Li J-L, Sun X, Chen L, Guo L-D (2016) Community structure of endophytic fungi of four mangrove species in Southern China. Mycology 7(4):180–190. https://doi.org/10.1080/21501203.2016.1258439
Locquin M (1947) Études sur le genre Coprinus I. – Quelques coprins fimicoles. Bull Soc mycol Fr 63(1-2):75–88
Löytynoja A (2014) Phylogeny-aware alignment with PRANK. Methods Mol Biol 1079:155–170. https://doi.org/10.1007/978-1-62703-646-7_10
Löytynoja A, Goldman N (2005) An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA 102:10557–10562. https://doi.org/10.1073/pnas.0409137102
Löytynoja A, Goldman N (2008a) A model of evolution and structure for multiple sequence alignment. Philos Trans R Soc Lond B Biol Sci 363:3913–3919. https://doi.org/10.1098/rstb.2008.0170
Löytynoja A, Goldman N (2008b) Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320:1632–1635. https://doi.org/10.1126/science.1158395
Ludwig E (2007) Pilzkompendium Bd. 2, Beschreibungen. Fungicon, Berlin
Maniotis J (1964) The Coprinoid state of Rhacophyllus lilacinus. Am J Bot 51:485–494
Melzer A (2008) Neue Funde seltener Psathyrella-Arten. Boletus 30(2):89–94
Melzer A (2009a) Coprophile Tintlinge auf Alpaka-Dung. Österr Z Pilzk 18:15–24
Melzer A (2009b) Alpaka-Tintlinge. Der Tintling 59:36–40
Melzer A (2009c) Tintling auf Abwegen. Der Tintling 61:4–7
Melzer A (2010) Geisterpilze. Der Tintling 65:7–10
Melzer A (2017) Der vergessene Tintling. Der Tintling 108:7–13
Melzer A (2018) Zur Kenntnis der Psathyrella spadiceogrisea - Gruppe, Teil II. Z Mykol 84(1):3–28
Melzer A, Richter T, Schößler W (2016) Drei coprinoide Arten der Familie Psathyrellaceae neu in Deutschland. Z Mykol 82(2):333–348
Melzer A, Ferisin G, Dovana F (2017) Coprinopsis aesontiensis, a new species found in Friuli-Venezia Giulia, Italy. Micol Veget Medit 31(2):125–132
Melzer A, Kimani VW, Ullrich R (2019) Psathyrella aberdarensis, a new species of Psathyrella (Agaricales) from a Kenyan National Park. Österr Z Pilzk 27:23–30
Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E (1953) Equations of state calculations by fast computing machines. J Chem Phys 21:1087–1091. https://doi.org/10.1063/1.1699114
Mifsud S (2017) Contribution to the Mycobiota and Myxogastria of the Maltese islands. Part I (2014-2016). Micol Veget Medit 32(1):3–58
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA pp 1-8. doi: https://doi.org/10.1109/GCE.2010.5676129
Moreau PA, Durand M, Durand C (2002) Coprinus albidofloccosus Locquin – Une espèce méconnue de la section Micacei. Bull mycol bot Dauphiné-Savoie 165:19–24
Moreno G, Faus J (1984) Tres especies raras del genero Coprinus (Agaricales) de Cataluña, España. Cryptogamie, Mycologie 5:3–17
Moreno G, Manjón JL (2010) Guía de los hongos de la Península Ibérica. Edicione Omega, Barcelona
Moreno G, Heykoop M, Esqueda M, Olariaga I (2015) Another lineage of secotioid fungi is discovered: Psathyrella secotioides sp. nov. from Mexico. Mycol Progr 14:34. https://doi.org/10.1007/s11557-015-1057-8
Morgan AP (1908) North American species of Agaricaceae (Continued). J Mycol 14(2):64–75
Müller K (2005) SeqState – primer design and sequence statistics for phylogenetic DNA data sets. Applied Bioinformatics 4:65–69
Müller K (2006) Incorporating information from length-mutational events into phylogenetic analysis. Mol Phyl Evol 38:667–676. https://doi.org/10.1016/j.ympev.2005.07.011
