Abstract
Duosporium is a monotypic genus including only the type species Duosporium yamadanum which has been treated in the literature as “anamorphic Pezizomycotina.” Nevertheless, this is just a conjecture since its true phylogenetic affinities remain unknown. This fungus is known to cause leaf spots on several members of Cyperus spp. It has been intensively investigated in the 1990s as a potential candidate for use as an inundative biocontrol agent against purple nutsedge—a major tropical weed. Its morphology was recognized, as somewhat close to those of Curvularia and Bipolaris. Nevertheless, it was kept in a separate genus based on two distinctive morphological features: production of two kinds of spores (as indicated by its generic name) and straight versicolored macroconidia. No molecular studies have ever been made to elucidate the placement of Duosporium. In a relatively recent publication, Curvularia americana and C. chlamydospora were found to produce macro- and microconidia, as in Duosporium. Besides, there are several species of Curvularia known to have predominantly straight conidia. Here, a multilocus phylogenetic analysis including the internal transcribed spacer regions and intervening 5.8S nrRNA gene (ITS) and translation elongation factor 1-α (TEF1) sequences placed D. yamadanum within Curvularia, close to C. tuberculata, C. oryzae, and C. reesi. This led to the proposal of the new combination C. yamadana and the synonymization of Duosporium with Curvularia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Duosporium is a genus treated by Kirk et al. (2008) as “anamorphic Pezizomycotina.” Only its asexual dematiaceous hyphomycete stage is known. Its correct taxonomic placement is uncertain since no molecular study has ever been made to elucidate its true phylogenetic affinities. Duosporium was proposed by Thind and Rawla (1961) based on a fungus found infecting the leaves of Cyperus iria in India. Since its proposal, no additions were made to the genus which remains monotypic and only contains Duosporium yamadanum. Tsuda and Ueyama (1982) in their literature survey found that well before D. cyperi was proposed by Thind and Rawla (1961), Matsuura (1931) had described a highly morphologically similar fungus from Japan (Honshu). This was found attacking Cy. iwasakii. Matsuura named it Brachysporium yamadanum. Tsuda and Ueyama (1982) concluded upon this observation that the name D. cyperi was inadequate since B. yamadanum had priority over D. cyperi. The new combination D. yamadanum having B. yamadaeanum as basionym was then proposed by Tsuda and Ueyama (1982).
As pointed out by Thind and Rawla (1961) and Tsuda and Ueyama (1982), Duosporium appeared to be related to either Bipolaris or Curvularia. However, in these earlier publications, it was recognized that conidiogenous cells leading to the formation of macroconidia of Duosporium were mostly monotretic whereas those where microconidia originated were often polytretic. Conversely, the conidiogenous cells on both Bipolaris and Curvularia are typically polytretic. Also, as emphasized in its generic name, Duosporium was found to typically produce two kinds of conidia: macro and microconidia. Such a feature was, at the time, unknown for members of Bipolaris and Curvularia. This justified the proposal of Duosporium to accommodate the fungus on Cyperus spp.
Duosporium yamadanum is only known in association with members of the genus Cyperus. Four species of Cyperus have been reported as hosts of this fungus: Cyperus ferax in Venezuela (Urtiaga 1986), Cy. iria in India and Venezuela (Ellis 1971), and Cy. iwasakii Matsuura (1931) in Japan and Cy. rotundus in Brazil (Barreto and Evans 1995). Additional records of D. yamadanum have been published but without a full identification of their Cyperus host. These records are from Cuba (Mercado Sierra 1984) and West Indies (Minter et al. 2001).
This fungus attracted great interest when it was first collected in association with severe leaf spots and blight of foliage of Cy. rotundus in Brazil during surveys for potential weed biocontrol agents (Barreto and Evans 1995). Cyperus rotundus is considered as one of the worst tropical weeds worldwide (Holm et al. 1991) and a challenging target for mechanical or chemical management (Edenfield et al. 2005). A series of intensive studies on the biology and management of D. yamadanum was initiated at the Universidade Federal de Viçosa (state of Minas Gerais, Brazil) by Pomella (1999) and later retaken by Macedo (2006). Results of the attempts at developing a D. yamadanum-based mycoherbicide produced inconsistent results and this project was interrupted. Observations made during these years of study led to speculation that the taxonomic treatment for Duosporium might be incorrect and that its affinity with the Curvularia/Bipolaris complex should be reevaluated. A study including the use of molecular tools was then conducted in order to better clarify the taxonomic status of Duosporium and results are presented herein.
