Abstract
Anomoloma is a cosmopolitan white rot fungal genus, and it currently contains four species worldwide. In the present study, the species diversity of Anomoloma in China is investigated based on morphological characters, and relationships among the species are assessed with phylogenetic analysis based on the internal transcribed spacer (ITS) regions and the large subunit nuclear ribosomal RNA gene (nLSU) sequences. Two new species, A. luteoalba and A. submyceliosum, are described and illustrated as well as compared with related taxa from other regions of Asia, North America and Europe. Phylogenetic analysis showed that A. luteoalba and A. submyceliosum are clearly distinct species within Anomoloma. A key to all species in this genus is provided.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anomoloma Niemelä & K.H. Larss. was separated from Anomoporia Pouzar by Niemelä et al. (2007) on the basis of decay type and nuclear rDNA sequences data. It is typified by A. albolutescens (Romell) Niemelä & K.H. Larss. and currently contains four species: A. albolutescens, A. flavissimum (Niemelä) Niemelä & K.H. Larss., A. myceliosum (Niemelä) Niemelä & K.H. Larss. and A. rhizosum Y.C. Dai & Niemelä. The genus is characterized by resupinate, strongly rhizomorphic basidiocarps with white, cream or yellow surfaces; a monomitic hyphal system with clamped generative hyphae; elliptical and slightly thick-walled basidiospores; the species in this genus cause a white rot on woody debris of angiosperm and gymnosperm trees (Niemelä et al. 2007). Recent phylogenetic analysis indicated that Anomoloma belongs to the Amylocorticiales (Binder et al. 2010).
During the investigations on the diversity of wood-rotting fungi in subtropical areas of southern China, two new species of Anomoloma were identified based on morphological characters and phylogenetic analysis. Illustrated descriptions of the two new species and an identification key to worldwide species of Anomoloma and Anomoporia are provided.
Materials and methods
Morphological studies
The examined specimens were deposited in the herbarium of the Institute of Microbiology, Beijing Forestry University (BJFC). Macro-morphological descriptions were based on field notes. Special color terms followed Petersen (1996). Micro-morphological data were obtained from dried specimens, and observed under a light microscope following methods in Li et al. (2014). Sections were studied at a magnification of up to × 1000 using a Nikon E 80i microscope and phase contrast illumination. Drawings were made with the aid of a drawing tube. Microscopic features, measurements and drawings were made from slide preparations stained with cotton blue and Melzer’s reagent. Spores were measured from sections cut from the tubes. To represent variation in the size of spores, 5 % of measurements were excluded from each end of the range, and are given in parentheses. The following abbreviations are used: IKI = Melzer’s reagent, IKI– = neither amyloid nor dextrinoid, KOH = 5 % potassium hydroxide, CB = cotton blue, CB– = acyanophilous, L = mean spore length (arithmetic average of all spores), W = mean spore width (arithmetic average of all spores), Q = variation in the L/W ratios between the specimens studied, n (a/b) = number of spores (a) measured from given number (b) of specimens.
Molecular phylogeny
A CTAB (hexadecyltrimethylammonium bromide) rapid plant genome extraction kit (DN14, Aidlab Biotechnologies, Beijing) was used to extract total genomic DNA from dried specimens and to perform the polymerase chain reaction (PCR), according to the manufacturer’s instructions with some modifications (Chen et al. 2015). The ITS region was amplified with primer pair ITS5 (GGA AGT AAA AGT CGT AAC AAG G) and ITS4 (TCC TCC GCT TAT TGA TAT GC; White et al. 1990). For the nuclear LSU region, a new primer pair, LR0R-1 (GAC CGT GTA TAA GTT CTC CTG) and LR7-1 (GCT TCT TCA CTG ACC TCC), was designed based on sequences obtained from the National Center for Biotechnology Information (NCBI, GU187559) using Primer-Premier 5 (Premier Biosoft International, Palo Alto, CA, USA). The PCR procedure for ITS was as follows: initial denaturation at 95 °C for 3 min, followed by 35 cycles at 94 °C for 40 s, 54 °C for 45 s and 72 °C for 1 min, and a final extension at 72 °C for 10 min. The PCR procedure for nLSU was as follows: initial denaturation at 94 °C for 1 min, followed by 35 cycles at 94 °C for 30 s, 50 °C for 1 min and 72 °C for 1.5 min, and a final extension at 72 °C for 10 min. The PCR products were purified and sequenced at the Beijing Genomics Institute, China, with the same primers. All newly generated sequences were deposited at GenBank and listed in Table 1.