Muñoz G, Caballero A (2012) Contribución al conocimiento del género Psathyrella en la Península Ibérica (I). Bol Micol FAMCAL 7:37–74
Muñoz G, Caballero A (2013) Contribución al conocimiento del género Psathyrella (incluidos taxones ahora transferidos a los géneros Coprinopsis y Parasola) en la Península Ibérica (II). Bol Micol FAMCAL 8:17–46
Muñoz G, Sánchez L (2018) Contribución al conocimiento del género Psathyrella en la Península Ibérica (IV). Bol Micol FAMCAL 13:41–59
Nagy LG (2007) Notes on taxa of Coprinus subsection Alachuani from Hungary. Österr Z Pilzk 16:167–180
Nagy LG (2011) An investigation of the phylogeny and evolutionary processes of deliquescent fruiting bodies in the mushroom family Psathyrellaceae (Agaricales). Ph. D. Thesis, University of Szeged, Faculty of Science and Informatics, Department of Microbiology
Nagy LG, Kocsubé S, Papp T, Vágvölgyi C (2009) Phylogeny and character evolution of the coprinoid mushroom genus Parasola as inferred from LSU and ITS nrDNA sequence data. Persoonia 22:28–37. https://doi.org/10.3767/003158509X422434
Nagy LG, Urban A, Örstadius L, Papp T, Larsson E, Vágvölgyi C (2010a) The evolution of autodigestion in the mushroom family Psathyrellaceae (Agaricales) inferred from Maximum Likelihood and Bayesian methods. Mol Phylogenet Evol 57(3):1037–1048. https://doi.org/10.1016/j.ympev.2010.08.022
Nagy LG, Vágvölgyi C, Papp T (2010b) Type studies and nomenclatural revisions in Parasola (Psathyrellaceae) and related taxa. Mycotaxon 112:103–141
Nagy LG, Házi J, Vágvölgyi C, Papp T (2011a) Phylogeny and species delimitation in the genus Coprinellus with special emphasis on the haired species. Mycologia 104(1):254–275. https://doi.org/10.3852/11-149
Nagy LG, Walther G, Hazi J, Vágvölgyi C, Papp T (2011b) Understanding the evolutionary processes of fungal fruiting bodies: correlated evolution and divergence times in the Psathyrellaceae. Syst Biol 60(3):303–317. https://doi.org/10.1093/sysbio/syr005
Nagy LG, Házi J, Szappanos B, Kocsubé S, Bálint B, Rákhely G, Vágvölgyi C, Papp T (2012a) The evolution of defense mechanisms correlate with the explosive diversification of autodigesting Coprinellus mushrooms (Agaricales, Fungi). Syst Biol 61(4):595–607. https://doi.org/10.1093/sysbio/sys002
Nagy LG, Kocsubé S, Csanádi Z, Kovács GM, Petkovits T, Vágvölgyi C, Papp T (2012b) Re-mind the gap! Insertion – deletion data reveal neglected phylogenetic potential of the nuclear ribosomal internal transcribed spacer (ITS) of fungi. PloS One 7(11):e49794. https://doi.org/10.1371/journal.pone.0049794
Nagy LG, Desjardin DE, Vágvölgyi C, Kemp R, Papp T (2013a) Phylogenetic analyses of Coprinopsis sections Lanatuli and Atramentarii identify multiple species within morphologically defined taxa. Mycologia 105(1):112–124. https://doi.org/10.3852/12-136
Nagy LG, Vágvölgyi C, Papp T (2013b) Morphological characterization of clades of the Psathyrellaceae (Agaricales) inferred from a multigene phylogeny. Mycol Progress 2013(12):505–517. https://doi.org/10.1007/s11557-012-0857-3
Nakamura T, Yamada KD, Tomii K, Katoh K (2018) Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34:2490–2492. https://doi.org/10.1093/bioinformatics/bty121
Nylander JA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL (2004) Bayesian phylogenetic analysis of combined data. Syst Biol 53:47–67. https://doi.org/10.1080/10635150490264699
Örstadius L (2007) Studies on Psathyrella within the project Funga Nordica. Agarica 27:64–89
