Abstract
Next generation sequencing (NGS) technologies generate huge amounts of sequencing data. Several microbial genome projects, in particular for fungal whole genome sequencing, have used NGS techniques because of their cost efficiency. However, NGS techniques also require computational tools able to process and analyze huge datasets. Data processing steps, including quality and length filters, often lead to a remarkable improvement in the accuracy and quality of data analyses. Choosing appropriate parameters for this purpose is not always straightforward, as these will vary with the dataset. In this study we present the FastQFS (Fastq Quality Filtering and Statistics) tool, which can be used for both an assessment of filtering parameters and read filtering. There are several tools available, but an important asset of FastQFS is that it provides the information of filtering parameters that fit best to the raw dataset, prior to computationally expensive filtering. It generates statistics of reads meeting different quality and length thresholds, and also the expected coverage depth of the genome which would be achieved after applying different filtering parameters. Thus, the FastQFS tool will help researchers to make informed decisions on an NGS read filtering parameters, and avoiding time-consuming optimization of filtering criteria after initial analyses. The source code of the tool and related files are available from 10.12761/SGN.2015.4.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Next generation sequencing (NGS) technologies have revolutionized genomics and transcriptomics, with a wide range of applications in biological and medical sciences. Massive parallel sequencing technologies generate sequence data in short time frames and with low sequencing costs, compared to traditional sequencing methods. Thus, several whole genome and/or transcriptome sequencing projects have considered the benefits of NGS technologies for sequencing novel species (Kemen et al. 2011; Laurie et al. 2012; Quinn et al. 2013; Levesque et al. 2010; Jiang et al. 2013). An example for this is the 1000 Fungal Genomes project (http://1000.fungalgenomes.org/), which has the aim to sequence more than 1000 fungal genomes using NGS technologies. In addition to sequencing new genomes, NGS techniques have also been implemented to study the fungal communities in environmental samples (Meiser et al. 2013; Schmidt et al. 2013).
With the advent of NGS, many computational tools have been developed to analyze the huge amounts of sequencing data generated by NGS methods. Sequence read filtering is an important step before starting any analyses based on the read files. However, to perform and optimize these data filtering steps, several data processing parameters need to be considered, and the decision-making regarding the choice of values for these parameters is often not straightforward and will also depend on data availability and downstream analyses. Many tools have been developed to view the basic statistics of data reading and to perform filtering steps; these include FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), the Fastx-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), the NGS QC toolkit (Patel and Jain 2012), Trimmomatic (Bolger et al. 2014), RobiNA (Lohse et al. 2012), PRINSEQ (Schmieder and Edwards 2011), HTQC (Yang et al. 2013), NGSQC (Dai et al. 2010), RSeQC (Wang et al. 2012) and Sickle (https://github.com/najoshi/sickle). But not all of these packages consider the features of both reads of mate-pair or paired-end data simultaneously while generating the read quality/length statistics. Moreover, they do not provide any data read characteristics information prior to filtering, to evaluate the effect of changes in filtering parameters.
Often, it is very difficult to guess how much coverage depth could be achieved using certain filtering thresholds. Moreover, many tools do not consider the phred quality of individual bases, they rather consider the average quality of the whole read or an average quality within a certain window size. It has become a rule of thumb to first choose standard filtering parameters for data processing and to optimize these iteratively after evaluating the filtered reads and initial analyses, which is very time-consuming. There is always a subtle balance between keeping the coverage high enough for good assemblies and to remove data of suboptimal quality, which is not easily achieved by an iterative method.
Thus, a next-generation sequencing data filtering tool called “FastQFS” is presented here. This tool first provides the user an evaluation of the variation of data with different quality and length cutoff parameters. Afterwards, it generates coverage depth variation statistics for different filtering thresholds. FastQFS also performs data filtering steps, considering the following parameters: Reads containing Ns, reads which contain at least one base having a quality below a certain threshold, reads having an average read quality below a certain threshold and reads of a length below specified threshold values. Since the majority of sequenced fungal genomes is small in size compared to animal and plant genomes, fungal genome sequencing projects thus generate comparatively less data, which makes it easier to optimize read filtering. FastQFS has been successfully applied on plant and oomycete genomic data, but has been developed and extensively tested only for fungal genomic and community barcoding datasets. It is probably comparatively slow in handling huge datasets for mammalian sized genomes.
Implementation
FastQFS takes raw input files in fastq format for both forward and reverse reads. First, it parses the fastq format and calculates various parameters including lengths of both the forward and the reverse read, the average base quality of both read pairs, the lowest quality score of a single base within sequence of both mates and whether the read sequence contains ambiguous bases (Ns) or not. While running this tool, it asks whether the user wants to perform filtering or plotting the filtering statistics of data. The plotting of data statistics is useful to make a decision on the data filtering parameters. From the plots, the percentage of reads which would be passing the different filtering parameters discussed above can be obtained. Moreover, FastQFS generates a plot representing the variation of the expected coverage depth with different quality filtering parameters. These plots provide users information about which parameters can be applied to their dataset for retaining sufficient coverage, enabling an informed decision before performing time consuming data filtering steps.