Materials and method
Isolates and morphology
Samples deposited in the herbarium at the Universidade Federal de Viçosa (VIC) during the 1990s were re-examined under a dissecting microscope in the laboratory. Fungal structures growing externally on plant tissues were scraped from colonized plant surfaces and mounted in lactophenol or lactofuchsin. Observations of fungal structures were performed with an Olympus BX53 microscope adapted with differential interference contrast lighting and digital image capture system (Olympus Q-Color 3 ™). Biometric data was obtained from the measurement of at least 30 representative fungal structures.
Existing pure cultures from the studies performed in the 1990s and deposited in the culture collection of the Universidade Federal de Viçosa - Coleção Octávio de Almeida Drumond (COAD) in silica gel, as described in Dhingra and Sinclair (1995), were recovered by aseptically transferring fragments of filter paper carrying fungal structures onto Petri dishes containing potato carrot-agar (PCA) and maintained in a controlled temperature room at 25 °C under a 12-h daily light/12-h dark regime (light provided by two white and one near-UV lamps placed 35 cm above the plates).
Culture description
Colony descriptions were based on fungal growth on PDA and PCA, after 7 days. The fungus was grown either under a daily 12-h light regime as mentioned above, or in the dark (Petri dishes wrapped in aluminum foil). Color terminology followed Rayner (1970).
Molecular characterization and multilocus phylogenetic analysis
Genomic DNA was extracted from COAD 141, COAD 359, and COAD 375 grown in vegetable broth-agar (VBA)-medium described in Pereira et al. (2003) under a 12-h daily light regime for 2 weeks. Approximately 50 mg of mycelium was scraped from the surface of the colonized medium and placed inside sterile plastic tubes containing zirconium spheres and placed in a grinder (L-Beader-3, Loccus Biotecnologia). After 5-s grinding, the resulting suspension was drained into a sterile plastic tube and used for DNA extraction. This was performed with the Wizard Genomic DNA Purification Kit following the manufacturer’s protocol. The PCR reaction was performed as described by Pinho et al. (2012). The primers ITS4 and ITS5 (White et al. 1990) were used to amplify the ITS region and the 5.8S rRNA gene. A partial region of TEF1 was amplified using the primer pair EF1-983 and EF1-2218R (Schoch et al. 2009). PCR products were analyzed on GelRed ™ (Biotium Inc., Hayward, CA, E.U.A.) and visualized under UV light to verify the size and purity of amplification. The PCR products were sequenced by Macrogen Inc., South Korea (http://www.macrogen.com). The nucleotide sequences were edited with software SeqAssem ver. 07/2008 (Hepperle 2004). All sequences were manually verified and nucleotides with ambiguous positions were clarified using sequences from both directions.
The ITS and TEF1 consensus sequences were compared with others deposited in the GenBank database using the MegaBLAST program. Sequences from GenBank and Tan et al. (2018) were aligned using MUSCLE (Edgar 2004) and built in MEGA v.6 (Tamura et al. 2011). All of the ambiguously aligned regions within the dataset were excluded from the analyses. Gaps (insertions/deletions) were treated as missing data.
Bayesian inference (BI) analyses employing a Markov Chain Monte Carlo method were performed with all sequences, first with each locus separately and then with the concatenated sequences. Before launching the BI, the best nucleotide substitution models were determined for each gene with MrMODELTEST 2.3 (Posada and Buckley 2004). Once the likelihood scores were calculated, the models were selected according to the Akaike Information Criterion (AIC). The GTR + I + G model of evolution was used for ITS, whereas GTR + G was used for TEF1. One concatenated tree with ITS and TEF1 was generated with Mesquite v. 3.1 (Maddison and Maddison 2011) and estimated on the CIPRES web portal using MrBayes on XSEDE 3.2.6 (Miller et al. 2010).
Additionally, a maximum likelihood (ML) tree was generated with the nearest-neighbor-interchange (NNI) ML heuristic method and the Tamura-Nei substitution model as tree inference options, using CIPRES web portal. The chain stabilities of the phylogenetic tree were assessed by using the bootstrap re-sampling strategy with 1000 bootstrap test replicates. The resulting tree topologies using the two methods (ML and BI) were then compared and the phylogram layout (BI tree) was edited with CorelDRAW Graphics Suite 2017.