Örstadius L, Knudsen H (2008) Psathyrella. In: Knudsen H, Vesterholt J (eds) Funga Nordica. Nordsvamp, Copenhagen, pp 586–623
Örstadius L, Ryberg M, Larsson E (2015) Molecular phylogenetics and taxonomie in Psathyrellaceae (Agaricales) with focus on psathyrelloid species: introduction of three new genera and 18 new species. Mycol Prog 14(5) 25:1–42. https://doi.org/10.1007/s11557-015-1047-x
Orton PD (1957) Notes on British Agarics 1-5 (Observations on the genus Coprinus). Trans Brit mycol Soc 40(2):263–276
Orton PD (1960) New check list of British Agarics and Boleti. Part III. Notes on genera and species in the list. Tans Brit mycol Soc 43(2):159–439
Orton PD (1972) Notes on British Agarics: IV. Notes R bot Gdn Edinb 32(1):135–150
Orton PD (1988) Notes on British Agarics. IX. Trans Brit mycol Soc 91(4):545–571
Orton PD, Watling R (1979) British Fungus Flora. Part 2: Coprinaceae Part 1: Coprinus. R Bot Gard, Edinburgh
Pacioni G (1999) Psathyrella paecilosperma, una nuova specie palmicola della sezione Spintrigerae. Micol Veg Medit 13(2):149–152
Padamsee M, Matheny PB, Dentinger BT, McLaughlin DJ (2007) The mushroom family Psathyrellaceae: evidence for large-scale polyphyly of the genus Psathyrella. Mol Phylogenet Evol 46:415–429. https://doi.org/10.1016/j.ympev.2007.11.004
Peck CH (1872) Report of the Botanist (1869). A Rep NY St Mus nat Hist 24:41–108
Peck CH (1906) A new species of Galera. J Mycol 12(4):148–149
Pegler DN (1977) A preliminary Agaric flora of East Africa. Kew Bull Addit Ser 6:1–615
Pegler DN (1983) Agaric Flora of the Lesser Antilles. Kew Bull Addit Ser 9:1–668
Pegler DN (1987) A revision of the Agaricales of Cuba 2. Species described by Earle and Murill. Kew Bull 42(4):855–888
Pegler DN, Legon NW (1994) Profiles of fungi 57, Coprinus hiascens. Mycologist 8(1):12
Pennington LH (1918) Coprinus Pers. in: Kauffman CH: The Agaricaceae of Michigan. Vol. I, Text. Wynkoop, Hallenbeck Crawford Co., Lansing, pp. 206–236
Perez-Izquierdo L, Morin E, Maurice JP, Martin F, Rincon A, Buee M (2017) A new promising phylogenetic marker to study the diversity of fungal communities: The Glycoside Hydrolase 63 gene. Mol Ecol Resour 17(6):e1–e11. https://doi.org/10.1111/1755-0998.12678
Picón RM (2003) Coprinus lotinae. Une nouvelle espèce saprophyte, sur Eucalyptus, du littoral cantabrique. Docums Mycol 32(126):31–36
Piel, W. H., Chan, L., Dominus, M. J., Ruan, J., Vos, R. A., and V. Tannen 2009. TreeBASE v. 2: a database of phylogenetic knowledge. In: e-BioSphere (2009)
Pilát A, Svrček M (1967) Revisio specierum sectionis Herbicolae Pil. et Svr. generis Coprinus (Pers. ex) S.F. Gray. Ceská Mykol 21(3):136–145
Pittman YR, Valente L, Jeppesen MG, Andersen GR, Patel S, Kinzy TG (2006) Mg2+ and a Key Lysine Modulate Exchange Activity of Eukaryotic Translation Elongation Factor 1Bα. J Biol Chem 281(28):19457–19468. https://doi.org/10.1074/jbc.M601076200
Porras-Alfaro A, Herrera J, Sinsabaugh RL, Odenbach KJ, Lowrey T, Natvig DO (2008) Novel root fungal consortium associated with a dominant desert grass. Appl Environ Microbiol 74(9):2805–2813. https://doi.org/10.1128/AEM.02769-07
Prydiuk MP (2010) New records of dung inhabiting Coprinus species in Ukraine II. Section Coprinus. Czech Mycol 62(1):43–58
Rannala B (2002) Identifiability of parameters in MCMC Bayesian inference of phylogeny. Syst Biol 51:754–760. https://doi.org/10.1080/10635150290102429
