Abstract
The taxonomic and genetic diversity of Cortinarius section Calochroi in one of the most biodiversity-rich regions in Europe, the Iberian Peninsula, was investigated through morphological and phylogenetic methods. This combined methodological approach allowed the identification of 15 known species and one new species, Cortinarius ortegae, which is described here. A dichotomous key is provided for field recognition of Calochroi species inhabiting different Iberian and Balearic Island woodlands. Polymorphism analyses within some studied species showed that the distribution of intraspecific lineages is dependent on geography and, in some cases, tree host. Furthermore, the dating analysis suggested that diversification within Calochroi started in the Pliocene and most of the current genetic diversity originated in the Pleistocene. This temporal scenario supports hypotheses in which climatic oscillations in the Quaternary may have been driving the evolution of this group of ectomycorrhizal fungi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cortinarius is one of the most species-rich, abundant and widespread genera of ectomycorrhizal (ECM) fungi, as demonstrated by a number of taxonomic, phylogenetic and environmental DNA (meta)barcoding studies (Garnica et al. 2016; Horton et al. 2017; Liimatainen et al. 2014; Teasdale et al. 2013). In general, species in this genus associate with many woody plants in temperate to arctic-alpine ecosystems of both hemispheres (Geml et al. 2011; Nouhra et al. 2013; Pérez-Izquierdo et al. 2017), where they play important roles in nutrient mobilisation from organic pools (Bödeker et al. 2014). Within Cortinarius, section Calochroi comprises ca. 100 species distributed mainly in the Northern Hemisphere and that associate primarily with broad-leaved tree species of Fagaceae (Quercus, Fagus and Castanea) and Betulaceae (Carpinus and Corylus) and more rarely with coniferous trees (Picea, Pinus, Abies and Pseudotsuga) (Frøslev et al. 2007; Garnica et al. 2009; Ortega et al. 2008). Calochroid species produce conspicuous basidiomes with a generally well-developed gelatinous layer, stipe with a rounded or marginate bulb, and non-anthraquinonoid pigments which often change colour with the application of KOH (e.g. Bellanger 2015; Bidaud et al. 2001; Garnica et al. 2009). During the last decade, a number of works have aimed at exploring the taxonomic diversity of Calochroi in central and northern Europe, as well as in North America (e.g. Frøslev et al. 2006, 2007, 2017; Garnica et al. 2009, 2011; Harrower et al. 2011). In the Mediterranean region, however, a reliable assessment of the species diversity of this group of ECM fungi is still lacking, despite a few studies suggesting that the area may constitute a hot spot of diversity for this lineage (Clericuzio et al. 2017; Garrido-Benavent et al. 2015; Ortega et al. 2008).
Species concepts in Calochroi are still hotly debated (Bellanger 2015; Frøslev et al. 2007). The morphological, chemical and ecological characters historically used to describe these taxa have often proven to be misleading, making the taxonomy of the section extremely difficult. Fortunately, recent phylogenetic studies have built a more consistent framework for species circumscription (Frøslev et al. 2007; Garnica et al. 2009; Garrido-Benavent et al. 2015; Ortega et al. 2008). By combining traditional examinations of sporocarps with DNA barcoding methods, these studies have also contributed to increase the amount of molecular data stored in public databases and, more importantly, they improved their taxonomic reliability. In this context, for example, ECM fungal lineages detected in environmental DNA (meta)barcoding studies can be ascribed to known species with confidence (Truong et al. 2017). As climate change is predicted to modify temperature and precipitation regimes in the Mediterranean (Pachauri et al. 2014), it is expected that the diversity of the soil biota in forests, including ECM fungi, such as the Calochroi, changes as well (Flores-Rentería et al. 2015; Rodríguez et al. 2016). Documenting the current fungal diversity before these changes occur is therefore imperative.
Here, we study several collections made in the Iberian Peninsula and Balearic Islands using both DNA barcoding (Hebert et al. 2003) and traditional taxonomy techniques. Our primary goal is to shed light into the genetic diversity of this group of ECM fungi in this region based on nrITS data and to describe new taxa. Particular emphasis is given to a single collection made in a calcareous, Quercus ilex subsp. rotundifolia forest, which shows no morphological affinity with any known species in this section. We also present data on intraspecific variability and estimates of divergence times for discussing the evolution in space and time of the species considered here. Finally, a dichotomous key is provided to help recognise selected species in field inventories.
Material and methods
Sample collection and studies of morphology and microscopy
A total of 33 collections were included in this study (Table 1). Macroscopic descriptions were produced from fresh basidiomata and microscopic observations from dried material previously rehydrated with 1% NH4OH for at least 15 min. Handmade sections of pilei and cross-sections of lamellae were mounted in H2O, and 3% KOH or Congo red mixed with 1% NH4OH, and were observed under a Zeiss Axioplan 2 microscope fitted with “Nomarski” differential interference contrast (DIC). Photographs were acquired with a Zeiss AxioCam digital camera. Microscopic measurements were made on material mounted in water under the Zeiss Axiovision 4.8 imaging system. At least 20 spore measurements were made per sample. Values representing the average (in italics) and 10th and 90th percentiles are given, as well as the minimum and maximum measurements (in parentheses). The same calculations were done for the length/width ratio (Q = L/w). Macrochemical reactions were performed following standard protocols and using 20% KOH, phenolaniline (Ph.A) and guaiac tincture. Cailleux (Cx, 1981) has been followed for the colour names. A preliminary, morphology-based taxonomic hypothesis for our collections was constructed with data from the following works: Bidaud et al. (2001), Frøslev et al. (2006, 2007), Ortega et al. (2008), Oertel et al. (2009), Bellanger (2015) and Clericuzio et al. (2017). The studied material is deposited in the Royal Botanical Garden of Madrid (MA) (Table 1).
DNA extraction and PCR amplification
Genomic DNA was isolated from dried tissue using a modified protocol based on cetyltrimethylammonium bromide (CTAB): CTAB 2%, NaCl 1.4 M, EDTA pH 8.0 20 mM, Tris–HCl pH 8.0 100 mM (Doyle and Doyle 1987). The entire Internal Transcribed Spacer (ITS1 and ITS2, including 5.8S) region of the nuclear rDNA was amplified using the universal fungal primer pair ITS1F–ITS4 (Gardes and Bruns 1993; White et al. 1990). PCR reactions were performed in a total volume of 25 μl, containing 2.5 μl of reaction buffer (Biotools®), 5 μl of dNTPs (1 mM), 1.25 μl of each primer (10 μM), 1 μl of MgCl2 (50 mM), 0.5 U of DNA polymerase (Biotools®) and 1.5 μl of genomic DNA; final volume was reached by adding distilled water. The following reaction conditions were used: 95 °C hot start for 5 min, followed by 35 cycles for 45, 30 and 45 s at 94, 54 and 72 °C, respectively, with a final extension step at 72 °C for 10 min. PCR products were checked in a 1% agarose gel stained with GelRed™ (Biotium) for visualisation of the bands. Positive reactions were purified and sequenced at Macrogen Europe (The Netherlands) using the same primers. Chromatograms were checked and assembled using SeqmanII v.5.07© (Dnastar Inc.). Accession numbers corresponding to the new sequences produced in this study are in Table 1.
Sequence alignment and phylogenetic analysis
The 33 newly produced sequences were checked for possible PCR-product contamination against the GenBank nucleotide database (http://www.ncbi.nlm.nih.gov/) with the BLAST online tool (Altschul et al. 1990). This strategy also allowed for selecting and retrieving available, highly similar Cortinarius nrITS sequences to our queries to be used in subsequent phylogenetic analyses (Online Resource 1). Moreover, most sequences of calochroid-type specimens were selected and downloaded following results of recent phylogenetic studies (Garnica et al. 2009; Clericuzio et al. 2017). Cortinarius rapaceoides Bidaud, G. Riousset & Riousset and C. evosmus Joachim ex Bidaud & Reumaux were selected as outgroup taxa.