If only the filtering option is chosen (e.g., for data that has been plotted previously), data filtering is done without generating data statistics plots.
While processing a raw dataset, FastQFS considers features of both read pairs. If at least one read fails to meet the specified thresholds, then the whole read pair will be dropped out from the paired end file. This dropped pair is again scanned if any individual read matches the provided cutoffs, in which case the read is listed in a singleton file. The workflow of the tool is shown in Fig. 1.
The FastQFS.pl (Supplementary file 1) script can be used for plotting and filtering paired-end data. The two main features of FastQFS, plotting and filtering, can be used simultaneously or one after the other. The following commands demonstrate the usage of these modules.
Plotting variation of data/coverage depth with filtering parameters
perl FastQFS.pl -plotting Yes -fw demoR1.fq -rw demoR2.fq -prefix Prefix -sc 33 -gsize 20 -l 100
The above command will generate two different files, “Prefix_File_for_plotting_coverage.txt” and “Prefix_File_for_plotting_reads_percentages.txt”, containing the information regarding variation of read coverage depth and percentages of reads retained after applying the filtering parameters, respectively. These output files can further be imported to the R scripts “Plotting_Coverage_depth.R” (Supplementary file 2) and “Plotting_read_Percentages.R” (Supplementary file 3) for plotting coverage depth and percentage variations, respectively. All input parameters are briefly explained in the help section of the FastQFS script.
Plotting coverage depth variation
Rscript Plotting_Coverage_depth.R Prefix_File_for_plotting_coverage.txt
Plotting read percentage variation
Rscript Plotting_read_Percentages.R Prefix_File_for_plotting_reads_percentages.txt
Performing read filtering
perl FastQFS.pl -filtering Yes -fw demoR1.fq -rw demoR2.fq -prefix Prefix -sc 33 -mq 10 -q 26 -l 100
Forward and reverse filtered reads will be written in files “Prefix_R1.fq” and “Prefix_R2.fq”, respectively. Singletons will be written in file “Prefix_Singltons.fq”.
Performing read filtering and plotting
perl FastQFS.pl -filtering Yes -fw demoR1.fq -rw demoR2.fq -prefix Prefix -sc 33 -mq 10 -q 26 -l 100 -plotting Yes -gsize 20
Running the FastQFS script without any input parameter will generate a help message, this help message explains all input parameters required for this script in detail.
Results
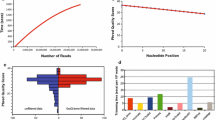
For demonstration purpose, FastQFS was used on a fungal genomic dataset. This dataset had three different insert size libraries. Figure 2 shows the percentage of reads from the 3 kbp insert size library meeting different length and quality cutoffs. The dataset was tested with various average read quality cutoffs from phred scores of 18 to 30 (Fig. 2a–d), with an increment of 4, length cutoffs from 50 to 100 bp with an increment of 10 bp and phred quality cutoffs of individual bases from 3 to 18 with an increment of 5. It was revealed that the read filtering output is highly influenced by length cutoffs exerted on both of the reads, i.e., that a filtering parameter which might seem applicable when considering only the aggregate statistics of either read is potentially not useful when both reads are considered. The average phred score quality cutoff does not show much impact on filtering paired reads, but as expected, the impact of individual base quality cutoffs in read filtering was higher. A plot showing the variation of coverage depths of the 3 kbp library according to different filtering parameters is shown in Fig. 3.
Exemplary plots for the percentage of data left after applying different read filtering parameters to reads from a 3 kbp library. Plots have been generated using average read quality cutoffs of 18, 22, 26, and 30, in A, B, C, and D, respectively, and using length cutoffs from 100 to 150 bp for both reads, with an increment of 10 bp. Minimum base quality (MBQ) was set to phred scores of 3 to 18 with an increment of 5
Coverage depth variation with different quality filtering parameters applied to reads from a 3 kbp library. Plots have been generated using average read quality cutoffs of 18, 22, 26, and 30, in A, B, C and D, respectively, using length cutoffs from 100 to 150 bp for both reads, with an increment of 10 bp. Minimum base quality (MBQ) was set to phred scores of 3 to 18 with an increment of 5
Similar plots using the 250 bp insert library (Supplementary Figs. 1–2) and the 8 kbp library were generated (Supplementary Figs. 3–4). It became apparent that the long distance libraries are more influenced by changes in data filtering parameters than the shorter insert libraries. Figure 4 illustrates the percentage of data left after applying different length cutoffs to three different libraries.