Raut JK, Suzuki A, Fukiharu T, Shimizu K, Kawamoto S, Tanaka C (2011) Coprinopsis neophlyctidospora sp. nov., a new ammonia fungus from boreal forest in Canada. Mycotaxon 115:227–238
Raut JK, Fukiharu T, Shimizu K, Kawamoto S, Takeshige S, Tanaka C, Yamanaka T, Suzuki A (2015) Coprinopsis novorugosobispora (Basidiomycota, Agaricales), an ammonia fungus new to Canada. Mycosphere 6(5):612–619
Redhead SA, Traquair JA (1981) Coprinus sect. Herbicolae from Canada. Mycotaxon 13(2):373–404
Redhead SA, Smith AH (1986) Two new genera of agarics based on Psilocybe corneipes and Phaeocollybia perplexa. Can. J. Bot. 64(3):643–647
Redhead SA, Vilgalys R, Moncalvo JM, Johnson J, Hopple JS Jr (2001) Coprinus Pers. and the disposition of Coprinus species sensu lato. Taxon 50:203–241
Reid DA (1958) New or interesting records of British hymenomycetes. II. Trans Brit Mycol Soc 41(4):419–445
Rejinders AFM (1979) Developmental anatomy of Coprinus. Persoonia 10(3):383–424
Robinson DR, Foulds LR (1981) Comparison of phylogenetic trees. Math Biosci 53:131–147. https://doi.org/10.1016/0025-5564(81)90043-2
Romagnesi H (1944) Classification du genre Drosophila Quélet. Bull mens Soc linn Lyon 13(4):51–54
Romagnesi H (1951) Étude de quelques Coprins (3° série). Rev Mycol 16:108–128
Romagnesi H (1952) Species et formae novae ex genere Drosophila Quélet. Bull mens Soc linn Lyon 21:151–156
Romagnesi H (1975) Description de quelques espèces de Drosophila Quél. (Psathyrella ss. dilat.). Bull Soc mycol Fr 91(2):137–224
Romagnesi H (1976) Quelques espèces rares ou nouvelles de macromycètes 1 – Coprinacées. Bull Soc mycol Fr 92(2):198–206
Romagnesi H (1982) Études complémentaires de quelques espèces de Psathyrella ss. lato (Drosophila Quélet). Bull Soc mycol Fr 98(1):5–68
Romero-Olivares AL, Baptista-Rosas RC, Escalante AE, Bullock SH, Riquelme M (2013) Distribution patterns of Dikarya in arid and semiarid soils of Baja California, Mexico. Fungal Ecology 6(1):92–101. https://doi.org/10.1016/j.funeco.2012.09.004
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Roshan U, Livesay DR (2006) Probalign: multiple sequence alignment using partition function posterior probabilities. Bioinformatics 22(22):2715–2721. https://doi.org/10.1093/bioinformatics/btl472
Ruiz Mateo A (2012) Coprinellus sassii una especie con poca citas mundiale presente en la peninsula Ibérica. Bol Soc Micol Madrid 36:135–140
Ruiz Mateo A (2013) Aportaciones al conocimiento de la micoflora en la Comunidad de Navarra. Coprinopsis xantholepis, una especie a diferenciar de Coprinopsis phaeospora, nueva cita peninsular. Errotari 9:14–17
Ruiz Mateo A, Cerdán D (2016) Aportaciones al conocimiento de la micoflora en la comunidad de Navarra, Tres especies interesantes de Coprinopsis sección Narcoticae. Errotari 13:44–56
Ruiz Mateo A, Garcia Murilo S (2012) Aportaciones al conocimiento de la micoflora en la Comunidad de Navarra. Coprinellus callinus, presente an la Peninula Ibérica. Errotari 9:16–21
Ruiz Mateo A, Casas R, Muñoz González G (2011) Coprinus lotinae Picón, una especie a integar es Psathyrella? Bull Soc Micol Madrid 34:21–28
Ruiz Mateo A, Iglesias P, Rodriquez B, Muñoz G (2013) Coprinopsis xenobia, descripción y primeras localizaciones en España. Comparación filogenética con Coprinopsis luteocephala. Bol Micológ FAMCAL 8:63–70
Rundell SM, Spakowicz DJ, Narváez-Trujillo A, Strobel SA (2015) The Biological Diversity and Production of Volatile Organic Compounds by Stem-Inhabiting Endophytic fungi of Ecuador. J Fungi 1(3):384–396. https://doi.org/10.3390/jof1030384