Besides the sequences generated from this study, other reference taxa for our phylogenetic analysis were selected from GenBank, and the original publications of the phylogenetic analyses contributing the sequences were referenced in Table 1. Sequences were aligned in MAFFT 6 (Katoh and Toh 2008; http://mafft.cbrc.jp/alignment/server/) using the “G-INS-I” strategy and manually adjusted in BioEdit (Hall 1999). Sequence alignments were deposited at TreeBase (submission ID 18429; www.treebase.org). Sequences of Jaapia argillacea Bres. and J. ochroleuca (Bres.) Nannf. & J. Erikss. obtained from GenBank were used as outgroups to root trees following Binder et al. (2010).
Maximum parsimony (MP) analysis was applied to the combined dataset of ITS and nLSU sequences. Tree construction was performed in PAUP* version 4.0b10 (Swofford 2002). All characters were equally weighted and gaps were treated as missing data. Trees were inferred using the heuristic search option with tree bisection and reconnection (TBR) branch swapping and 1000 random sequence additions. Max-trees were set to 5000, branches of zero length were collapsed and all parsimonious trees were saved. Clade robustness was assessed using a bootstrap (BT) analysis with 1000 replicates (Felsenstein 1985). Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for each maximum parsimonious tree (MPT) generated. DNA sequence data was also analyzed using maximum likelihood (ML) analysis; ML analysis was conducted with RAxML-HPC2 on Abe through the Cipres Science Gateway (www.phylo.org) involving 1000 replicates under the GTRGAMMA model, with all model parameters estimated by the program. In addition, 1000 rapid bootstrap replicates were run with the GTRCAT model. Phylogenetic trees were visualized using Treeview (Page 1996).
Bayesian inference (BI) was also applied to the combined dataset. Substitution models suitable for each partition in the dataset were determined using Akaike information criterion (AIC) implemented in MrModeltest2.3 (Posada and Crandall 1998; Nylander 2004). The general time reversible (GTR) model was estimated as the best-fit evolution model for all partitions in the combined dataset. BI was calculated with MrBayes3.1.2 (Ronquist and Huelsenbeck 2003) with a GTR model of DNA substitution and an invgamma distribution rate variation across sites. Four Markov chains were run for two runs from random starting trees for 2 million generations of the combined ITS and nLSU dataset and sampled every 100 generations. The burn-in was set to discard the first 25 % of the trees. A majority rule consensus tree of all remaining trees was calculated. Branches that received bootstrap support for maximum parsimony (MP), maximum likelihood (BS) and Bayesian posterior probabilities (BPP) greater than or equal to 75 % (MP/BS) and 0.95 (BPP) were considered as significantly supported, respectively.
Results
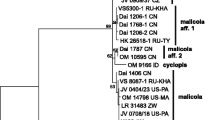
The combined ITS and nLSU dataset included sequences from 53 fungal samples representing 29 taxa. The dataset had an aligned length of 1587 characters, including gaps, of which 954 characters are constant, 127 are variable and parsimony-uninformative, and 506 are parsimony-informative. MP analysis yielded 40 equally parsimonious trees (length = 1860, CI = 0.537, RI = 0.762, RC = 0.409, HI = 0.463), and a strict consensus tree of these trees is shown in Fig. 1. The best model for the combined ITS and nLSU partition was a GTR + I + G model. BI analysis resulted in a similar topology.
The phylogenetic tree (Fig. 1) inferred from the combined ITS and nLSU sequences demonstrates two new lineages; collections from subtropical areas in southern China on angiosperm wood formed one lineage (MP = 100 %, BS = 100 %, BPP = 1.00), while collections from subtropical areas in southern China on gymnosperm wood formed another lineage (MP = 99 %, BS = 99 %, BPP = 0.99). They are considered as distinct phylogenetic species.
Taxonomy
Anomoloma luteoalba J. Song & B.K. Cui, sp. nov. (Figs. 2a and 3)
MycoBank no.: MB 815368
Anomoloma luteoalba is characterized by the following combined characters: cream to yellowish pore surface, 5–6 angular pores per mm, yellow rhizomorphs, ellipsoid basidiospores of 3–3.5 × 2–2.5 μm, and absence of cystidia.
Holotype. CHINA. Zhejiang Prov., Lin’an, Tianmushan Nature Reserve, on fallen trunk of Pinus, 11 Ocotober 2005, Cui 2687 (BJFC 000044).
Etymology. luteoalba (Lat.): referring to the cream to yellowish pore surface.
Fruitbody. Basidiomata annual, resupinate, felty or softly corky, without odor or taste when fresh, corky upon drying, up to 20 cm long, 5 cm wide, and 1 mm thick at centre. Sterile margin irregular, up to 1 cm wide, thinning out, cinnamon buff when dry; yellow rhizomorphs arising from margin and penetrating into decayed wood. Pore surface cream to yellowish when dry; pores angular, 5–6 per mm; dissepiments thin, entire. Subiculum cream to pale buff when dry, felty, up to 0.7 mm thick. Tubes concolorous with pore surface, softly corky, up to 0.3 mm long.