Sequences derived from this study were deposited in GenBank (http://www.ncbi.nlm.nih.gov/genbank) (Table 1). The alignment and tree were deposited in TreeBASE (http://www.treebase.org) (study number S25966).
Results
Phylogenetic analyses
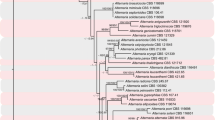
The alignment to construct phylogenetic trees included 52 strains (Table 1), representing many of the known Curvularia species, some isolates of Bipolaris, three isolates of Duosporium yamadanum, and the outgroup taxon (Alternaria alternata). The combined matrix consisted of 1911 characters including alignment gaps (ITS: 802 and EF1: 1109). The number of conserved sites was 1155 (ITS: 582 and EF1: 573). The number of variable and parsimony uninformative sites was 745 (ITS: 210 and EF1: 535) and 571 sites were variable and parsimony informative (ITS: 141 and EF1: 430). The trees obtained with ML and BI agreed on topology. The phylogenetic analyses inferred from the combined dataset (Fig. 1) indicated that the three strains of the fungus on Cy. rotundus (COAD 141, COAD 359, and COAD 375) clustered together with 100% (ML) and 1.0 (BI) support. These isolates of D. yamadanum clustered together with three Curvularia species: C. oryzae, C. reesii, and C. tuberculata. Additionally, D. yamadanum formed a distinct lineage within Curvularia. Based on the results of the molecular study, the status of Duosporium as a synonym of Curvularia became clear. The new combination C. yamadana was then proposed.
Phylogeny based on Bayesian inference inferred of combined ITS and TEF1 showing the relationship of Curvularia yamadana with other species within Curvularia and Bipolaris. Bootstrap support values (ML) or Bayesian posterior probabilities higher than 70% or 0.90 are indicated above or below thickened branches (– indicates lack of support). Isolates from this study are indicated by bold text
Taxonomy
Curvularia Boedijn, Bulletin du Jardin Botanique de Buitenzorg 13 (1): 123 (1933).
= Duosporium K.S. Thind & Rawla, American Journal of Botany 48 (10): 862 (1961). Syn. nov.
Curvularia yamadana (Matsuura) B.W. Ferreira and R.W. Barreto, comb. nov. (Fig. 2)
Curvularia yamadana (COAD 141). a Microconidiophores producing warted microconidia. b Collapsed macroconidium germinating to produce microconidia. c Macroconidium attached to the conidiogenous cell. d Mature macroconidia. e Colony on PCA after 7 days (incubation at 25 °C in 12-h light/dark cycle). f Colony on PDA after 7 days (incubation at 25 °C in 12-h light/dark cycle). Bars = 10 μm
Basionym. Brachysporium yamadanum Matsuura, Byochu-gai Zasshi [J. Pl. Prot. Tokyo] 17: 419, 1931 (as “yamadaeanum”). = Duosporium cyperi Thind & Rawla, American Journal of Botany 48: 862, 1961.
≡ Duosporium yamadanum (Matsuura) Tsuda and Ueyama, Mycotaxon 14: 145, 1982.
Lesions on living leaves; starting as dark brown, linear necrosis at apex or in the middle of the midrib, expanding to cover large portions of leaves, causing the blight of leaves and, sometimes, to death of all aerial part of plants. Internal mycelium intracellular, 1–6-μm diam, branched, septate, constricted at septae, concentrated adaxially under the epidermis, hyaline. External mycelium absent. Stromata absent. Macroconidiophores arising through stomata, amphigenous, mostly in loose fascicles of few conidiophores, rarely solitary, cylindrical, 43–132 × 4–7 μm, straight to slightly curved, inflated at the base and apex, septate, unbranched, smoky brown, smooth. Conidiogenous cells terminal, integrated, monotretic or polytretic, proliferating sympodially, or with percurrent proliferation with enteroblastic regenerative growth, cylindrical, 10–45 × 7 μm, inflated apical portion up to 11 μm wide, smoky brown becoming paler towards the apices. Conidiogenous loci indistinct to darkened. Macroconidia dry, solitary, tretic, oblong, 29–43 × 15–25 μm, apex and base rounded, hilum mostly indistinct, but visible in some spores as a darkened basal area, 3–4 μm wide, 3-septate, eguttulate, central cells dark brown to brown, end cells hyaline to subhyaline, smooth. Microconidiophores only seen in culture, arising from macroconidia or vegetative hyphae, cylindrical, strongly geniculate, straight to flexuous, mononematous, macronematous, 11–86 × 4–5 μm, walls thicker than on vegetative hyphae, brown, paler towards apex. Conidiogenous cells integrated, terminal or intercalary, cylindrical, proliferating sympodially, 6–11 × 3–5 μm, pale brown to brown, smooth, mono- or polytretic. Microconidia globose to subglobose to ellipsoidal, 5–13 × 5–9 μm, aseptate, brown to dark brown, warted, warts prominent, 3 × 2.5 μm.