We used MAFFT v. 7.308 (Katoh et al. 2002; Katoh and Standley 2013) as implemented in Geneious v. 9.0.2 to generate a multiple sequence alignment (MSA). The following parameters were selected to minimise homoplasy in an alignment containing very divergent nrITS sequences: the FFT-NS-I ×1000 algorithm, the 200PAM/k = 2 scoring matrix, a gap open penalty of 1.5 and an offset value of 0.123. Manual optimization of the resulting alignment was carried out in Geneious v. 9.0.2, and it consisted in either trimming alignment ends of longer sequences that included part of the 18S–28S ribosomal subunits, or replacing gaps at the ends of shorter sequences with an IUPAC base representing any base (N). Ambiguously aligned regions were dealt with automatically by using the software GBlocks v. 0.91b (Castresana 2000), implementing the least stringent parameters but allowing gaps in 50% of the sequences. The alignments are deposited in TreeBase under S21800 (http://purl.org/phylo/treebase/phylows/study/TB2:S21800). Subsequently, a phylogeny was estimated in a Maximum Likelihood (ML) framework with the online version of RAxML-HPC2 hosted at the CIPRES Science Gateway (Miller et al. 2010; Stamatakis 2006; Stamatakis et al. 2008). The analysis used the GTRGAMMA substitution model for two delimited partitions within the nrITS (ITS1 + ITS2 and 5.8S), and 1000 bootstrap pseudoreplicates were conducted to evaluate nodal support. Additionally, a Bayesian phylogenetic MCMC analysis was implemented using MrBayes v. 3.2.6 (Ronquist et al. 2012). Optimal substitution models for the above two nrITS partitions were inferred with PartitionFinder v. 1.1.1 (Lanfear et al. 2012) considering a model with linked branch lengths and the Bayesian Information Criterion (BIC). This analysis favoured the HKY + I + Γ model for the ITS1 + ITS2 partition, and the K80 for the 5.8S. Then, the MrBayes analysis was conducted with two parallel, simultaneous four-chain runs executed over 2.5 × 107 generations starting with a random tree, and sampling after every 250th step. The first 25% of data was discarded as burn-in. The 50% majority-rule consensus tree and corresponding posterior probabilities were calculated from the remaining trees. Chain convergence was assessed by ensuring that average standard deviation of split frequencies (ASDSF) values were below 0.01 and potential scale reduction factor (PSRF) values approached 1.00.
We further inferred evolutionary relationships among our selected taxa with an alternative method that allows for iteratively estimating multiple sequence alignments and phylogenetic trees. It was implemented in the program SATé v. 2.2.7 (Liu et al. 2009, 2011) and used the original MAFFT MSA. This approach can boost alignment accuracy substantially and is especially desirable when using difficult-to-align regions, as the nrITS (Leavitt et al. 2012a). SATé v. 2.2.7 uses Maximum Likelihood methods in a divide-and-conquer technique consisting in three steps: (1) to divide the set of sequences into subsets, (2) to re-align each subset and (3) to merge the resulting alignments into an alignment of the full set of sequences. The following options were selected when running the analysis: “Aligner” = MAFFT, “Merger” = MUSCLE, “Tree Estimator” = FASTTREE, and “Model” = GTR + CAT, a 20% value for Max. Subproblem, and 500 iterations following the final improvement under the remaining default SATé settings.
Species delimitation
The topology-independent, Automatic Barcode Gap Discovery (ABGD, Puillandre et al. 2012) approach was used to identify species boundaries in the nrITS data set. This method automatically detects a “barcode gap” that delimits candidate species based on non-overlapping values of intra- and interspecific genetic distances, and it has been shown to be accurate in inferring species limits across several fungal groups (Garrido-Benavent et al. 2016; Leavitt et al. 2015). Analyses used two versions of the nrITS alignment: the GBlocks-trimmed alignment and the initial, full alignment generated in MAFFT. Runs were remotely carried out at http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html, setting the Pmax value to 0.01 (Puillandre et al. 2012), the relative gap width (X) to 1 and the remaining model parameters to default. The Kimura two parameters (K2P) model was used to calculate genetic distances between individuals. Transition/transversion ratio (TS/TV) values obtained in MEGA 5.2 (Tamura et al. 2011) for the above-mentioned alignments were 1.99 and 1.87, respectively.
Intraspecific genetic diversity
Fourteen species with collections in the Iberian Peninsula were selected for analysing intraspecific genetic diversity: Cortinarius albertii, C. cisticola, C. corrosus clade I, C. fulvocitrinus, C. haasii, C. insignibulbus, C. molochinus, C. piceae, C. platypus, C. sancti-felicis, C. splendidior, C. subgracilis, C. sublilacinopes and C. violaceipes. First, newly produced sequences (Table 1) were used as queries in blastn (Altschul et al. 1990) searches and the retrieved sequences (including uncultured/environmental sample sequences; Online Resource 1) that displayed a query coverage between 90 and 100% and a similarity threshold ≥ 99% (Garnica et al. 2016) were downloaded and aligned with MAFFT v. 7.308 in Geneious v. 9.0.2. Ends of sequences that showed unreliable nucleotide substitutions as well as columns with ambiguous positions were removed, and final alignments were trimmed to the shortest sequence. The UNITE database (http://unite.ut.ee/) was also surveyed to look for absent sequences in GenBank and also for retrieving information about geographical locations and associated plants for each Cortinarius collection (Online Resource 2). Second, DNA polymorphism levels were evaluated after excluding gaps by calculating segregating sites (s), nucleotide diversity (π), average number of nucleotide differences (k) and number of haplotypes (h) with DnaSP v.5.10 (Librado and Rozas 2009). Results are displayed in Table 2.
Dating analysis
Divergence age estimates for the calochroid taxa considered in the present study were inferred to set a time frame for the diversification of this species-rich group of Cortinarius. Because this genus lacks a suitable fossil record, a secondary calibration was imposed on the nrITS substitution rate using the software BEAST v. 1.8.1 (Drummond et al. 2012). In particular, the analysis was run using an average nrITS rate of 5.84 × 10−3 substitutions per site per million years as inferred for Cortinarius s.l. in Ryberg and Matheny (2011) along with an uncorrelated lognormal relaxed molecular clock and a birth–death process tree prior. Before running the analysis, the GBlocks-trimmed nrITS alignment was further edited to avoid sequence redundancies, and therefore, only 63 sequences were finally included. Moreover, to assess the effect of keeping ambiguously aligned regions on the inference of divergence ages, the same analysis was re-run with the original GBlocks-unprocessed nrITS alignment and the same BEAST parameter settings.
Overall, running instructions included chain lengths of 2.5 × 107 steps, saving always 10,000 trees and removing the first 25% of them as burn-in. The programs Tracer v. 1.5, TreeAnnotator v. 1.8.1 and FigTree v. 1.4 (all available at: http://tree.bio.ed.ac.uk/) were employed to check for convergence, annotate the median heights of the post-burn-in tree samples, and construct 50% majority rule consensus trees, respectively.
Results
Alignments, phylogenies and species delimitation
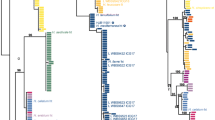
The ingroup data set considered in the present study comprised 82 sequences of which 43 were obtained from type material. The MAFFT algorithm produced an alignment of 632 nucleotides in length. After the automatic removal of ambiguously aligned positions in GBlocks v. 0.91b, 91% (580 nucleotides) of the original length was kept in 27 selected blocks. The final alignment included 185 variable positions, of which 136 were parsimony informative and 49 corresponded to singleton sites. The ML analysis resulted in a single best tree of Ln = − 3366.8483. The MrBayes analysis reached an average standard deviation of split frequencies of 0.01 after 3.517 × 106 generations. No statistically supported conflict was observed among the topologies obtained with these two methods, and the RAxML-resulting topology is presented in Fig. 1. The analysis run with SATé v. 2.2.7, which iteratively estimates the MSA and a phylogenetic tree in a ML framework, produced a tree with a score of Ln = − 3529.5043 (Online Resource 3).
