Abstract
A new monotypic Beltrania-like genus, Subsessila, with its type species S. turbinata, is described, illustrated and compared with similar genera. The new genus is introduced in the family Beltraniaceae based on phylogenetic analysis and morphological characters. Subsessila can be easily distinguished from other Beltrania-like genera by dark setae arising from radially lobed basal cells, mostly lacking macronematous conidiophores. Conidiogenous cells are ampulliform or doliiform and produce turbinate to clavate conidia with rostrate proximal end and rounded distal end. Evidence for establishment of the new genus is provided based on morphological comparison and DNA sequence data analyses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Xylariales is a large order of perithecial ascomycetes with eight-spored unitunicate asci, usually with a J+ apical ring and ascospores with a prominent germ pore or germ slit and is accommodated in the subclass Xylariomycetidae (Smith et al. 2003; Senanayake et al. 2015; Maharachchikumbura et al. 2016). Asexual morphs of the Xylariales are usually hyphomycetous, with holoblastic conidiogenesis (Maharachchikumbura et al. 2015, 2016). Presently, there are 22 accepted families in the Xylariales, viz. Amphisphaeriaceae, Apiosporaceae, Bartaliniaceae, Beltraniaceae, Cainiaceae, Clypeosphaeriaceae, Coniocessiaceae, Diatrypaceae, Hyponectriaceae, Iodosphaeriaceae, Lopadostomaceae, Melogrammataceae, Microdochiaceae, Myelospermataceae, Pestalotiopsidaceae, Phlogicylindriaceae, Pseudomassariaceae, Requienellaceae, Robillardaceae, Sporocadaceae, Vialaeaceae and Xylariaceae (Maharachchikumbura et al. 2015, 2016).
A fungal tribe named Beltranieae Sacc. was established by Saccardo (1886) to accommodate a single genus Beltrania Penzig. Nannizzi (1934) introduced Beltraniaceae Nann. to accommodate Beltrania and some similar genera, and the tribe Beltranieae was treated as a synonym of this family. Crous et al. (2015b) emended the family Beltraniaceae and accepted Beltrania, Beltraniella Subram., Beltraniopsis Bat. & J.L. Bezerra, Parapleurotheciopsis P.M. Kirk and Pseudobeltrania Henn. They provided DNA sequence data to support the family. Crous et al. (2015b) proposed that three more genera, Beltraniomyces Manohar., D.K. Agarwal & Rao, Porobeltraniella Gusmão and Subramaniomyces Varghese & V.G. Rao, should be accepted in this family. Due to the lack of reliable strains and sequence data, they could not confirm the familial relationships of these genera within the Beltraniaceae. Maharachchikumbura et al. (2016) accepted the genus Subramaniomyces within the family Beltraniaceae based on phylogenetic analysis. Rajeshkumar et al. (2016a) accepted the genera Hemibeltrania and Porobeltraniella and preliminarily confirmed the monophyly of all of the recognised genera based on phylogenetic analysis.
The asexual morphs in Beltraniaceae are hyphomycetous (Seifert et al. 2011; Crous et al. 2015b). Stromata are usually present. Setae are present or absent, branched or unbranched and usually with a radially lobed basal cell. The conidiophores are branched or unbranched, arising from the base of setae or separate, sometimes arising from radially lobed basal cells. The conidiogenous cells are monoblastic or polyblastic, sympodial, integrated or discrete and denticulate. Separating cells are present or absent, oval to subglobose, also with one to several denticles. Conidia are biconic, lageniform to navicular, hyaline to red-brown, generally with a lightly pigmented transverse band at the widest part of the conidium, rounded or 1-denticulate or rostrate at the base and spicate or apiculate or truncate at the apex (Crous et al. 2015b).
During a survey of hyphomycetes in Karst areas of Thailand, a Beltrania-like species was collected. It was shown to belong to a new genus in Beltraniaceae based on morphology and analyses of ITS and LSU sequence data. The natural classification of this new taxon is determined based on phylogenetic analysis and morphology.