Culture characters—slow-growing (3-cm diam on PDA and 3.4-cm diam on PCA after 7 days), flat to raised centrally with depressed marginal ring, undulate, edges entire, felty, center pale mouse gray becoming olivaceous gray, margin white, reverse fuscous black and white margin (PDA); center pale mouse gray, scarlet near the edge, border orange, reverse bay with orange margin (PCA). Not sporulating except on PCA in the dark.
Known distribution—Brazil, Cuba, India, Japan, Venezuela, West Indies.
Material examined: Brazil: Bahia, Itabuna, CEPLAC Brasil, on Cyperus rotundus, 11 Apr 2000, A. W. V. Pomella (VIC 27784F–culture COAD 141, MBT390712); Minas Gerais, Viçosa, Chácara Cristal, on Cyperus rotundus, 11 Apr 1998, R. W. Barreto, (culture COAD 375); Rio de Janeiro, Carmo, Fazenda São José, on Cyperus rotundus, 20 Jan 1998, R. W. Barreto, (culture COAD 359).
Notes: Duosporium was originaly described from a specimen collected in Punjab, India, on living leaves of Cyperus iria, but collected earlier in Honshu, Japan, on leaves of Cyperus iwasakii and mistakenly placed in Brachysporium, as B. yamadanum. No herbarium specimen or ex-type culture were designated by these authors.
The multilocus phylogenetic analyses indicated C. yamadana to be close to C. tuberculata, C. oryzae, and C. reesii. Curvularia reesii was recently described from colonies obtained from an air sample (Tan et al. 2018) and has not known to have Cyperus spp. as a substrate. Nevertheless, Farr and Rossman (2020) lists C. tuberculata and C. oryzae on Cyperus spp. C. oryzae was reported on Cy. rotundus from West Indies (Minter et al. 2001) and C. tuberculata was reported on Cy. malaccensis from Taiwan (Matsushima 1980). Curvularia yamadana has conidia similar in size to C. tuberculata (23–52 × 13–20 μm), C. oryzae (24–40 × 12–22 μm) and C. reesii (31–35 × 12–13 μm). However, conidia of C. tuberculata are sometimes curved, have 3–8-distoseptae and these are tuberculate at maturity. Conidia in C. oryzae and C. reesii are obclavate to ellipsoidal, whereas in C. yamadana are oblong having a rounded apex and base. Two other Curvularia species have been listed by Farr and Rossman (2020) on Cyperus spp. Curvularia aeria on Cy. rotundus from West Indies (Minter et al. 2001) and Curvularia pallescens on Cy. antillanus from Cuba (Mercado Sierra 1984). Both are morphologically rather different from C. yamadana. Curvularia aeria has conidia which are straight or curved, ellipsoidal, obovoid or clavate, 18–32 × 8–16. Curvularia pallescens has ellipsoidal to fusiform conidia, usually slightly curved, 17–32 × 7–12 μm. In the phylogenetic tree, both C. aeria and C. pallescens were phylogenetically distant from C. yamadana.