Phylogram depicting phylogenetic relationships among members of Cortinarius section Calochroi obtained with RAxML and based on nrITS data. For each terminal, the nrITS accession number, species name, collection number and origin of sample (type material, T) are given. Support values are given for each node (ML and BI analyses, on the left and right, respectively). Branches showing strong support (BP ≥ 70% and/or PP ≥ 0.95) are in bold. Columns on the right represent the proposed phylogenetic species inferred from ML and BI analyses (first column) and results of ABGD species delimitation using the original (second column) and the GBlocks-trimmed (third column) nrITS alignments. The two shades of grey in each column represent the assignment to a different species. The asterisk symbol in the second and third columns is meant to highlight a likely erroneous inclusion of C. insignibulbus, C. parasuaveolens-pseudogracilior, C. spectabilis (DQ663426) and JB-5939-07 within the same ABGD partition due to the existence of abundant missing data in some of the original sequences
In general, the phylogenetic relationships inferred with RAxML and MrBayes are in agreement with results of previous works using a similar taxon sampling (Bellanger 2015; Clericuzio et al. 2017). A high clade support (BP > 70%, PP > 0.95; Fig. 1) was mainly inferred for sister taxa relationships, such as C. violaceipes and C. insignibulbus, C. largentii and C. corrosus clade I-calojanthinus, C. haasii, C. haasii var. quercus-ilicicola (hereafter C. haasii s.l.) and C. splendidior, C. molochinus and C. “nymphicolor”, and C. cobaltinus and C. caesiocinctus. A supported relationship between C. sancti-felicis and a clade containing C. violaceipes and C. insignibulbus was also recovered. In contrast, the tree backbone (inner relationships) showed no support in either the ML or BI inference methods. The phylogenetic tree obtained with SATé v. 2.2.7 showed moderate to high values of bootstrap support (BP > 70%) for inner evolutionary relationships, thus improving the resolution of the tree backbone (Online Resource 3). Relationships between sister taxa and outer clades also received increased support when implementing this alternative method of phylogenetic inference. No supported topological conflict was observed between this phylogeny and those inferred with RAxML and MrBayes.
The new species Cortinarius ortegae (see the “Taxonomy” section) was found to be close to a clade comprising C. splendidior, C. haasii s.l., C. sublilacinopes, C. subgracilis and C. chailluzii (Fig. 1), but this relationship was not statistically supported (BP < 70%, PP < 0.95). However, the phylogenetic tree obtained with SATé gave support to a clade including C. ortegae, C. subgracilis, C. chailluzii, C. haasii s.l. and C. splendidior (BP = 84%, Online Resource 3). Both studied samples of C. ortegae, one from the eastern Iberian Peninsula and the other from Mallorca (Balearic Islands), showed no differences at the molecular level. Furthermore, phylogenetic analyses together with evidences from the morphological study of sporocarps allowed the ascription of most of the studied collections to known calochroid taxa, e.g. C. violaceipes, C. insignibulbus, C. sancti-felicis, C. albertii, C. platypus, C. haasii s.l., C. splendidior, C. subgracilis, C. molochinus, C. fulvocitrinus, C. cisticola, C. barbaricus, C. piceae and C. sublilacinopes (Fig. 2). Three collections morphologically resembling C. corrosus were nested in two distinct clades. Thus, MES-3612-00 and JB-6765-09 were included within a C. corrosus clade I, together with C. calojanthinus type and a further C. corrosus accession from GenBank (EU057057), whereas MES-4688 defined, together with an Iberian Peninsula collection morphologically similar to C. spectabilis sensu Bidaud et al. 2001 (MES-4313-05), the C. corrosus clade II. Despite the type of C. corrosus has not been sequenced yet, the working hypothesis here is that C. corrosus clade I represents C. corrosus. A second Iberian Peninsula collection morphologically akin to C. spectabilis, JB-5939-07, clustered into an evolutionarily unrelated clade, together with a sequence from GenBank annotated as C. spectabilis (DQ663426). The SATé results gave low support (BP = 69%) to a clade containing samples MES-4315-05 and MES-4688, C. largentii and C. corrosus clade I-calojanthinus (Online Resource 3). Support (BP = 90%) was given to another clade containing the sample JB-5939-07 together with C. spectabilis (DQ663426), C. magnivelatus and the type sequence of C. bigelowii. Finally, the close phylogenetic relationships between the sequences of the types of C. parasuaveolens and C. pseudogracilior, C. platypus and C. frondosophilus, C. haasii and C. aurantiorufus, and C. albovestitus and C. ochraceopallescens point to the synonymy of these pairs of taxa as far as the nrITS locus alone is concerned (Bellanger 2015; Liimatainen et al. 2014).
a Cortinarius cisticola (JB-9114-16, in a Quercus ilex forest, Spain). b Cortinarius corrosus (JB-6765-09, in an Abies alba forest, Spain). c Cortinarius fulvocitrinus (JB-8465-14, in a Fagus sylvatica forest, Spain). d Cortinarius haasii (JB-8420-14, in an Abies alba forest, Spain). e Cortinarius insignibulbus (JB-8966-16, in a Fagus sylvatica forest, Spain). f Cortinarius molochinus (MES-3968-03, in a Quercus ilex subsp. rotundifolia and Pinus halepensis mixed forest, Spain). g Cortinarius piceae (JB-8676-15, in an Abies alba and Fagus sylvatica mixed forest, Spain). h Cortinarius platypus (JB-9040-16, in a Quercus ilex forest, Spain). i Cortinarius sancti-felicis (MES-4490-14, in a Quercus ilex subsp. rotundifolia and Pinus sylvestris mixed forest, Spain). j Cortinarius subgracilis (JB-9144-16, in a Quercus ilex and Pinus nigra mixed forest, Spain). k Cortinarius sublilacinopes (JB-6844-09, in a Dryas octopetala-dominated heath community, Spain). l Cortinarius violaceipes (JB-8476-14, in a Quercus ilex forest, Spain). Photo credits: J. Ballarà (a–e, g–h, j–l), R. Mahiques (f, i)
ABGD analyses of the GBlocks-trimmed and full nrITS-rooted datasets yielded similar results (Fig. 1). In the former analysis, initial and recursive partitions converged on 41 partitions (P = 0.0022–0.0077), while in the second analysis, they converged on 42 partitions (P = 0.0022–0.0060). In both cases, the new species C. ortegae was allocated to an independent partition or putative species. A separate partition was also given to the sample pair MES-4313-05 and MES-4688, as well as JB-5939-07 and C. spectabilis (DQ663426). Moreover, the ABGD species delimitation algorithm supported the ascription of the remaining sequences produced in the present study to known calochroid taxa, and results were mostly in agreement with those based on the phylogenetic results (Fig. 1). However, the allocation of the three sequences of C. parasuaveolens–pseudogracilior and two sequences of C. insignibulbus (initial, full nrITS alignment) together with JB-5939-07 and C. spectabilis (DQ663426) (GBlocks-trimmed nrITS alignment) within the same partition or putative species was unexpectedly found. This could be an artefact due to the large amount of missing data included in some of these sequences (e.g. C. insignibulbus).