Hyphal structure. Hyphal system monomitic; generative hyphae with clamp connections, CB–, IKI–; tissues unchanged in KOH.
Subiculum. Generative hyphae hyaline, thin- to slightly thick-walled with a wide lumen, frequently branched, interwoven, 2–5 μm in diam. Hyphae close to the substrate often covered with minute hyaline crystals.
Tubes. Generative hyphae hyaline, thin-walled, frequently branched, interwoven, 2–3 μm in diam. Cystidia and cystidioles absent. Basidia clavate, bearing four sterigmata and a basal clamp connection, 20–25 × 4–5 μm; basidioles in shape similar to basidia, but smaller.
Spores. Basidiospores ellipsoid, hyaline, slightly thick-walled, smooth, amyloid, CB–, 3–3.5 × 2–2.5 μm, L = 3.2 μm, W = 2.18 μm, Q = 1.41–1.49 (n = 40/2).
Type of rot. A white rot.
Additional specimen (paratype) examined: CHINA. Anhui Prov., She County, Qingliangfeng Nature Reserve, on fallen trunk of Pinus, 14 December 2009, Cui 8686 (BJFC 007175).
Anomoloma submyceliosum J. Song & B.K. Cui, sp. nov. (Figs. 2b and 4)
MycoBank no.: MB 815369
Anomoloma submyceliosum is characterized by the following combined characters: white pore surface and rhizomorphs, 2–3 angular pores per mm, ellipsoid basidiospores of 3–4.2 × 2–2.7 μm, and growth on angiosperm wood in subtropical areas.
Holotype. CHINA. Fujian Prov., Wuyishan, Longfeng Valley, on angiosperm stump, 17 October 2005, Cui 2942 (holotype, BJFC 000070).
Etymology. submyceliosum (Lat.): referring to the morphological similarity to Anomoloma myceliosum.
Fruitbody. Basidiomata annual, resupinate, felty or softly corky, without odor or taste when fresh, up to 15 cm long, 4–8 cm wide, and 1.2 mm thick at centre. Sterile margin irregular, white, thinning out, up to 1.5 cm wide; radiciform and thread-like, white rhizomorphs arising from margin and penetrating into decayed wood. Pore surface white when fresh, becoming cream to cream buff when dry; pores angular, 2–3 per mm; dissepiments thin, entire. Subiculum white when fresh, becoming cream to buff when dry, felty, up to 0.8 mm thick. Tubes concolorous with pore surface, softly corky, up to 0.4 mm long.
Hyphal structure. Hyphal system monomitic; generative hyphae with clamp connections, IKI–, CB–; tissues unchanged in KOH.
Subiculum. Generative hyphae hyaline, slightly thick-walled with a wide lumen, frequently branched, interwoven, 2.2–4.5 μm in diam. Hyphae close to the substrate often covered with minute hyaline crystals.
Tubes. Generative hyphae hyaline, thin- to slightly thick-walled, frequently branched, interwoven, 2–3 μm in diam. Cystidia and cystidioles absent. Basidia clavate, bearing four sterigmata and a basal clamp connection, 23–30 × 5–6 μm; basidioles in shape similar to basidia, but smaller.
Spores. Basidiospores ellipsoid, hyaline, slightly thick-walled, smooth, amyloid, CB–, 3–4.2(−4.5) × 2–2.7 μm, L = 3.74 μm, W = 2.32 μm, Q = 1.54–1.69 (n = 60/2).
Type of rot. A white rot.
Additional specimens (paratypes) examined: CHINA. Fujian Prov., Wuyishan, Wuyishan Nature Reserve, Taoyuanyu, on rotten angiosperm wood, 22 Ocotober 2005, Dai 7402 (BJFC 000067), Dai 7402–2 (BJFC 000071).
Discussion
Previously, species of Anomoloma were treated in Anomoporia (Niemelä 1994). The two genera were separated by phylogenetic analysis based on rDNA sequences data (Niemelä et al. 2007). In the present study, phylogenetic analyses of Anomoloma using ITS and nLSU sequences reflect the overall structure of the Amylocorticiales as defined by Binder et al. (2010). The phylogeny shows that Anomoloma forms a monophyletic clade and is clearly separated from Anomoporia; the species with a yellowish pore surface gather together and form a weakly supported group; two new species in China were identified and strongly supported (Fig. 1).