Discussion
The genus Curvularia includes species associated with plant diseases worldwide (Sivanesan 1987; Manamgoda et al. 2012a, b; da Cunha et al. 2013; Hyde et al. 2014; Manamgoda et al. 2015). It is characterized by the production of sympodial conidiophores with tretic, terminal, and intercalary conidiogenous cells and elongate, transversely septate conidia with a dark basal scar. Conidia are often curved because of it having asymmetrically swollen intermediate cells, (Sivanesan 1987; Manamgoda et al. 2015). For a long time, curvature of conidia was used as the key feature to differentiate fungi in the genus Curvularia from species belonging to related genera. However, it is now known that some Curvularia species such as C. cymbopogonis, C. oryzae-sativae, C. protuberata, and C. ryleyi have predominantly straight conidia (Manamgoda et al. 2015). Species delimitation in Bipolaris and Curvularia remained problematic for taxonomists for very long due to the overlapping morphological characters among many species (Manamgoda et al. 2014, 2015; Sivanesan 1987).
Such a subjectivity of the morphological distinction of taxa in Bipolaris and Curvularia was only resolved through the use of molecular data. In addition to ITS, other loci were found to be of high informative value in the phylogenetic analyses of sequence data from species belonging to these two genera. Particularly, the protein-coding loci of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), TEF1, and RNA polymerase II second largest subunit (RPB2) which became critical for such analyses (Hernández-Restrepo et al. 2018; Manamgoda et al. 2014; Marin-Felix et al. 2017).
According to Thind and Rawla (1961) and Tsuda and Ueyama (1982), Duosporium was considered distinct from Curvularia, because conidiogenous cells generating macroconidia of Duosporium are mostly monotretic and those generating microconidia are polytretic whereas conidiogenous cells of Curvularia are, predominantly, polytretic. However, C. muehlenbeckiae, C. beerburrumensis, C. boeremae, C. coatesiae, C. colbranii, C. kenpeggii, C. mebaldsii, C. petersonii, C. platzii, C. reesii, and C. warraberensis, along with other species, were recognized to have either mono- or polytretic conidiogenous cells (Tan et al. 2018). Duosporium was also occasionally seen to produce polytretic macroconidiophores, as mentioned above and in the published description of Barreto and Evans (1995).
Another feature which led Duosporium to be treated as distinct from Curvularia, is the production of microconidia in Duosporium. This remains a partially valid difference, since it is now known that some species of Curvularia do in fact produce secondary conidia. Among them are C. americana and C. chlamydospora (Madrid et al. 2014). Nevertheless, such secondary conidia in those two species are only formed directly from macroconidia and not from conidiophores as seen regularly in vitro for C. yamadana. In C. americana, microconidia are aseptate, pale brown, globose, and 5–6 μm wide. C. chlamydospora produce microconidia which are 1–2 celled, pale brown, globose to subglobose, and 4–6 μm diam. On the other hand, in C. yamadana, microconidia are aseptate, warted, brown to dark brown, globose to subglobose to ellipsoidal, and 5–13 × 5–9 μm. Microconidia remain a puzzle in the life cycle of C. yamadana. These are produced in large quantities in vitro in older cultures, but it was never seen in nature. Only a very small proportion of the microconidia were found to germinate, even after a series of treatments were attempted to break their dormancy (Pomella 1999). This led to the speculation that these structures would function as resting spores for C. yamadana, a hypothesis which remains to be better investigated.
The multilocus phylogenetic analyses clearly indicated that the fungus on Cyperus spp. belongs to Curvularia. Curvularia yamadana is close to C. tuberculata, C. oryzae, and C. reesii. Some species of Curvularia have some morphological resemblance to C. yamadana but the combination of oblong, straight versicolored 3-septate conidia and production of warted microconidia in culture is exclusive to the species formerly placed in Duosporium.
The need for epitipification of plant pathogenic fungi was emphasized by Cai et al. (2011) as the way forward towards clarification of the taxonomy and phylogeny of such taxa and towards the stability in the application of names. A recent case in point is that of the dematiaceous hyphomycete Acroconidiella, a genus which we recently “debunked” (Ferreira and Barreto 2019) showing its type species to be an “unusually shaped” Alternaria. Here, we provided a small contribution to this formidable task by showing that Duosporium is an artificial genus which needs to be recognized as a late synonym of Curvularia. Nevertheless, the designation of an epitype, ideally a specimen from the type locality (Honshy, Japan) on Cy. iwasakii, would be of great value for a confirmation of our understanding on the taxonomy of C. yamadana.