DNA polymorphism at the species level
Intraspecific genetic diversity was found to be low or absent for most species, and it was greatly influenced by the number of sequences (n) used for each species and by the presence of numerous ambiguous base calls in most alignments (Table 2). Cortinarius corrosus clade I-calojanthinus, C. haasii s.l., C. piceae, C. subgracilis and C. violaceipes showed the highest values for the number of polymorphic sites (s) and haplotypes (h) and nucleotide diversity (π). Sequences of C. corrosus clade I-calojanthinus, which associates with coniferous trees (Picea, Abies and Pinus, Table 2, Online Resource 2), segregated in two distinct groups overlapping with geography (North America and Europe). Data of C. haasii s.l. also split in two groups, one of them including collections mostly found in southern Europe associated with several species of Quercus (C. haasii var. quercus-ilicicola sensu Ortega et al. 2008), and the other containing collections from the Iberian Peninsula to Sweden, all associated with conifers, except for one French sample collected under Q. ilex (KY290683). A subset of sequences of C. subgracilis was apparently restricted to the Mediterranean Basin, particularly the Iberian Peninsula, and associated with Q. ilex, while a second formed a widespread group in Europe associated to either Quercus or Picea. Other taxa also presented significant levels of polymorphism, but these occurred in positions with ambiguous base calls which were removed prior to the analyses. For example, six out of the ten columns with ambiguous base calls in the C. platypus alignment showed nucleotide substitutions. These would have rendered at least five different haplotypes, thus increasing genetic diversity indices in this widespread European species (data not shown). In this hypothetical scenario, C. platypus would include two groups of sequences; one restricted to southern Europe (France, Iberian Peninsula) and associated mainly to Quercus, whereas the second would correspond to samples widely distributed across Europe, from the Iberian Peninsula to Denmark and associated with Fagus, Quercus and Corylus (Table 2, Online Resource 2).
Dating analyses
The topologies inferred with BEAST using the two nrITS datasets in the dating analyses were congruent with those obtained in previous ML and BI analyses. Minor topological differences between them were always unsupported (PP < 0.95). Similarly, the use of the full-length, unprocessed nrITS data set had no relevant effect on the estimated ages. Therefore, for simplicity, only the chronogram obtained with the GBlocks-trimmed data set is presented in Fig. 3. In general, diversification of the calochroid taxa considered in the present study started during the Pliocene, ca. 3.91 million years ago (Ma) (4.92–3.04 Ma, 95% highest posterior density, HPD). Most splits between sister species occurred from the Early to Middle Pleistocene, whereas splits at inner clades were dated back to the Late Pliocene to Early Pleistocene. The new species C. ortegae diverged from a supported clade containing C. splendidior and C. haasii s.l. ca. 2 Ma (2.74–1.26 Ma, 95% HPD). However, this evolutionary relationship was not supported (PP < 0.95). Diversification within C. haasii s.l. started between 0.61 and 0.05 Ma (95% HPD). Similar age estimates were inferred for diversification in other taxa for which intraspecific data was included in the analysis, e.g. C. subgracilis, C. ochraceopallescens-albovestitus, C. corrosus clade I-calojanthinus and C. platypus-frondosophilus.
BEAST time-calibrated MCC tree of calochroid taxa estimated from a nrITS data set using a secondary calibration on the average nrITS substitution rate of Cortinarius inferred in Ryberg and Matheny (2011). Median age estimates are provided for selected nodes representing the diversification of calochroid taxa, the divergence of the new species C. ortegae, and the intraspecific diversification of selected species. Grey bars show the 95% highest posterior density intervals (HPD) of divergence age estimates for each node. Black dots on nodes represent significant statistical nodal support (PP ≥ 0.95)
Taxonomy
Cortinarius ortegae Mahiques, Ballarà, Salom, Bellanger & Garrido-Benavent, sp. nov.
Mycobank No.: MB 823766 (Fig. 4).
Cortinarius ortegae. a–b Habitat and habitus (holotype, MA-91185 collected in a Quercus ilex subsp. rotundifolia in the Valencian Country, eastern Iberian Peninsula). c–d Habitat and habitus (MA-91186 collected in a Q. ilex subsp. ilex forest in the Balearic Islands). e–g Spores, basidium and cellular elements in lamella edge respectively (holotype). h–i Spores and cellular elements in lamellae edge in Congo Red (MA-91186). Scale bars 10 μm. Photographs a–b by R. Mahiques; c–d, h–i by J.C. Salom; e–g by I. Garrido-Benavent. f–g “Nomarsky” differential contrast
Diagnosis: Pileus 30–55 mm diam., brunneo-ochraceo-aurantiacus; lamellae griseo-roseae versus lilacinas; stipes 30–50 × 6–12 (22–26) mm, albescens vel pallide ochraceus, longitudo stipitis brevior quam diameter pilei, bulbo lato, planatus, circumciso margine praeditus; ope KOH supra pilei superficiem, partim spadiceo (CxP39) partim brunneo-ochraceo vel brunneo-aurantiaco; in pilei caro grisescente, in bulbi caro et eo margine grisescente vel brunneo-aurantiaco. Sporae amygdaliformes vel ovatae, rare subpapillatae, 8–8.8–10.1 (11) × 5–5.5–5.6 (6) μm. Q = (1.4) 1.5–1.6–1.7 (2). Habitatione in mediterraneensibus sclerophyllis Quercubus ilicibus silvis, in calcareo solo.
Holotype: SPAIN, Valencian Country, Vall d’Albaida region, Llutxent, Els Surars, el Racó de les Pereres, 38° 59′ 00.48″ N, 0° 18′ 00.24″ W, 613 m a.s.l., in a dense, small forest of Q. ilex subsp. rotundifolia with an understorey composed of young Q. coccifera, Pteridium aquilinum, Ulex parviflorus, Rubus ulmifolius and Cistus spp., on calcareous soil, 26.10.2015, leg. R. Mahiques. Holotype MA-91185 (MA). GenBank: MG696273.
Etymology: named after our friend and mycologist Antonio Ortega Díaz (1954–2014).
Subgenus Phlegmacium (Fr.) Trog, section Calochroi M.M. Moser & E. Horak.
Pileus 30–55 mm diam., at first hemispheric, later plano-convex, homogenously ochraceous-brown with some faint orange hues (Cx K80), and later mottled; margin undulate, sometimes incurved; surface glutinous, not innately fibrillose, slightly velvety to the pileus centre, later becoming somewhat scaly with minute scales first whitish and then brownish.
Lamellae adnate to sinuate, moderately crowded (7–8 gills per cm), ca. 5 mm broad, greyish pink to pinkish (Cx L51), later with a lilac hue; lamellar edges slightly serrate and of the same colour of the pleura.
Stipe 30–50 × 6–12 (22–26) mm, shorter than pileus diameter, stout, cylindrical, ending with a marginate bulb; surface whitish, later pale ochraceous, longitudinally fibrillose, with scarce whitish universal veil remnants; bulb broad with a sharp edge that originates a flattened upper side, which is covered with veil remnants; bulb surface whitish with ochraceous spots; bulb base rounded showing a whitish mycelium.
Context of pileus and stipe firm, whitish with ochraceous and orange hues as well as some rusty-coloured spots; smell and taste indistinct or ± earthy.
Macrochemical reactions: KOH moderately reddish brown (Cx P39), ochraceous brown to orange brown on pileus cuticle; greyish on pileus context; greyish or orange brown on bulb context and bulb edge surface. Ph.A and Guaiac Tincture negative even after 30 min after exposure.
Spores amygdaliform to subovoid, rarely subpapillate, with warts of a moderate size that sometimes appear connected, generating a granular spore outline; MA-91185 (Holotype): 8–8.8–10.1 (11) × 5–5.5–5.6 (6) μm, and Q = (1.4) 1.5–1.6–1.7 (2); MA-91186 (JCS-770B): (8) 8.5–9.9–12 (13) × (5) 5.3–5.7–6 (7) μm, and Q = (1.4) 1.5–1.7–2 (2.2).
Basidia 19–30 × 8–10 μm, 4-spored, slightly congophilic and containing granules; lamellar edge with basidia and chains of catenulate cells ending with a congophilic, subglobose to sphaero-pedunculate cell, 14–23 × 7–10 μm in size.
Pileipellis an ixocutis, composed of a thick layer of interwoven and gelatinized, 1–5 μm wide hyphae with attenuated ends and with a dense granular wall pigmentation; subcutis indistinct, with 3–10 μm wide hyphae showing granular wall pigments.
Ecology and distribution: In a dense, small forest of Q. ilex subsp. rotundifolia, on calcareous soil.