The new taxon is morphologically similar to Beltrania and some similar genera, but its conidiophores and conidiogenous cells differ from all previously described genera of this group. We propose a new genus to accommodate the new fungus which is introduced here.
Materials and methods
Collection and isolation of fungi
Dead materials (stem, wood and leaves) from a variety of plants were randomly collected during July to August 2015 from Karst areas at Khao Lom Muak (11°47′3.96″–11.24″N, 99°48′49.13″–49′0.63″E), Prachuap Khiri Khan in Thailand. Samples were taken to the laboratory in zip-lock plastic bags for examination. The specimens were incubated in sterile moist chambers and examined using a Motic SMZ-168 series microscope (Speed Fair Co., China). Fungi were removed with a needle and placed in a drop of distilled water on a slide for morphological study. Photomicrographs of fungal structures were captured using a Nikon Eclipse 80i compound microscope (Nikon Co., Japan) with a Canon 450D digital camera (Canon Co., Japan). All measurements were made by the Tarosoft® Image FrameWork program (Tarosoft, Thailand). Photo-plates were made with Adobe Photoshop CS6 Extended version 13.0.1 (Adobe Systems, USA). Isolation onto potato dextrose agar (PDA) or malt extract agar (MEA) was performed by the single spore isolation method (Chomnunti et al. 2014; Dai et al. 2017). The herbarium material is deposited in the herbarium of Mae Fah Luang University (MFLU), Chiang Rai, Thailand and the herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (HKAS), Kunming, China. Cultures are deposited at the Mae Fah Luang University Culture Collection (MFLUCC), Chiang Rai, Thailand and Kunming Institute of Botany, Chinese Academy of Sciences (KUMCC), Kunming, China. Faces of Fungi and Index Fungorum numbers are registered (Jayasiri et al. 2015; Index Fungorum 2016). Colours and colour codes were determined according to Kornerup and Wanscher (1978).
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from fungal mycelium grown on PDA or MEA at room temperature with the Fungal gDNA Kit (BioMIGA, USA), according to the manufacturer’s instructions. The internal transcribed spacer region of ribosomal DNA (ITS) and large subunit nuclear ribosomal DNA (LSU) genes were amplified via polymerase chain reaction (PCR) using the following primers: ITS5 and ITS4 (White et al. 1990) for ITS and LROR and LR5 (Vilgalys and Hester 1990) for LSU. The PCR products were sequenced with the same primers.
Phylogenetic analyses
Original sequences were checked using BioEdit version 7.0.5.3 (Hall 1999) and most reference sequences originated from previous publications, viz. Crous and Groenewald (2013), Crous et al. (2014, 2015a, b) and Maharachchikumbura et al. (2015). The remaining homologous sequences were obtained by BLAST searches (Altschul et al. 1990) from GenBank. All sequences used in this study are listed in Table 1. Alignments for each locus were done in MAFFT v7.212 (Katoh and Standley 2013) and manually verified in MEGA 6.06 (Tamura et al. 2013). Conserved blocks were selected from the initial alignments with Gblocks 0.91b (Castresana 2000). The interleaved NEXUS files were formatted with PAUP*4.0b10 (Swofford 2002) and manually formatted for Bayesian inference analyses. Bayesian inference (BI), maximum parsimony (MP) and maximum likelihood (ML) were used in this study for phylogenetic analyses. For Bayesian inference analysis, the best model of evolution was determined using MrModeltest v2 (Nylander 2004). Bayesian inference analysis was done with MrBayes v3.2.5 (Ronquist et al. 2012). Maximum parsimony analysis was performed in PAUP*4.0b10 (Swofford 2002). Maximum likelihood analysis was performed in raxmlGUI v1.3.1 (Silvestro and Michalak 2012). Phylogenetic trees were drawn with TreeView 1.6.6 (Page 1996).