The taxonomic placement of the fungus on Cyperus spp. has, hopefully, been finally resolved, but an important pending challenge remains. That of determining its true potential as a practical tool to be deployed against the “tropical scourge” Cy. rotundus, as placed by William (1976). Although the theme has been shelved long ago, improvements in tools for the mass production, formulation, and application of biological control agents have occurred along the years and may justify revisiting C. yamadana as an “ecologically benign” weed biocontrol product (mycoherbicide).
References
Barreto RW, Evans HC (1995) Mycobiota of the weed Cyperus rotundus in the state of Rio de Janeiro, with an elucidation of its associated Puccinia complex. Mycol Res 98:1107–1116. https://doi.org/10.1016/s0953-7562(09)80638-2
Cai L, Udayanga D, Manamgoda DS, Maharachchikumbura SS et al (2011) The need to carry out re-inventory of plant pathogenic fungi. Trop Plant Pathol 36:205–213. https://doi.org/10.1590/S1982-56762011000400001
da Cunha KC, Sutton DA, Fothergill AW, Gene J et al (2013) In vitro antifungal susceptibility and molecular identity of 99 clinical isolates of the opportunistic fungal genus Curvularia. Diagn Microbiol Infect Dis 76:168–174. https://doi.org/10.1016/j.diagmicrobio.2013.02.034
Dhingra OD, Sinclair JB (1995) Basic plant pathology methods. CRC Press, Boca Raton
Edenfield MW, Brecke BJ, Colvin DL, Dusky JA et al (2005) Purple nutsedge (Cyperus rotundus) control with glyphosate in soybean and cotton. Weed Technol 19:947–953. https://doi.org/10.1614/WT-03-232R1.1
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Ellis MB (1971) Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Kew
Farr DF, Rossman AY (2020) Fungal databases, systematic mycology and microbiology laboratory, ARS, USDA Available at: http://ntars-gringov/fungaldatabases/Accessed 10 Jan 2020
Ferreira BW, Barreto RW (2019) Debunking Acroconidiella. Mycol Prog 18:1303–1315. https://doi.org/10.1007/s11557-019-01525-y
Hepperle D (2004) SeqAssem©. Win32–Version. A sequence analysis tool contig assembler and trace data visualization tool for molecular sequences. Available at: http://www.sequentix.de. Accessed 3 Jan 2020
Hernández-Restrepo M, Madrid H, Tan YP, da Cunha KC et al (2018) Multi-locus phylogeny and taxonomy of Exserohilum. Persoonia 41:71–108. https://doi.org/10.3767/persoonia.2018.41.05
Holm LG, Plucknett DL, Pancho JV, Herberger JP (1991) The world’s worst weeds: distribution and biology. Krieger Publishing Company, Malabar
Hyde KD, Nilsson RH, Alias SA, Ariyawansa HA et al (2014) One stop shop: backbone trees for important phytopathogenic genera: I. Fungal Divers 67:21–125. https://doi.org/10.1007/s13225-014-0298-1
Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008) Dictionary of the fungi. CAB International, Wallinford
Macedo DM (2006) Duosporium yamadanum: Produção massal, formulação e associação com herbicidas para o controle de tiririca. MSc thesis. Universidade Federal de Viçosa
Maddison WP, Maddison DR (2011) Mesquite: a modular system for evolutionary analysis. Version 3.51 http://www.mesquiteproject.org. Accessed 3 Jan 2020
Madrid H, da Cunha KC, Gené J, Dijksterhuis J et al (2014) Novel Curvularia species from clinical specimens. Persoonia 33:48–60. https://doi.org/10.3767/003158514X683538
Manamgoda DS, Cai L, McKenzie EHC, Crous PW et al (2012a) A phylogenetic and taxonomic re-evaluation of the Bipolaris–Cochliobolus–Curvularia complex. Fungal Divers 56:131–144. https://doi.org/10.1007/s13225-012-0189-2
Manamgoda DS, Cai L, McKenzie EHC, Chukeatirote Eet al. (2012b) Two new Curvularia species from northern Thailand. Sydowia 64:255–266
Manamgoda DS, Rossman AY, Castlebury LA, Crous PW et al (2014) The genus Bipolaris. Stud Mycol 79:221–288. https://doi.org/10.1016/j.simyco.2014.10.002