Additional material examined: SPAIN, Balearic Islands, Mallorca Island, Campanet, Monument Natural de ses Fonts Ufanes (Gabellí Petit), 39° 48′ 14.35″ N, 2° 57′ 51.68″ E, 75 m. a.s.l., under Q. ilex subsp. ilex, on calcareous soil, 28.11.2008, leg. S. Pinya, F. Dubon & J.C. Salom, JCS-770-B, MA-91186.
Comments: The overall habitus of C. ortegae resembles that of C. platypus (Fig. 2h), but the latter usually shows a pileus with pale yellowish to ochraceous colours, sometimes with mauve or pinkish hues, and without orange tinges, a slightly more flattened stipe bulb, a reddish KOH reaction on pileus cuticle, and positive reactions to Ph.A and Guaiac Tincture (Bidaud et al. 2001: f. 526, Pl. 336). Cortinarius haasii s.l. and C. splendidior, which often fruit in Mediterranean Q. ilex forests on calcareous soil, show slightly larger spores and yellowish mycelia (Bidaud et al. 2001; Bellanger 2015; Ortega et al. 2008). Cortinarius subgracilis (Fig. 2j), which seems to associate with either Q. ilex or several coniferous trees, is a polymorphic species that can display lilac hues in pileus cuticle, lamellae and stipe, turns red when exposed to KOH, and has larger spores than C. ortegae (Ortega et al. 2008). Cortinarius chailluzii differs from the new species in showing a heterogeneously-tinged pileus (yellowish brown in the centre and pale yellowish towards the margin), lamellae and upper stipe purplish grey, and has smaller spores (Frøslev et al. 2006). Cortinarius sublilacinopes (Fig. 2k) produces slender basidiomata and pilei are pale ochraceous to yellowish, the stipe and lamellae have lilac hues, and reacts deep red to KOH on pileus cuticle (Bidaud et al. 2001). Cortinarius ochraceopallescens produces stout and larger sporocarps, usually exceeding 80 mm in width, pilei are brownish to yellowish brown, and it shows different reactions to KOH compared with C. ortegae (Bidaud et al. 2001; Frøslev et al. 2006). The dichotomous key provided below shows further differences of the new species when compared with other calochroid taxa. Finally, C. ortegae could be confused with C. rapaceotomentosus, which falls outside section Calochroi, but the latter has a more whitish pileus at first, later with bluish spots, and a pinkish brown reaction to KOH on pileus cuticle (Delaporte et al. 2002).
Key to the Cortinarius sect. Calochroi taxa present in the Iberian Peninsula supported by phylogenetic inference
-
1 Stipe base showing a yellowish mycelium ..........................2
-
1* Stipe base showing a whitish mycelium ............................4
-
2 Pileus yellowish orange, without olivaceous tinges; apparently associated with Mediterranean sclerophyllous trees .................................................C. haasii var. quercus-ilicicola
-
2* Pileus often with olivaceous tinges ....................................3
-
3 Pileus from yellowish brown to olivaceous yellow; in coniferous forests .............................................C. haasii var. haasii
-
3* Pileus ochraceous, somewhat lilaceous to bluish, with or without apparent olivaceous tinges, and reacting weakly to KOH; in coniferous forests ...............................C. splendidior
-
4 Growing mainly in calcareous, coniferous forests ..............5
-
4* Growing mainly in calcareous, broad-leaved forests .............9
-
5 Sporocarps without bluish tinges; pileus pale ochraceous, with a more or less deep red reaction to KOH C. corrosus clade I (= C. calojanthinus M.M. Moser & Ammirati)
-
5* Bluish tinges present in some sporocarp regions ..............6
-
6 Overall stipe cuticle with a very weak to null reaction to KOH. Pileus yellowish gold, innately fibrillose, with darker scales in the centre; pinkish brown reaction to KOH on pileus cuticle, null on pileus and stipe context and on the basal felt of the C. piceae (= C. calochrous var. coniferarum (M.M. Moser) Nezdojm. ss. CFP)
-
6* Bulb surface with a reddish pink reaction to KOH ..........7
-
7 Sporocarps slender, relatively small (diameter of pileus approx. 50 mm or smaller). Pileus with clashing colours, usually ochraceous brown with yellowish and with or without lilaceous tinges; red reaction to KOH on cuticle of pileus and stipe bulb, more pinkish on the basal felt of the stipe ............................................................................C. subgracilis
-
7* Sporocarps stout, relatively big (diameter of pileus approx. 80 mm or greater) ...................................................................8
-
8 Pileus pale ochraceous with slight mauve tinges; KOH reaction red in pileus cuticle, pinkish on pileus context, stipe bulb surface and mycelium, and negative on lamellae and bulb context ...........................................................C. cf. spectabilis
-
8* Pileus yellowish; stipe with lilaceous tinges; vivid reddish pink reaction to KOH on pileus cuticle and stipe base ............................................................................C. barbaricus
-
9 Growing in Mediterranean sclerophyllous forests, mainly composed of broad-leaved tree species ...............................10
-
9* Growing in areas with a different type of vegetation .........14
-
10 Pileus first homogenously ochraceous-brown with some faint orange hues, then with some darker mottles; KOH moderately reddish brown, ochraceous brown to orange brown on pileus cuticle; greyish on pileus context; greyish or orange brown on bulb context and bulb edge surface ..................................................................................C. ortegae
-
10* Pileus mostly without orange tinges ..............................11
-
11 KOH reaction pale pink to vivid red on pileus and bulb cuticle; pileus, lamellae an upper part of stipe pale violaceous, with whitish veil remnants in the pileus surface ....................................................................C. molochinus
-
11* Less vivid KOH reaction on pileus cuticle ...................12
-
12 Pinkish KOH reaction only on areas of pileus cuticle with lilaceous tinges; pileus first mauve to lilaceous, then ochraceous with whitish, ochraceous to yellowish veil remnants; lamellae and stipe with mauve tinges .....................................................................C. violaceipes
-
12* Reacting differently to KOH ..........................................13
-
13 Maroon red reaction to KOH on pileus and bulb cuticle; pileus, lamellae and upper stipe with persistent mauve tinges .........................................................................C. sancti-felicis
-
13* Deep red reaction to KOH on bulb cuticle, and weak in other parts of the sporocarp; pileus homogenously yellowish, with whitish veil remnants ....................................................C. cisticola
-
14 Associated with diverse broad-leaved trees. Deep red reaction to KOH on pileus cuticle; pileus pale ochraceous to yellowish; lamellae and stipe with mauve-lilac tinges .......................................................................C. sublilacinopes
-
14* Preferentially associated with Fagus sylvatica in central and northern Europe, but also with Quercus spp. in the Mediterranean area; different reaction to KOH on pileus cuticle .........................................................................................15
-
15 Pinkish to vinaceous reaction to KOH on pileus cuticle, and weakly pinkish or null on pileus context; habitus squat ................................................................................C. platypus
-
15* Null or weak reaction to KOH; habitus different .............16
-
16 Absence of bluish tinges in sporocarps; greenish yellow colours on pileus and bulb context, and fulvous brown colours on lamellae; null reaction to KOH everywhere but for the veil, which shows a red reaction .............................C. fulvocitrinus
-
16* Presence of bluish tinges in sporocarps .........................17
-
17 Sporocarps with disproportionate large stipe bulb, and medium sized pilei; null KOH reaction on pileus cuticle and context, and pale pink on bulb cuticle ....................C. insignibulbus
-
17* Sporocarps with smaller stipe bulb; positive KOH reaction on pileus cuticle and/or stipe .........................................18
-
18 Sporocarps with pileus on average 50–80 mm wide; pileus ochraceous to beige with olivaceous or pinkish tinges, sometimes with abundant pale beige, violaceous to brownish veil remnants in the centre; null or weakly pinkish reaction to KOH on pileus cuticle, and eosin red on the basal felt of the stipe ....................................................................C. selandicus
-
18* Sporocarps stout, larger, usually exceeding 80 mm in width. Pileus whitish in young sporocarps, with saffron yellow spots; KOH reaction first pinkish and then reddish on pileus and bulb cuticle, greyish brown with orange tinges on pileus and bulb context, and null on stipe context ...................................................................................C. albertii
Discussion
By using DNA barcoding and morphological data, 33 Cortinarius collections from the Iberian Peninsula could be assigned to 15 known species and one, C. ortegae, is described as new. In general, this study provides additional support for using this combined approach for species discovery and delimitation in this genus of ECM fungi (Bellanger 2015; Frøslev et al. 2007; Liimatainen et al. 2014, 2017). This is particularly relevant in the context of the Iberian Peninsula where many studies during the last decade have documented a high taxonomic and genetic diversity in Cortinarius (Ballarà et al. 2016; Garrido-Benavent et al. 2015, 2016; Niskanen et al. 2011; Ortega et al. 2008) and confirmed the occurrence of taxa known only from high-latitude European regions in the Iberian woodlands (Ballarà et al. 2016, 2017; Ortega et al. 2008). So far, the number of Calochroi species reported in the peninsula as well as the Balearic Islands in morphology-based studies was 29 (Ballarà et al. 2014; Cadiñanos Aguirre 2004; Mahiques 2011; Salom et al. 2015). The present study adds four species to the checklist: C. fulvocitrinus, C. insignibulbus, C. ortegae and C. violaceipes. Moreover, molecular data is provided for the first time to Iberian collections of the following species: C. albertii, C. barbaricus, C. cisticola, C. corrosus, C. molochinus, C. piceae and C. sublilacinopes. Cortinarius corrosus clade II and C. cf. spectabilis collections (MES-4313-05, MES-4688, JB-5939-07) could not be assigned a name with confidence due to the absence of publically available sequences of type specimens. More collections and genetic data are needed to investigate whether these constitute new species. Similarly, a phylogenetic assessment of collections corresponding to the 29 initially reported calochroid species may be useful for providing a more exact figure of species diversity in this group of Cortinarius in the Iberian Peninsula as well as the Balearic Islands.