Russo P, Juuti JT, Raudaskoski M (1992) Cloning, sequence and expression of a β-tubulin-encoding gene in the homobasidiomycete Schizophyllum commune. Gene 119(2):175–182. https://doi.org/10.1016/0378-1119(92)90269-U
Saccardo PA (1887) Sylloge Fungorum, vol 5. Agaricineae. P. A, Saccardo, Padua
Sammut C, Melzer A (2010) Psathyrellaceae from Malta, a preliminary survey. Micol Veget Medit 27(1):33–44
Schafer DJ (2012a) Keys to sections of Parasola, Coprinellus, Coprinopsis and Coprinus in Britain. Field Mycology 11(2):44–51
Schafer DJ (2012b) Coprinellus heterothrix and C. cinnamomeotinctus. Field Mycology 13(3):99–104
Schwarz G (1978) Estimating the dimension of a model. Ann Stat 6:461–464. https://doi.org/10.1214/aos/1176344136
Seok SJ, Kim YS, Kim WG, Kwon SW, Park IC (2010) Notes on Some New Species of Psathyrella. Mycobiology 38(4):323–327
Shipunov A, Newcombe G, Raghavendra AK, Anderson CL (2008) Hidden diversity of endophytic fungi in an invasive plant. Am J Bot 95(9):1096–1108
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49:369–381. https://doi.org/10.1093/sysbio/49.2.369
Simmons MP, Müller K, Norton AP (2007) The relative performance of indel-coding methods in simulations. Mol Phylogenet Evol 44(2):724–740. https://doi.org/10.1016/j.ympev.2007.04.001
Singer R (1948) Diagnoses fungorum novorum Agaricalium. Sydowia 2(1-6):26–42
Singer R (1951, “1949”) The Agaricales in modern taxonomy. Lilloa 22:5–832
Singer R (1959) New and interesting species of Basidiomycetes. VI. Mycologia 51(3):375–400
Singer R (1962a) The Agaricales in modern taxonomy, 2th edn. Cramer, Weinheim
Singer R (1962b, “1961”) Diagnoses Fungorum novorum Agaricalium II. Sydowia 15:45–83
Singer R (1975) The Agaricales in modern taxonomy, 3th edn. Cramer, Vaduz
Singer R (1986) The Agaricales in modern taxonomy, 4th edn. Koeltz Scientific Books, Koenigstein
Smith AH (1941) Studies of North American Agarics – I. Contr Univ Mich Herb 5:1–73
Smith AH (1948) Studies in the dark-spored Agarics. Mycologia 40(6):669–707
Smith AH (1972) The North American species of Psathyrella. Mem N Y bot Gdn 24:1–633
Smith AH, Hesler LR (1946) New and unusual dark-spored Agarics from North America. J Elisha Mitchell Sci Soc 62:177–200
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. https://doi.org/10.1093/bioinformatics/btl446
Stamatakis A (2014) RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics, open access link: http://bioinformatics.oxfordjournals.org/content/early/2014/01/21/bioinformatics.btu033.abstract?keytype=ref&ijkey=VTEqgUJYCDcf0kP. doi: https://doi.org/10.1093/bioinformatics/btu033
Stöver BC, Müller KF (2010) TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7. https://doi.org/10.1186/1471-2105-11-7
Strittmatter E, Obenauer H (2013) Ein Fund des Hornstieligen Scheinschwefelkopfes Mythicomyces corneipes (Fr.) Redhead & A.H. Sm. in Südwestdeutschland. Z Mykol 79(2):337–349
Sugiura N (1978) Further analysis of the data by akaike’s information criterion and the finite corrections. Commun Stat Theory Methods A7:13–26. https://doi.org/10.1080/03610927808827599
Suzuki A, Tsuchida S, Fukada J, Tanaka C, Tsuda M, Oda T, Bougher NL, Tommerup IC, Buchanan PK, Fukiharu T, Sagara N (2002) ITS rDNA variation of the Coprinopsis phlyctidospora (syn.: Coprinus phlyctidosporus) complex in the Northern and the Southern Hemispheres. Mycoscience 43(3):229–238. https://doi.org/10.1007/S102670200033