Manamgoda DS, Rossman AY, Castlebury LA, Chukeatirote Eet al. (2015) A taxonomic and phylogenetic re-appraisal of the genus Curvularia (Pleosporaceae): human and plant pathogens. Phytotaxa 212:175–198. https://doi.org/10.11646/phytotaxa.212.3.1
Marin-Felix Y, Senwanna C, Cheewangkoon R, Crous PW (2017) New species and records of Bipolaris and Curvularia from Thailand. Mycosphere 8:1556–1574. https://doi.org/10.5943/mycosphere/8/9/11
Matsushima T (1980) Saprophytic microfungi from Taiwan, part 1. Hyphomycetes. Matsushima Mycological Memoirs 1:1–82
Matsuura I (1931) Studies on the plant diseases caused by Brachysporium. A new disease, leaf blight of Cyperus iwasakii. Journal of Plant Protection 17:413–419
Mercado Sierra A (1984) Hifomicetes Demaciaceos de Sierra del Rosario, Cuba. Editorial Academica, Havana
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, pp 1–8
Minter DW, Rodriguez-Hernandez M, Mena-Portales J (2001) Fungi of the Caribbean: an annotated checklist. PDMS Publishing
Pereira JM, Barreto RW, Ellison CA, Maffia LA (2003) Corynespora cassiicola f. sp. lantanae: a potential biocontrol agent for Lantana camara from Brazil. Biol Control 26:21–31. https://doi.org/10.1016/S1049-9644(02)00112-3
Pinho DB, Firmino AL, Pereira OL, Ferreira Junior WG (2012) An efficient protocol for DNA extraction from Meliolales and the description of Meliola centellae sp. nov. Mycotaxon 122:333–345. https://doi.org/10.5248/122.333
Pomella AWV (1999) Avaliação do fungo Duosporium yamadanum no controle biológico da tiririca (Cyperus rotundus). DSc thesis. Universidade Federal de Viçosa
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808. https://doi.org/10.1080/10635150490522304
Rayner RW (1970) A mycological colour chart. CMI and British Mycological Society, Kew
Raza M, Zhang ZF, Hyde KD, Diao YZ, Cai L (2019) Culturable plant pathogenic fungi associated with sugarcane in southern China. Fungal Divers 99:1–104. https://doi.org/10.1007/s13225-019-00434-5
Schoch C, Crous PW, Groenewald J, Boehm E et al (2009) A class-wide phylogenetic assessment of Dothideomycetes. Stud Mycol 64:1–15. https://doi.org/10.3114/sim.2009.64.01
Sivanesan A (1987) Graminicolous species of Bipolaris, Curvularia, Drechslera, Exserohilum and their teleomorphs. Myc Papers 158:1–261. https://doi.org/10.2307/3759472
Tamura K, Peterson D, Peterson N, Stecher G et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. https://doi.org/10.1093/molbev/msr121
Tan YP, Crous PW, Shivas RG (2016) Eight novel Bipolaris species identified from John L. Alcorn’s collections at the Queensland Plant Pathology Herbarium (BRIP). Mycol Prog 15:1203–1214. https://doi.org/10.1007/s11557-016-1240-6
Tan YP, Crous PW, Shivas RG (2018) Cryptic species of Curvularia in the culture collection of the Queensland Plant Pathology Herbarium. MycoKeys 35:1. https://doi.org/10.3897/mycokeys.35.25665
Thind KN, Rawla GS (1961) A new fungus on Cyperus iria. Am J Bot 48:859–862. https://doi.org/10.1002/j.1537-2197.1961.tb11722.x
Tsuda M, Ueyama A (1982) Duosporium yamadanum, a pathogen of Cyperus spp. Mycotaxon 14:145–148
Urtiaga R (1986) Indice de enfermedades en plantas de Venezuela y Cuba. Nuevo Siglo, Venezuela
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
William RD (1976) Purple nutsedge: tropical scourge. Hortscience 11:357–364
Woudenberg JH, Groenewald JZ, Binder M, Crous PW (2013) Alternaria redefined. Stud Mycol 75:171–212. https://doi.org/10.3114/sim0015
Funding
This study received financial support from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Marc Stadler
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ferreira, B.W., Barreto, R.W. Debunking Duosporium. Mycol Progress 19, 715–723 (2020). https://doi.org/10.1007/s11557-020-01592-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-020-01592-6