While care must be taken when interpreting date estimates of analyses based on secondary calibrations (Schenk 2016), the estimated Pliocene diversification of calochroid taxa found in the present study agrees to some extent with results of Ryberg and Matheny (2011). These authors found that most taxa in Calochroi originated between the Middle Miocene and Pliocene. Species diversification during this time interval has been also inferred for members of Cortinarius sect. Cortinarius (Harrower et al. 2015), as well as for other ECM fungi, such as members of Amanita sect. Caesareae (Sánchez-Ramírez et al. 2015), and even some lichen-forming fungi (Leavitt et al. 2012b). In Europe, and especially in the Mediterranean area, it is likely that climatic oscillations during that period (Zachos et al. 2001) forced calochroid taxa to co-migrate with their host plants (Rodríguez-Sánchez et al. 2010). Subsequent fragmentation of populations by either biotic or abiotic factors could have ultimately led to speciation (Garnica et al. 2011). Allopatry or parapatry has been suggested as responsible for diversification in several groups of ECM fungi belonging to the Agaricales (Ryberg and Matheny 2011).
Polymorphism statistics revealed the influence of geography in the distribution of genetic diversity within calochroid species. The Iberian Peninsula is shown to host all intraspecific lineages in most of the studied species (e.g. C. corrosus clade I-calojanthinus, C. haasii s.l., C. platypus and C. subgracilis). In fact, the Iberian Peninsula is part of the Mediterranean biodiversity hotspot (Fig. 1 in Myers et al. 2000), and it has been shown to act as a refuge in the last glaciations for many organisms (Hewitt 2011), including woody plant species to which calochroid taxa associate (e.g. Quercus spp., Rodríguez-Sánchez et al. 2010). It is therefore possible that these ECM fungi also used this region as a refuge during the last glaciations, maintaining a higher genetic diversity, and subsequently, some lineages followed their plant hosts in the colonisation of more northerly latitudes. The age estimates for intraspecific diversification in the studied taxa agree with this scenario. Moreover, the available data also indicates that many species associate with different host species in the Mediterranean area as compared with other northern European regions. Thus, C. albertii, C. cisticola, C. haasii s.l. and C. platypus form ectomycorrhizae mainly with Quercus spp. in the Iberian Peninsula, southern France and Italy, whereas in Germany, Sweden and Denmark, they associate mainly with several deciduous trees, such as F. sylvatica, Carpinus betulus and Corylus avellana (Frøslev et al. 2006, 2007; Garnica et al. 2009), or even with coniferous trees, as is the case of C. haasii s.l. and C. subgracilis (Ortega et al. 2008). Cortinarius sublilacinopes associates with Fagus and Carpinus in central Europe while it associates with Quercus and, most interestingly, Dryas octopetala in high-altitude habitats of the Iberian Peninsula (Ballarà and Mahiques 2010). The latter constitutes a widespread dwarf shrub in alpine and arctic regions that has been shown to associate with many ECM fungi, including Cortinarius (Bjorbækmo et al. 2010; Ryberg et al. 2009). Host switching and expansion have been postulated to boost diversification rates in several groups of ECM fungi (Han et al. 2017; Looney et al. 2016). On the other hand, C. fulvocitrinus and C. violaceipes are very selective to their plant host even in climatically different regions in Europe. An increased sampling at the spatial and molecular levels is needed before proposing further explanations on the mechanisms controlling intraspecific diversification and host-association patterns in this group of ECM fungi.
In conclusion, the present work highlights the high inter- and intraspecific genetic variability of Cortinarius sect. Calochroi in the Iberian Peninsula woodlands and establishes a methodological framework for future studies documenting its taxonomic and genetic diversity in this region. Furthermore, the provided molecular data may assist environmental DNA (meta)barcoding studies in identifying particular lineages inhabiting Mediterranean forests. This basic information is pivotal for assessing, for example, species-specific responses to changes in climate (Fernández et al. 2017), and forest management (Kyaschenko et al. 2017) in a scenario of global change.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Ballarà J, Cadiñanos Aguirre JA, Campos JC, Escánez LL, Fernández Sasia R, Gutiérrez C, Macau N, Mahiques R, Mateos A, Meléndez A, Pérez A, Pérez-de-Gregorio MÀ, Reyes JD, Salom JC, Santamaría N, Serrano S, Suárez E (2014) Cortinarius ibero-insulares 4. Fungi non Delineati, Pars LXXI–LXXII. Edizioni Candusso, Alassio
Ballarà J, Mahiques R (2010) Phlegmacium raros o nuevos asociados a Dryas octopetala L. J JEC 12:57–68
Ballarà J, Mahiques R, Garrido-Benavent I (2016) Estudi de Cortinariaceae del Parc natural del Cadí-Moixeró (III). Moixeró 8:20–50
Ballarà J, Mahiques R, Garrido-Benavent I (2017) Estudi de Cortinariaceae del Parc natural del Cadí-Moixeró (IV). Moixeró 9:20–49
Bellanger JM (2015) Les cortinaires calochroïdes: une mise au point taxinomique. Documents Mycologiques 36:3–34
Bidaud A, Moënne-Loccoz P, Reumaux P (2001) Atlas des Cortinaires. Pars XI (2), sous-genre Phlegmacium (Fr.) Trog, section Calochroi Moser & Horak. Éditions Fédération Mycologique Dauphiné-Savoie, Annecy, France
Bjorbækmo M, Carlsen T, Brysting A et al (2010) High diversity of root associated fungi in both alpine and arctic Dryas octopetala. BMC Plant Biol 10:244. https://doi.org/10.1186/1471-2229-10-244
Bödeker ITM, Clemmensen KE, de Boer W, Martin F, Olson Å, Lindahl BD (2014) Ectomycorrhizal Cortinarius species participate in enzymatic oxidation of humus in northern forest ecosystems. New Phytol 203:245–256. https://doi.org/10.1111/nph.12791
Cadiñanos Aguirre JA (2004) Cortinarius subgen. Phlegmacium raros o interesantes. Fungi non delineati, Pars XXIX. Edizioni Candusso. Alassio, Italy
Cailleux A (1981) Code des couleurs des sols. Boubée, France
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. https://doi.org/10.1093/oxfordjournals.molbev.a026334
Clericuzio M, Dovana F, Bellanger J-M, Brandrud TE, Dima B, Frøslev TG, Boccardo F, Jeppesen TS, Vizzini A (2017) Cortinarius parasuaveolens (= C. pseudogracilior): new data and a synonymy of a very poorly known species of section Calochroi. Sydowia 69. https://doi.org/10.12905/0380.sydowia69-2017-0213
Delaporte A, Eyssartier G, Moenne-Loccoz P (2002) Cortinarius rapaceotomentosus sp. nov., un nouveau cortinaire proche de Cortinarius europaeus. Bulletin de la Société Mycologique de France 118:11–18
Doyle JJ, Doyle JL (1987) A rapid procedure for DNA purification from small quantities of fresh leaf tissue. Phytochemical Bulletin 19:11–15