Szarkándi JG, Schmidt-Stohn G, Dima B, Hussain S, Kocsubé S, Papp T, Vágvölgyi C, Nagy LG (2017) The genus Parasola: phylogeny of the genus and the description of three new species. Mycologia 109(4):620–629. https://doi.org/10.1080/00275514.2017.1386526
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Tan G, Muffato M, Ledergerber C, Herrero J, Goldman N, Gil M, Dessimoz C (2015) Current methods for automated filtering of multiple sequence alignments frequently worsen single-gene phylogenetic inference. Syst Biol 64(5):778–791. https://doi.org/10.1093/sysbio/syv033
Tavare S (1986) Some probabilistic and statisical problems on the analysis of DNA sequences. Lect Math Life Sci 17(2):57–86
Tibpromma S, Hyde KD, Jeewon R et al (2017) Fungal diversity notes 491-602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 83:1–261
Tóth A, Hausknecht A, Krisai-Greilhuber I, Papp T, Vágvölgyi C, Nagy LG (2013) Iteratively refined guide trees help improving alignment and phylogenetic inference in the mushroom family bolbitiaceae. PLoS ONE 8(2):e56143. https://doi.org/10.1371/journal.pone.0056143
Uljé CB (1984) Coprinus amphithallus, weinig bekend en toch zo gemakkelijk. Coolia 27(4):82–83
Uljé CB (1988) Over de Coprinus hemerobius - Groep. Coolia 29(2):25–31
Uljé CB, Bas C (1991) Studies in Coprinus II. Subsection Setulosi of section Pseudocoprinus. Persoonia 14(3):275–339
Uljé CB, Noordeloos ME (1993) Studies in Coprinus III. Coprinus section Veliformis, Subdivision and revision of subsection Nivei emend. Persoonia 15(3):257–301
Uljé CB, Noordeloos ME (1997) Studies in Coprinus IV. Coprinus section Coprinus. Subdivision and revision of subsection Alachuani. Persoonia 16(3):265–333
Uljé CB, Noordeloos ME (1999) Studies in Coprinus V. Coprinus section Coprinus. Revision of subsection Lanatuli Sing. Persoonia 17(2):165–199
Uljé CB, Noordeloos ME (2003) Notulae ad floram agaricinam Neerlandicam XLII, additions to Coprinus subsection Setulosi. Persoonia 18(2):259–264
Uljé CB, Verbeken A (2002) A new species in Coprinus subsection Setulosi. Persoonia 18(1):143–145
Uljé CB, Doveri F, Noordeloos ME (2000) Additions to Coprinus subsection Lanatuli. Persoonia 17(3):465–471
Vašutová M, Antonin V, Urban A (2008) Phylogenetic studies in Psathyrella focusing on sections Pennatae and Spadiceae – new evidence for the paraphyly of the genus. Mycol Res 112:1153–1164. https://doi.org/10.1016/j.mycres.2008.04.005
Versper A, Melzer A (2015) Coprinellus fuscocystidiatus L. Nagy, Házi, Papp & Vágvölgyi in Deutschland. Z Mykol 81(1):41–47
Vila J, Rocabruna A (1996) Aportación al conocimiento del género Coprinus Pers. en Cataluña. II. Revista Soc Catalana Micol 19:73–90
Vila J, Rocabruna A (2002) Aportación al conocimiento del género Coprinus Pers. en Cataluña IV. C. cardiasporus Bender. Revista Soc Catalana Micol 24:131–134
Vizzini A, Consiglio G, Marchetti M (2019) Mythicomycetaceae fam. nov. (Agaricineae, Agaricales) for accommodating the genera Mythicomyces and Stagnicola, and Simocybe parvispora reconsidered. FUSE 3:41-56 doi.org/10.3114/fuse.2019.03.05
Von Bonsdorff T, Kytovuori I, Vauras J, Huhtinen S, Halme P, Rama T, Kosonen L, Jakobsson S (2014) Sienet ja Metsien Luontoarvot (Mushrooms and the Natural Value of Forests). Norrlinia 27:1–272