Results
Molecular phylogeny
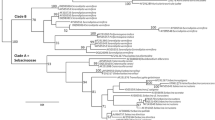
The aligned sequence matrix comprises LSU (848 bp) and ITS (633 bp) sequence data for 40 taxa and one outgroup taxon for a total of 1481 characters, of which 267 were parsimony-informative, 100 were parsimony-uninformative and 1114 characters were constant. The result of maximum likelihood (ML) analysis based on combined LSU and ITS sequence data consisting of five families (Amphisphaeriaceae, Apiosporaceae, Beltraniaceae, Pestalotiopsidaceae and Robillardaceae) within the order Xylariales is shown in Fig. 1.
Phylogenetic tree generated from maximum likelihood (ML) analysis based on combined LSU and ITS sequence data for some selected families within the order Xylariales. Bootstrap support values for maximum likelihood (ML) and maximum parsimony (MP) greater than 50% and Bayesian posterior probabilities greater than 0.8 are indicated above or below the nodes as MLBS/MPBS/PP. The ex-type strains are in bold and the new isolate is in bold and red. The tree is rooted with Anthostomella leucospermi (CBS 110126)
In the present study, we found that the strain of Subsessila turbinata (MFLUCC 15-0831) grouped together with Hemibeltrania sp. (CL12WA) and Hemibeltrania cinnamomi (NFCCI 3695 and NFCCI 3997) with 68% ML bootstrap support, 63% MP bootstrap support and 99% Bayesian posterior probabilities within the family Beltraniaceae (Fig. 1). The genera Hemibeltrania, Porobeltraniella, Pseudobeltrania and Subsessila clustered together with 63% ML bootstrap support, 89% MP bootstrap support and 100% Bayesian posterior probabilities.
Taxonomy
Subsessila C.G. Lin & K.D. Hyde, gen. nov.
Index Fungorum number: IF552504; Facesoffungi number: FoF 02613
Etymology: In reference to the conidiophores which are mostly absent or reduced to conidiogenous cells.
Type species: Subsessila turbinata C.G. Lin & K.D. Hyde
Saprobic on plant host. Asexual morph: Colonies on plant substrate effuse, pale brown, hairy, velvety. Mycelium partly superficial and partly immersed. Stroma absent. Setae numerous, erect, arising from radially lobed basal cells, straight or slightly flexuous, unbranched, 2–4-septate, thick-walled, verrucose, pale to dark brown, swollen at the base, tapering towards the apex. Conidiophores mostly absent or reduced to conidiogenous cells; when present, arising from the basal cells of setae, simple, aseptate, subcylindrical, pale- to mid-brown, smooth. Conidiogenous cells polyblastic, discrete, determinate, ampulliform, doliiform, hyaline. Separating cells absent. Conidia aggregated, acrogenous, simple, dry, straight, smooth, thin-walled, turbinate to clavate, rostrate at proximal end, rounded at distal end, hyaline. Sexual morph: Undetermined.
Subsessila turbinata C.G. Lin & K.D. Hyde, sp. nov. (Fig. 2)
Subsessila turbinata (holotype MFLU 15-3271). a Leaf material. b, c Conidiophores on the host surface. d Conidiophores, seta and conidia. e Short conidiophores with conidiogenous cell. f, g Conidiogenous cell and conidia. h–j Conidia. k Germinating conidium. l, m Colonies on MEA culture, l from above, m from below. Scale bars: b = 500 μm, c = 200 μm, d = 50 μm, e–k = 10 μm
Index Fungorum number: IF552503; Facesoffungi number: FoF 02614
Etymology: In reference to the turbinate conidia.
Holotype: MFLU 15-3271
Saprobic on plant host. Asexual morph: Colonies on plant substrate effuse, pale brown (6C2), hairy, velvety. Mycelium partly superficial and partly immersed. Stroma absent. Setae numerous, erect, arising from radially lobed basal cells, straight or slightly flexuous, unbranched, 2–4-septate, thick-walled, verrucose, medium to dark brown (6C4 to 6F8), 50–280 μm long, swollen at the base and 4.5–13 μm wide, 2–7 μm wide just above the swollen base, tapering to a pointed apex. Conidiophores mostly absent and reduced to conidiogenous cells; if present, arising from basal cells of setae, simple, aseptate, subcylindrical, pale- to mid-brown (6C2 to 6C4), up to 6.5 μm long, 3–7.5 μm wide. Conidiogenous cells polyblastic, discrete, determinate, ampulliform, doliiform, hyaline, smooth, 4.5–6.5 μm (\( \overline{x} \) = 5.5 μm, n = 30) long, 4–6 μm (\( \overline{x} \) = 5.1 μm, n = 30) wide in the broadest part. Separating cells absent. Conidia aggregated, acrogenous, simple, dry, straight, smooth, thin-walled, turbinate to clavate, rostrate to pointed at proximal end, broadly rounded at distal end, hyaline, 14.5–20.5 μm (\( \overline{x} \) = 17.70 μm, n = 50) long, 4.5–6.5 μm (\( \overline{x} \) = 5.50 μm, n = 50) wide in the broadest part. Sexual morph: Undetermined.