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. https://doi.org/10.1093/molbev/mss075
Fernández CW, Nguyen NH, Stefanski A, Han Y, Hobbie SE, Montgomery RA, Reich PB, Kennedy PG (2017) Ectomycorrhizal fungal response to warming is linked to poor host performance at the boreal-temperate ecotone. Glob Chang Biol 23:1598–1609. https://doi.org/10.1111/gcb.13510
Flores-Rentería D, Curiel Yuste J, Rincón A, Brearley FQ, García-Gil JC, Valladares F (2015) Habitat fragmentation can modulate drought effects on the plant–soil–microbial system in Mediterranean Holm oak (Quercus ilex) forests. Microb Ecol 69:798–812. https://doi.org/10.1007/s00248-015-0584-9
Frøslev TG, Brandrud TE, Dima B (2017) Cortinarius stjernegaardii and C. kristinae (Basidiomycota, Agaricales), two new European species with a mainly northern distribution. Mycol Prog 16:145–153. https://doi.org/10.1007/s11557-016-1264-y
Frøslev TG, Jeppesen TS, Læssøe T (2006) Seven new calochroid and fulvoid species of Cortinarius. Mycol Res 110:1046–1058. https://doi.org/10.1016/j.mycres.2006.05.012
Frøslev TG, Jeppesen TS, Læssøe T, Kjøller R (2007) Molecular phylogenetics and delimitation of species in Cortinarius section Calochroi (Basidiomycota, Agaricales) in Europe. Mol Phylogenet Evol 44:217–227. https://doi.org/10.1016/j.ympev.2006.11.013
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes—application for the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. https://doi.org/10.1111/j.1365-294x.1993.tb00005.x
Garnica S, Schön ME, Abarenkov K et al (2016) Determining threshold values for barcoding fungi: lessons from Cortinarius (Basidiomycota), a highly diverse and widespread ectomycorrhizal genus. FEMS Microbiol Ecol 92:fiw045. https://doi.org/10.1093/femsec/fiw045
Garnica S, Spahn P, Oertel B, Ammirati J, Oberwinkler F (2011) Tracking the evolutionary history of Cortinarius species in section Calochroi, with transoceanic disjunct distributions. BMC Evol Biol 11:213. https://doi.org/10.1186/1471-2148-11-213
Garnica S, Weiß M, Oertel B, Ammirati J, Oberwinkler F (2009) Phylogenetic relationships in Cortinarius, section Calochroi, inferred from nuclear DNA sequences. BMC Evol Biol 9(1). https://doi.org/10.1186/1471-2148-9-1
Garrido-Benavent I, Ballarà J, Mahiques R (2015) New insights into subg. Phlegmacium sect. Calochroi: adding morphological and molecular data from Mediterranean representatives, with special regard to Cortinarius prasinus, C. natalis and C. murellensis species complexes. Journal des J.E.C. 17:8–78
Garrido-Benavent I, Ballarà J, Mahiques R (2016) Dos nuevas especies de Cortinarius, subgénero Telamonia, del Parque Natural del Cadí-Moixeró (noreste de la Península Ibérica), basadas en caracteres morfológicos y moleculares. Journal des J.E.C. 18:66–85
Geml J, Timling I, Robinson CH, Lennon N, Nusbaum HC, Brochmann C, Noordeloos ME, Taylor DL (2011) An arctic community of symbiotic fungi assembled by long-distance dispersers: phylogenetic diversity of ectomycorrhizal basidiomycetes in Svalbard based on soil and sporocarp DNA. J Biogeogr 39:74–88. https://doi.org/10.1111/j.1365-2699.2011.02588.x
Han LH, Feng B, Wu G et al (2017) African origin and global distribution patterns: evidence inferred from phylogenetic and biogeographical analyses of ectomycorrhizal fungal genus Strobilomyces. J Biogeogr. https://doi.org/10.1111/jbi.13094
Harrower E, Ammirati JF, Cappuccino AA et al (2011) Cortinarius species diversity in British Columbia and molecular phylogenetic comparison with European specimen sequences. Botany 89:799–810. https://doi.org/10.1139/b11-065
Harrower E, Bougher NL, Henkel TW, Horak E, Matheny PB (2015) Long-distance dispersal and speciation of Australasian and American species of Cortinarius sect. Cortinarius. Mycologia 107:697–709. https://doi.org/10.3852/14-182
Hebert PD, Cywinska A, Ball SL (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci 270:313–321. https://doi.org/10.1098/rspb.2002.2218
Hewitt GM (2011) Mediterranean peninsulas: the evolution of hotspots. In: Zachos F, Habel J (eds) Biodiversity hotspots. Springer, Berlin, Heidelberg, pp 123–147. https://doi.org/10.1007/978-3-642-20992-5_7
Horton BM, Morag G, Davidson NJ, Ratkowsky DA, Close DC, Wardlaw TJ, Mohammed C (2017) An assessment of ectomycorrhizal fungal communities in Tasmanian temperate high-altitude Eucalyptus delegatensis forest reveals a dominance of the Cortinariaceae. Mycorrhiza 27:67–74. https://doi.org/10.1007/s00572-016-0725-0
Katoh K, Standley DM (2013) MAFFT: iterative refinement and additional methods. Methods Mol Biol 1079:131–146. https://doi.org/10.1007/978-1-62703-646-7_8
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. https://doi.org/10.1093/nar/gkf43
Kyaschenko J, Clemmensen KE, Hagenbo A, Karltun E, Lindahl BD (2017) Shift in fungal communities and associated enzyme activities along an age gradient of managed Pinus sylvestris stands. ISME J 11:863–874. https://doi.org/10.1038/ismej.2016.184
Lanfear R, Calcott B, Ho SYW, Guindon S (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29:1695–1701. https://doi.org/10.1093/molbev/mss020
Leavitt SD, Divakar PK, Ohmura Y, Wang L, Esslinger TL, Lumbsch HT (2015) Who’s getting around? Assessing species diversity and phylogeography in the widely distributed lichen-forming fungal genus Montanelia (Parmeliaceae, Ascomycota). Mol Phylogenet Evol 90:85–96. https://doi.org/10.1016/j.ympev.2015.04.029
Leavitt SD, Esslinger TL, Lumbsch HT (2012a) Neogene-dominated diversification in neotropical montane lichens: dating divergence events in the lichen-forming fungal genus Oropogon (Parmeliaceae). Am J Bot 99:1764–1777. https://doi.org/10.3732/ajb.1200146
Leavitt SD, Esslinger TL, Divakar PK, Lumbsch H (2012b) Miocene and Pliocene dominated diversification of the lichen-forming fungal genus Melanohalea (Parmeliaceae, Ascomycota) and Pleistocene population expansions. BMC Evol Biol 12:176. https://doi.org/10.1186/1471-2148-12-176
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Liimatainen K, Carteret X, Dima B, Kytövuori I, Bidaud A, Reumaux P, Niskanen T, Ammirati JF, Bellanger J-M (2017) Cortinarius section Bicolores and section Saturnini (Basidiomycota, Agaricales), a morphogenetic overview of European and North American species. Persoonia 39:175–200. https://doi.org/10.3767/persoonia.2017.39.08
Liimatainen K, Niskanen T, Dima B, Kytövuori I, Ammirati JF, Frøslev TG (2014) The largest type study of Agaricales species to date: bringing identification and nomenclature of Phlegmacium (Cortinarius) into the DNA era. Persoonia 33:98–140. https://doi.org/10.3767/003158514x684681
Liu K, Raghavan S, Nelesen S, Linder CR, Warnow T (2009) Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. Science 324:1561–1564. https://doi.org/10.1126/science.1171243