Vos RA, Balhoff JP, Caravas JA, Holder MT, Lapp H, Maddison WP, Midford PE, Priyam A, Sukumaran J, Xia X, Stoltzfus A (2012) NeXML: rich, extensible, and verifiable representation of comparative data and metadata. Systematic Biology 61(4):675–689
Vu D, Groenewald M, de Vries M, Gehrmann T, Stielow B, Eberhardt U, Al-Hatmi A, Groenewald JZ, Cardinali G, Houbraken J, Boekhout T, Crous PW, Robert V, Verkley GJM (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud Mycol 92:135–154. https://doi.org/10.1016/j.simyco.2018.05.001
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303. https://doi.org/10.1093/nar/gky427
Watling R (1967) Notes on some British Agarics. Notes R bot Gdn Edinb 28(1):39–56
Wilhelm M (2017) Pilze in der Masaola-Halle des Züricher Zoos. Folge 16: Dunkelsporige Blätterpilze. Der Tintling 107:29–34
Yagame T, Funabiki E, Nagasawa E, Fukiharu T, Iwase K (2013) Identification and symbiotic ability of Psathyrellaceae fungi isolated from a photosynthetic orchid, Cremastra appendiculata (Orchidaceae). Am J Bot 100(9):1823–1830
Yamada KD, Tomii K, Katoh K (2016) Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees, additional information. Bioinformatics 32:3246–3251. https://doi.org/10.1093/bioinformatics/btw412
Yan JQ, Bau T (2017) New and newly recorded species of Psathyrella (Psathyrellaceae, Agaricales) from northeast China. Phytotaxa 321(1):139-150. doi: https://doi.org/10.11646/phytotaxa.321.1.7
Yan JQ, Bau T (2018) The Northeast Chinese species of Psathyrella (Agaricales, Psathyrellaceae). MycoKeys 33:85–102. https://doi.org/10.3897/mycokeys.33.24704
Yang Z (1993) Maximum likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol Biol Evol 10:1396–1401. https://doi.org/10.1093/oxfordjournals.molbev.a040082
Yang Z (1994) Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J Mol Evol 39:306–314. https://doi.org/10.1007/BF00160154
Yang J, Wang Y, Wang T, Jiang J, Botting CH, Liu H, Chen Q, Yang J, Naismith JH, Zhu X, Chen L (2016) Pironetin reacts covalently with cysteine-316 of α-tubulin to destabilize microtubule. Nat Commun 7:12103. https://doi.org/10.1038/ncomms12103
Acknowledgments
We are thankful for the information, photographs (also in Supplement S2), fresh or dry material and friendly help from “Abeja”, Claudio Angelini, Ditte Bandini, Michael Beeckmann, Hans Bender, Konstanze Bensch, Julian Branscombe, Micheline Broussal, Klaus Büchler, Emanuele Campo, David Dandria, Daniel Deschuyteneer, Matthias Dondl, Giuliano Ferisin, Gernot Friebes, Andreas Gminder, Pérez-De-Gregorio, Anton Hausknecht, Norbert Heine, Michel Heykoop, Shah Hussain, Alexander Karisch, Amandeep Kaur, Lothar Kreuer, Lothar Krieglsteiner, Irmgard Krisai-Greilhuber, Steffen Lorenz, Regina Lüdecke, Rudi Markones, Jürgen Marqua, Antonio Ruiz Mateo, Andgelo Mombert, Gabriel Moreno, Bernd Mühler, Guillermo Muñoz, Andrea Oppolzer, Leif Örstadius, Shaun Pennycook, Marcus Rave, Matthias Reul, Torsten Richter, Antonio Ruiz, Carmel Sammut, Pablo Schäfer, Robert Schaike, M. Schönfeld, Wolfgang Schößler, Rika Seibert, Karl Soop, Muhammad Usman, Andreas Vesper, Heidrun Wawrok, Karl Wehr, Gerhard Wührleitner, Jun-Qing Yan, Rainer Ziehbart, Helmut Zitzmann and Stefan Zinke. Thankfully, Pablo Alvarado performed several sequencing sessions. We are grateful for the support for program-specific problems by Kazutaka Katoh, Ari Löytynoja, Bernd Kretschmer, Rob Lanfear, Anders Larsson, Mark Miller, Sonja J. Prohaska, Alexandros Stamatakis and Ben Stöver.