Culture characteristics: Conidia germinating on MEA within 12 h. Colonies on MEA effuse, greyish white (−B1) from above, greyish yellow (2B5) from below, reaching a diam. of 2–3 cm in 20 days at 25 °C.
Material examined: THAILAND, Prachuap Khiri Khan, Khao Lom Muak, 11°47′3.96″–11.24″N, 99°48′49.13″–49′0.63″E, on unidentified decaying leaf, 29 July 2015, Chuan-Gen Lin, KLM 8-1 (MFLU 15-3271, holotype; HKAS 96229, isotype), ex-type living culture MFLUCC 15-0831, KUMCC 16-0126.
Discussion
In the tree generated from the maximum likelihood (ML) analysis based on combined ITS and LSU sequence data for the order Xylariales, Subsessila turbinata (MFLUCC 15-0831) grouped together with Hemibeltrania sp. (CL12WA) and Hemibeltrania cinnamomi (NFCCI 3695 and NFCCI 3997) with 100% ML bootstrap support, 99% MP bootstrap support and 100% Bayesian posterior probabilities within the family Beltraniaceae (Fig. 1). Subsessila differs from Hemibeltrania and Pseudobeltrania by the absence of distinct conidiophores, which, whenever present, are very short and arise from the basal cells of setae; terminal, ampulliform, doliiform, hyaline conidiogenous cells and turbinate to clavate, subhyaline, smooth conidia which are rostrate to pointed at the proximal end and broadly rounded at the rostrate distal end. In addition, Subsessila has numerous, unbranched setae arising from radially lobed basal cells. Stroma and separating cells are not observed in these three genera.
Morphologically, Subsessila is similar to several genera within the family Beltraniaceae, viz. Beltrania, Beltraniella, Beltraniopsis and Porobeltraniella, in having dark setae and conidiophores arising from radially lobed basal cells (Fig. 2). However, Beltrania, Beltraniella, Beltraniopsis and Porobeltraniella have distinct, swollen separating cells, and conidia of these four genera are turbinate or biconic, with a hyaline transverse band or several equatorial hyaline pores, whereas separating cells are not present in Subsessila and the conidia are turbinate to clavate, with a rostrate to pointed proximal end and rounded distal end. The most distinguishable characters that separate Subsessila from Beltrania and other similar genera are the absence of distinct conidiophores and conidia without a hyaline transverse band in the new genus.
With the above combination of morphological features and phylogenetic analysis, we place the new genus Subsessila within the family Beltraniaceae.
Presently, 14 genera, including our new genus, have some similar characters that are present in the “Beltrania complex”, viz. Beltrania Penzig, Beltraniella Subram., Beltraniomyces Manohar., D.K. Agarwal & Rao, Beltraniopsis Bat. & J.L. Bezerra, Beltramono Dubey, Pandey & Manohar., Hemibeltrania Piroz., Kiliophora Kuthub. & Nawawi, Maxibeltrania Rambelli, Parabeltrania Rambelli, Porobeltraniella Gusmão, Pseudobeltrania Henn., Rhombostilbella Zimm., Scolecobeltrania Iturr., R.F. Castañeda & R. Fernández and Subsessila. A synopsis of Beltrania and similar genera is provided in Table 2. These genera have unbranched or branched conidiophores and/or setae arising from radially lobed basal cells, with or without swollen separating cells, and biconic conidia with or without a hyaline equatorial or sub- or supraequatorial band. Presently, six of them, viz. Beltrania, Beltraniella, Beltraniopsis, Hemibeltrania, Porobeltraniella and Pseudobeltrania, are accepted in the family Beltraniaceae.