Liu K, Warnow TJ, Holder MT, Nelesen SM, Yu J, Stamatakis AP, Linder CR (2011) SATe-II: very fast and accurate simultaneous estimation of multiple sequence alignments and phylogenetic trees. Syst Biol 61:90–106. https://doi.org/10.1093/sysbio/syr095
Looney BP, Ryberg M, Hampe F, Sánchez-García M, Matheny PB (2016) Into and out of the tropics: global diversification patterns in a hyperdiverse clade of ectomycorrhizal fungi. Mol Ecol 25:630–647. https://doi.org/10.1111/mec.13506
Mahiques R (2011) Flora corològica i bibliogràfica dels Cortinarius Ibero-Insulars. Butlletí de la Societat Micològica Valenciana 16:121–162
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop (GCE), pp 1–8 New Orleans
Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GA, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858. https://doi.org/10.1038/35002501
Niskanen T, Liimatainen K, Mahiques R, Ballarà J, Kytövuori I (2011) Cortinarius badiolaevis, a new conifer-associated, darkening species in the subgenus Telamonia (Basidiomycota, Agaricales). Mycol Prog 10:101–105. https://doi.org/10.1007/s11557-010-0680-7
Nouhra E, Urcelay C, Longo S, Tedersoo L (2013) Ectomycorrhizal fungal communities associated to Nothofagus species in Northern Patagonia. Mycorrhiza 23:487–496. https://doi.org/10.1007/s00572-013-0490-2
Oertel B, Schmidt-Stohn G, Saar G (2009) Die Laugenreaktion am Stielbasisfilz bei Fruchtkörpern von Cortinarius, Subgen. Phlegmacium—Eine Bestandsaufnahme 23 Jahre nach Entdeckung dieser neuartigen Reaktion. Journal des J.E.C. 11:20–31
Ortega A, Suárez-Santiago VN, Reyes JD (2008) Morphological and ITS identification of Cortinarius species (section Calochroi) collected in Mediterranean Quercus woodlands. Fungal Divers 29:73–88
Pachauri RK, Allen MR, Barros VR et al (2014) Climate change 2014: synthesis report. Contribution of working groups I, II and III to the fifth assessment report of the Intergovernmental Panel on Climate Change. IPCC
Pérez-Izquierdo L, Zabal-Aguirre M, Flores-Rentería D, González-Martínez S, Buée M, Rincón A (2017) Functional outcomes of fungal community shifts driven by tree genotype and spatial–temporal factors in Mediterranean pine forests. Environ Microbiol 19:1639–1652. https://doi.org/10.1111/1462-2920.13690
Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, automatic barcode gap discovery for primary species delimitation. Mol Ecol 21:1864–1877. https://doi.org/10.1111/j.1365-294x.2011.05239.x
Rodríguez A, Curiel Yuste J, Rey A, Durán J, García-Camacho R, Gallardo A, Valladares F (2016) Holm oak decline triggers changes in plant succession and microbial communities, with implications for ecosystem C and N cycling. Plant Soil 414:247–263. https://doi.org/10.1007/s11104-016-3118-4
Rodríguez-Sánchez F, Hampe A, Jordano P, Arroyo J (2010) Past tree range dynamics in the Iberian Peninsula inferred through phylogeography and palaeodistribution modelling: a review. Rev Palaeobot Palynol 162:507–521. https://doi.org/10.1016/j.revpalbo.2010.03.008
Ronquist F, Teslenko M, van der Mark P et al (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Ryberg M, Matheny PB (2011) Asynchronous origins of ectomycorrhizal clades of Agaricales. P R Soc Lond B Biol 279:2003–2011. https://doi.org/10.1098/rspb.2011.2428
Ryberg M, Larsson E, Molau U (2009) Ectomycorrhizal diversity on Dryas octopetala and Salix reticulata in an alpine cliff ecosystem. Arct Antarct Alp Res 41:506–514. https://doi.org/10.1657/1938-4246-41.4.506
Salom JC, Siquier JL, Mahiques R (2015) El gènere Cortinarius a les Illes Balears (Espanya) I. Revista Catalana de Micologia 36:11–27
Sánchez-Ramírez S, Tulloss RE, Amalfi M, Moncalvo J-M (2015) Palaeotropical origins, boreotropical distribution and increased rates of diversification in a clade of edible ectomycorrhizal mushrooms (Amanita section Caesareae). J Biogeogr 42:351–363. https://doi.org/10.1111/jbi.12402
Schenk JJ (2016) Consequences of secondary calibrations on divergence time estimates. PLoS One 11:e0148228. https://doi.org/10.1371/journal.pone.0148228
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. https://doi.org/10.1093/bioinformatics/btl446
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57:758–771. https://doi.org/10.1080/10635150802429642
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. https://doi.org/10.1093/molbev/msr121
Teasdale SE, Beulke AK, Guy PL, Orlovich DA (2013) Environmental barcoding of the ectomycorrhizal fungal genus Cortinarius. Fungal Divers 58:299–310. https://doi.org/10.1007/s13225-012-0218-1
Truong C, Mujic AB, Healy R, Kuhar F, Furci G, Torres D, Niskanen T et al (2017) How to know the fungi: combining field inventories and DNA-barcoding to document fungal diversity. New Phytol 214:913–919. https://doi.org/10.1111/nph.14509
White TJ, Bruns TD, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols. Academic Press, San Diego, pp 315–322
Zachos J, Pagani M, Sloan L, Thomas E, Billups K (2001) Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292:686–693. https://doi.org/10.1126/science.1059412
Acknowledgements
We are very grateful to J.A. Cadiñanos Aguirre, J.D. Reyes and E. Suárez for sharing some Calochroi collections. We also thank S. García and P. Alvarado for collaborating in the production of molecular data. Asunción de los Ríos (Dept. of Biogeochemistry and Microbial Ecology, National Museum of Natural Sciences) is also thanked for providing necessary resources for conducting this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Zhu-Liang Yang
Electronic supplementary material
Online Resource 1
GenBank accession numbers of nrITS sequences used in phylogenetic and population genetic analyses (PDF 294 kb)
Online Resource 2
Calochroid species for which polymorphism statistics were calculated (see the “Material and methods” section), and associated plant species and source (in square brackets). Specimens studied in the present work are highlighted in bold. The type locality is indicated when known. Codes correspond with different countries (see www.countrycode.org). “m.f.”, mixed forest (PDF 301 kb)
Online Resource 3
Phylogram depicting the evolutionary relationships of members in Cortinarius section Calochroi obtained with the software SATé v. 2.2.7 and based on nrITS data. Terminals belonging to one phylogenetic species and represented by two or more sequences are collapsed for better visualisation. Species found in the Iberian Peninsula are highlighted, including the new species reported in the present study. Bootstrap support values are given for each node, and branches showing strong support (BP ≥ 70%) are in bold (PDF 439 kb)
Rights and permissions
About this article
Cite this article
Mahiques, R., Ballarà, J., Salom, J.C. et al. Morphogenetic diversity of the ectomycorrhizal genus Cortinarius section Calochroi in the Iberian Peninsula. Mycol Progress 17, 815–831 (2018). https://doi.org/10.1007/s11557-018-1394-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-018-1394-5