Special thanks to Matthias Reul for the comprehensive help with the translation.
Author information
Authors and Affiliations
Additional information
Section Editor: Zhu-Liang Yang
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix. Preliminary key to the genera
Appendix. Preliminary key to the genera
Below, the design of a key based on the natural system is shown. The characteristics are often not restricted to one genus. Sometimes there are overlaps with features of other genera. For the determination of the species, a key based on distinctive features, regardless of the system would be more appropriate.
1Pileipellis a cutis ...........................................Coprinopsis
1*Pileipellis a hymeniderm ............................................... 2
2(1) Pileus surface totally naked ...................................... 3
2*Pileus surface not totally naked; veil, pileocystidia, setae or hairs present .............................................................. 4
3(1) Pleurocystidia thick-walled, spores pale and ellipsoid ..................................................................... Homophron
3*Pleurocystidia thin-walled, spores dark and lentiform .................................................................... Parasola p.p.
4(2) Veil always absent .................................................... 5
4*Veil always present, but sometimes fugacious ........................................................................................ 6
5(4) Pileipellis with long brown hairs, no other elements present ....................................................... Parasola p.p.
5*Pileipellis without such hairs, but pileocystidia or setae may be present ........................................ Tulosesus p.p.
6(4) Veil not wipeable .................................................... 7
6*Veil wipeable ................................................................. 8
7(6) Spores warty ........................................... Lacrymaria
7*Spores not warty ...................................... Cystoagaricus
8(6) Veil consisting at least partially of spherical cells ....9
8*Veil without spherical cells ......................................... 15
9(8) Pileocystidia present .............................................. 10
9*Pileocystdia absent ...................................................... 13
10(9) Pileus with greenish tones .......................... Punjabia
10*Pileus without greenish tones ...................................... 11
11(10) Spores rounded-angular ................... Tulosesus p.p.
11*Spores otherwise ......................................................... 12
12(11) Pileus plicate or furrowed ............ Coprinellus p.p.
12*Pileus at most translucent striated ..................................... ................................... Psathyrella sect. Cystopsathyra p.p.
13(9) Spores strongly flattened, ouline tri- to polygonal ......................................................................... Narcissea
13*Spores otherwise ......................................................... 14
14(13) Pileus plicate or furrowed ............ Coprinellus p.p.
14*Pileus at most translucent striated ............................ .............................. Psathyrella sect. Cystopsathyra p.p.
15(8) Pileus plicate or furrowed ..................................... 16
15*Pileus at most translucent striated ............................... 17
16(15) Pileocystidia present ........................ Tulosesus p.p.
16*Pileocystidia absent .................................. Hausknechtia
17(15) Pleurocystidia absent .................... Candolleomyces
17*Pleurocystidia present ................................................. 18
18(17) Pleurocystidia with large, refractive globules .......................................................................... Typhrasa
18*Pleurocystidia otherwise .............................................. 19
19(18) Pleurocystidia predominantly spatula-shaped and strongly pediculated, often slightly thick-walled ............................................................................... Olotia
19*Pleurocystidia predominantly otherwise ..................... 20
20(19) Stipe distinctly rooting, cystidia with greenish deposits, pileocystidia or similar elements present ................................................................... Britzelmayria
20*Features not in this combination .................................. 21
21(20) Basidiocarps large, spores about 10 μm long, pale, germ pore absent or tiny .............................. Kauffmania
21*Features not in this combination .................. Psathyrella
Rights and permissions
About this article
Cite this article
Wächter, D., Melzer, A. Proposal for a subdivision of the family Psathyrellaceae based on a taxon-rich phylogenetic analysis with iterative multigene guide tree. Mycol Progress 19, 1151–1265 (2020). https://doi.org/10.1007/s11557-020-01606-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-020-01606-3