Kendrick (1980) proposed that the genera in the Beltrania complex must possess any three of the five following features: (1) dark setae; (2) setae or conidiophores with radially lobed bases; (3) swollen separating cells; (4) biconic conidia; (5) conidia with a hyaline equatorial band. There are nine genera showing Beltrania-like morphological features (Table 3), viz. Beltrania, Beltraniella, Beltraniopsis, Beltramono, Maxibeltrania, Parabeltrania, Porobeltraniella, Pseudobeltrania and Scolecobeltrania. Within this, Beltraniomyces, Hemibeltrania, Kiliophora, Rhombostilbella and Subsessila show some differences in that Beltraniomyces and Subsessila have only two of those characteristics, whereas Hemibeltrania, Kiliophora and Rhombostilbella have only one of those characteristics.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17(4):540–552
Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MK, Alias AS, Xu J, Liu X, Stadler M, Hyde KD (2014) The sooty moulds. Fungal Divers 66:1–36. doi:10.1007/s13225-014-0278-5
Crous PW, Groenewald JZ (2013) A phylogenetic re-evaluation of Arthrinium. IMA Fungus 4(1):133–154. doi:10.5598/imafungus.2013.04.01.13
Crous PW, Braun U, Schubert K, Groenewald JZ (2007) Delimiting Cladosporium from morphologically similar genera. Stud Mycol 58:33–56. doi:10.3114/sim.2007.58.02
Crous PW, Shivas RG, Quaedvlieg W, van der Bank M, Zhang Y, Summerell BA, Guarro J, Wingfield MJ, Wood AR, Alfenas AC, Braun U, Cano-Lira JF, García D, Marin-Felix Y, Alvarado P, Andrade JP, Armengol J, Assefa A, den Breeÿen A, Camele I, Cheewangkoon R, De Souza JT, Duong TA, Esteve-Raventós F, Fournier J, Frisullo S, García-Jiménez J, Gardiennet A, Gené J, Hernández-Restrepo M, Hirooka Y, Hospenthal DR, King A, Lechat C, Lombard L, Mang SM, Marbach PAS, Marincowitz S, Marin-Felix Y, Montaño-Mata NJ, Moreno G, Perez CA, Sierra AMP, Robertson JL, Roux J, Rubio E, Schumacher RK, Stchigel AM, Sutton DA, Tan YP, Thompson EH, van der Linde E, Walker AK, Walker DM, Wickes BL, Wong PTW, Groenewald JZ (2014) Fungal Planet description sheets: 214–280. Persoonia 32:184–306. doi:10.3767/003158514x682395
Crous PW, Carris LM, Giraldo A, Groenewald JZ, Hawksworth DL, Hernández-Restrepo M, Jaklitsch WM, Lebrun MH, Schumacher RK, Stielow JB, van der Linde EJ, Vilcāne J, Voglmayr H, Wood AR (2015a) The Genera of Fungi—fixing the application of the type species of generic names—G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia. IMA Fungus 6(1):163–198. doi:10.5598/imafungus.2015.06.01.11
Crous PW, Wingfield MJ, Le Roux JJ, Richardson DM, Strasberg D, Shivas RG, Alvarado P, Edwards J, Moreno G, Sharma R, Sonawane MS, Tan YP, Altés A, Barasubiye T, Barnes CW, Blanchette RA, Boertmann D, Bogo A, Carlavilla JR, Cheewangkoon R, Daniel R, de Beer ZW, de Jesus Y-MM, Duong TA, Fernández-Vicente J, Geering ADW, Guest DI, Held BW, Heykoop M, Hubka V, Ismail AM, Kajale SC, Khemmuk W, Kolařik M, Kurli R, Lebeuf R, Lévesque CA, Lombard L, Magista D, Manjón JL, Marincowitz S, Mohedano JM, Novákova A, Oberlies NH, Otto EC, Paguigan ND, Pascoe IG, Pérez-Butrón JL, Perrone G, Rahi P, Raja HA, Rintoul T, Sanhueza RMV, Scarlett K, Shouche YS, Shuttleworth LA, Taylor PWJ, Thorn RG, Vawdrey LL, Solano-Vidal R, Voitk A, Wong PTW, Wood AR, Zamora JC, Groenewald JZ (2015b) Fungal Planet description sheets: 371–399. Persoonia 35:264–327. doi:10.3767/003158515x690269
Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E (2017) Bambusicolous fungi. Fungal Divers 82:1–105. doi:10.1007/s13225-016-0367-8
Dubey R, Pandey AK, Manoharachary C (2011) Beltramono—a hitherto undescribed hyphomycetes genus. J Mycopathol Res 49(2):321–324
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Index Fungorum (2016) Home page at: http://www.indexfungorum.org/names/Names.asp.
Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat J, Buyck B, Cai L, Dai Y-C, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R, Jones EBG, Bahkali AH, Karunarathna SC, Liu J-K, Luangsa-ard JJ, Lumbsch HT, Maharachchikumbura SSN, McKenzie EHC, Moncalvo J-M, Ghobad-Nejhad M, Nilsson H, Pang K-L, Pereira OL, Phillips AJL, Raspé O, Rollins AW, Romero AI, Etayo J, Selçuk F, Stephenson SL, Suetrong S, Taylor JE, Tsui CKM, Vizzini A, Abdel-Wahab MA, Wen T-C, Boonmee S, Dai DQ, Daranagama DA, Dissanayake AJ, Ekanayaka AH, Fryar SC, Hongsanan S, Jayawardena RS, Li W-J, Perera RH, Phookamsak R, de Silva NI, Thambugala KM, Tian Q, Wijayawardene NN, Zhao R-L, Zhao Q, Kang J-C, Promputtha I (2015) The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers 74:3–18. doi:10.1007/s13225-015-0351-8
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780
Kendrick B (1980) The generic concept in hyphomycetes—a reappraisal. Mycotaxon 11(1):339–364
Kornerup A, Wanscher JH (1978) Methuen handbook of colour. Methuen, London, England
Maharachchikumbura SSN, Hyde KD, Groenewald JZ, Xu J, Crous PW (2014) Pestalotiopsis revisited. Stud Mycol 79:121–186. doi:10.1016/j.simyco.2014.09.005
Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Huang S-K, Abdel-Wahab MA, Daranagama DA, Dayarathne M, D’Souza MJ, Goonasekara ID, Hongsanan S, Jayawardena RS, Kirk PM, Konta S, Liu J-K, Liu Z-Y, Norphanphoun C, Pang K-L, Perera RH, Senanayake IC, Shang QJ, Shenoy BD, Xiao YP, Bahkali AH, Kang JC, Somrothipol S, Suetrong S, Wen TC, Xu JC (2015) Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers 72:199–301. doi:10.1007/s13225-015-0331-z
Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Bhat JD, Dayarathne MC, Huang S-K, Norphanphoun C, Senanayake IC, Perera RH, Shang Q-J, Xiao Y, D’souza MJ, Hongsanan S, Jayawardena RS, Daranagama DA, Konta S, Goonasekara ID, Zhuang W-Y, Jeewon R, Phillips AJL, Abdel-Wahab MA, Al-Sadi AM, Bahkali AH, Boonmee S, Boonyuen N, Cheewangkoon R, Dissanayake AJ, Kang J, Li Q-R, Liu JK, Liu XZ, Liu Z-Y, Luangsa-ard JJ, Pang K-L, Phookamsak R, Promputtha I, Suetrong S, Stadler M, Wen T, Wijayawardene NN (2016) Families of Sordariomycetes. Fungal Divers 79:1–317. doi:10.1007/s13225-016-0369-6
Manoharachary C, Agarwal DK, Rao NK (2003) Beltraniomyces, a new genus of Dematiaceous Hyphomycetes from India. Indian Phytopathol 56(4):418–421
Marincowitz S, Crous PW, Groenewald JZ, Wingfield MJ (2008) Microfungi occurring on Proteaceae in the fynbos. CBS Biodiversity Series, vol 7. CBS Fungal Biodiversity Centre, The Netherlands
Nannizzi A (1934) Repertorio sistematico dei miceti dell’uomo e degli animali. Trattato di micopatologia umana, vol 4. Poligrafica Meini, Siena, Italy
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12(4):357–358
Rajeshkumar KC, Crous PW, Groenewald JZ, Seifert KA (2016a) Resolving the phylogenetic placement of Porobeltraniella and allied genera in the Beltraniaceae. Mycol Prog 15(10–11):1119–1136. doi:10.1007/s11557-016-1234-4