Anomoloma flavissimum clusters together with A. luteoalba phylogenetically (Fig. 1), and it is similar to A. luteoalba by its yellow rhizomorphs and by growing on gymnosperm wood; however, A. flavissimum differs from A. luteoalba in producing a bright chrome or sulphur yellow pore surface, vesicular cystidia, slightly larger pores (4–5 per mm) and basidiospores (3–4 × 2–3 μm, Núñez and Ryvarden 2001). Anomoloma rhizosum also resembles A. luteoalba by its yellowish buff, pinkish buff or buff yellow pore surface and yellow rhizomorphs; however, A. rhizosum differs from A. luteoalba by producing larger pores (4–5 per mm) and basidiospores (4.1–5.3 × 3–4 μm), and thin- to thick-walled generative hyphae in the subiculum (Niemelä et al. 2007). Anomoloma myceliosum may be confused with A. submyceliosum by its white pore surface and rhizomorphs; nevertheless, A. myceliosum is distinguished from A. submyceliosum by its larger basidiospores (3.5–4.5 × 2.5–3 μm), growing on gymnosperm wood and circumboreal temperate distribution (Núñez and Ryvarden 2001).
Key to accepted species of Anomoloma and Anomoporia worldwide
1 Basidiospores usually > 5 μm in length 2
1* Basidiospores usually < 5 μm in length 3
2 Basidiocarps salmon pink, vesicular cells present in the context Anomoporia versiculosa
2* Basidiocarps violet-brown or lavender, vesicular cells absent in the context Anomoporia bombycina
3 Rhizomorphs present 4
3* Rhizomorphs absent Anomoporia kamtschatica
4 Pore surface and rhizomorphs white 5
4* Pore surface and rhizomorphs cream buff to yellow 6
5 Basidiospores 3–4.2 × 2–2.7 μm; mainly growing on angiosperm wood Anomoloma submyceliosum
5* Basidiospores 3.5–4.5 × 2.5–3 μm; mainly growing on gymnosperm wood Anomoloma myceliosum
6 Pores < 4 per mm Anomoloma albolutescens
6* Pores > 4 per mm 7
7 Basidiospores usually > 4 μm in length Anomoloma rhizosum
7* Basidiospores usually < 4 μm in length 8
8 Pore surface bright chrome or sulphur yellow; vesicular cystidia present Anomoloma flavissimum
8* Pore surface cream to yellowish; vesicular cystidia absent Anomoloma luteoalba
References
Binder M, Larsson KH, Matheny PB, Hibbett DS (2010) Amylocorticiales ord. nov and Jaapiales ord. nov.: early diverging clades of Agaricomycetidae dominated by corticioid forms. Mycologia 102:865–880. doi:10.3852/09-288
Chen JJ, Cui BK, Zhou LW, Korhonen K, Dai YC (2015) Phylogeny, divergence time estimation, and biogeography of the genus Heterobasidion (Basidiomycota, Russulales). Fungal Divers 71:185–200. doi:10.1007/s13225-014-0317-2
Felsenstein J (1985) Confidence intervals on phylogenetics: an approach using bootstrap. Evolution 39:783–791. doi:10.2307/2408678
Hall TA (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298. doi:10.1093/bib/bbn013
Li HJ, Cui BK, Dai YC (2014) Taxonomy and multi-gene phylogeny of Datronia (Polyporales, Basidiomycota). Persoonia 32:170–182. doi:10.3767/003158514X681828
Niemelä T (1994) Five species of Anomoporia, rare polypores of old forests. Ann Bot Fenn 31:93–115
Niemelä T, Larsson KH, Dai YC, Larsson E (2007) Anomoloma, a new genus separated from Anomoporia on the basis of decay type and nuclear rDNA sequence data. Mycotaxon 100:305–317
Núñez M, Ryvarden L (2001) East Asia polypores 2. Polyporaceae s. lato. Syn Fungorum 14:341–342
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
Petersen JH (1996) Farvekort. The Danish Mycological Society’s colour-chart. Foreningen til Svampekundskabens Fremme, Greve
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818. doi:10.1093/bioinformatics/14.9.817
RDM Page (1996) TREEVIEW: application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi:10.1093/bioinformatics/btg180
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer Associates, Sunderland
White TJ, Bruns TD, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic, San Diego, pp 315–322
Acknowledgments
We express our gratitude to Prof. Yu-Cheng Dai (BJFC, China) for help in field collections. The research was financed by the National Natural Science Foundation of China (project no. 31422001), the National Science and Technology Foundation Project of China (no. 2014FY210400) and the Beijing Higher Education Young Elite Teacher Project (YETP0774).
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Franz Oberwinkler
Rights and permissions
About this article
Cite this article
Song, J., Liu, XY., Wang, M. et al. Phylogeny and taxonomy of the genus Anomoloma (Amylocorticiales, Basidiomycota). Mycol Progress 15, 11 (2016). https://doi.org/10.1007/s11557-015-1155-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-015-1155-7