Rajeshkumar KC, Marathe SD, Madhusudhanan K, Castaneda-Ruiz RF (2016b) Taxonomic re-evaluation and phylogenetic position of Hemibeltrania cinnamomi within Xylariales. Mycotaxon 131(1):87–94. doi:10.5248/131.87
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542
Saccardo PA (1886) Syllogue fungorum omnium hucusque cognitorum IV. Padua
Seifert KA, Morgan-Jones G, Gams W, Kendrick B (2011) The genera of hyphomycetes. CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands
Senanayake IC, Maharachchikumbura SSN, Hyde KD, Bhat JD, Jones EBG, McKenzie EHC, Dai DQ, Daranagama DA, Dayarathne MC, Goonasekara ID, Konta S, Li WJ, Shang QJ, Stadler M, Wijayawardene NN, Xiao YP, Norphanphoun C, Li Q, Liu XZ, Bahkali AH, Kang JC, Wang Y, Wen TC, Wendt L, Xu JC, Camporesi E (2015) Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers 73:73–144. doi:10.1007/s13225-015-0340-y
Shirouzu T, Hirose D, Tokumasu S, To-Anun C, Maekawa N (2010) Host affinity and phylogenetic position of a new anamorphic fungus Beltraniella botryospora from living and fallen leaves of evergreen oaks. Fungal Divers 43:85–92. doi:10.1007/s13225-010-0037-1
Silvestro D, Michalak I (2012) raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12(4):335–337
Smith GJD, Liew ECY, Hyde KD (2003) The Xylariales: a monophyletic order containing 7 families. Fungal Divers 13:185–218
Swofford DL (2002) PAUP*: Phylogenetic Analysis Using Parsimony (and other methods). Version 4.0 b10. Sinauer Associates, Sunderland
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30(12):2725–2729. doi:10.1093/molbev/mst197
Tanaka K, Endo M, Hirayama K, Okane I, Hosoya T, Sato T (2011) Phylogeny of Discosia and Seimatosporium, and introduction of Adisciso and Immersidiscosia genera nova. Persoonia 26:85–98. doi:10.3767/003158511x576666
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172(8):4238–4246
White TJ, Bruns TD, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, California, pp 315–322
Acknowledgements
We would like to thank Shaun Pennycook (Landcare Research Manaaki Whenua, New Zealand) for advising on the fungal names and R.J. McGovern (Department of Entomology and Plant Pathology, Chiang Mai University, Thailand) for comments on the manuscript. The research is supported by the Key Laboratory of Yunnan Province Universities of the Diversity and Ecological Adaptive Evolution for Animals and Plants on Yun Gui Plateau and the National Natural Science Foundation of China (NSFC 31260087, NSFC 31460561). Gareth Jones is thanked for the award of Visiting Professor at Chiang Mai University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Marc Stadler and Kevin D. Hyde
This article is part of the “Special Issue in honour of the 70th birthday of Dr. Eric McKenzie.”
Rights and permissions
About this article
Cite this article
Lin, CG., Dai, DQ., Bhat, D.J. et al. Subsessila turbinata gen. et. sp. nov. (Beltraniaceae), a Beltrania-like fungus from Thailand. Mycol Progress 16, 393–401 (2017). https://doi.org/10.1007/s11557-017-1279-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-017-1279-z