The runtime of the script was calculated by performing data filtering of the three libraries differing in insert size. The three libraries, with insert sizes of 250 bp, 3 kbp and 8 kbp, were represented by around 123, 17 and 18 million raw reads, respectively. Filtering the three libraries using different length cutoffs required 7 h and 5 min, 49 min and 53 min, respectively.
For evaluating the variation of genome assembly quality parameters according to different filtering thresholds, i.e. the N50 scaffold size, the size of the largest scaffold, and the number of scaffolds, were compared after generating genome assemblies derived from different filtering thresholds. The three libraries of the test dataset were assembled using the velvet (Zerbino and Birney 2008) short read genome assembler. In these comparisons, a k-mer of size 45 was used to generate 6 different assemblies derived from the 6 filtered reads datasets, by using length thresholds from 50 to 100 bp. As expected, all parameters varied according to changes in length cutoffs (data not shown).
Discussion
Using NGS technologies, sequencing even a mammalian genome is a matter of a few weeks at sequencing costs that are affordable to many laboratories (Schatz et al. 2010). Due to these advantages, NGS technologies have quickly been implemented in various fields of life sciences (Metzker 2010), including de novo sequencing of whole genomes (Schatz et al. 2010; Sharma et al. 2015), genome re-sequencing (Stratton 2008), cDNA sequencing (Martin and Wang 2011; Ozsolak and Milos 2011; Wang et al. 2009), genotyping (Davey et al. 2011; Sharma et al. 2014; Yoshida et al. 2013), and community genomics analyses (Qin et al. 2010). Also, several filamentous organisms, including fungi and oomycetes have been sequenced over the last decade (Raffaele and Kamoun 2012). Due to the small genome sizes of most filamentous organisms, several studies in fungi and oomycetes have taken advantage of NGS technologies for whole genome sequencing (Quinn et al. 2013; Levesque et al. 2010; Laurie et al. 2012; Kemen et al. 2011; Jiang et al. 2013; Sharma et al. 2014).
Before starting any analysis on NGS data, it is important to perform data filtering, so the analyses do not suffer from low quality reads (Dai et al. 2010). Over the past few years, many filtering tools have been developed, which can process NGS data considering quality and length thresholds (Bolger et al. 2014; Schmieder and Edwards 2011). Applying different filtering parameters has a significant impact on downstream analyses, depending on the filtering method used (Del Fabbro et al. 2013). Filtering also has a pronounced impact on the amount of reads available for downstream analyses, as a too low coverage can have similar detrimental effects on downstream analyses as including data with low quality scores. Often, especially if funds are limited, a balance has to be sought between a quality filtering that will filter out reads of suboptimal quality and length on the one hand and the coverage retained on the other hand. To our knowledge, there is currently no tool which provides information about the coverage depth variation with different quality cutoffs prior to read filtering, providing straightforward way of choosing quality and length thresholds for filtering. Features of current NGS data processing tools, including FastQFS, are given in Supplementary Table 1.
An alternative or addition to filtering bad quality reads, which can be useful depending on the kind of analyses to be done, are error-correcting tools that help in correcting bad quality bases originating from wrong base-calls (Lim et al. 2014; Kelley et al. 2010). Such tools can help in correcting some bases that are generally trimmed out by the filtering tools. However, care should be taken while using error-correcting tools in studies where a major part of the study depends on the accuracy of a single base, for example studies including SNP detection or community barcoding. Otherwise, it can also be useful to employ error correcting tools prior to read filtering.
FastQFS generates estimated coverage depth plots after the filtering of reads with different quality and length cutoffs, using a user-provided estimated genome size. In case of RNA-Seq data this size could be the total length of protein coding genes. This information can be used to select the most stringent filtering parameters which generate a filtered dataset of the desired minimum coverage depth. Considering the quality of individual bases as available in FastQFS might be important, as in average-based filters, some reads will be retained that are having many bases with very high scores and some bases with very low quality scores. However, these low quality bases might be problematic in some downstream analyses, like variant detection, single nucleotide polymorphism (SNP) mining or genome assemblies.
FastQFS has been written in the Perl programming language (https://www.perl.org/), which is platform-independent and can be run on any Perl-supporting operating system. FastQFS does not depend on other Perl libraries or modules, which makes it user friendly also for biologists with limited bioinformatics knowledge.
Thus, we hope that FastQFS will prove useful for data filtering, especially with the aim to achieve an optimised balance between quality filtering and coverage.
References
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Dai M, Thompson RC, Maher C, Contreras-Galindo R, Kaplan MH, Markovitz DM, Omenn G, Meng F (2010) NGSQC: cross-platform quality analysis pipeline for deep sequencing data. BMC Genomics 11(Suppl 4):S7
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12:499–510
Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM (2013) An extensive evaluation of read trimming effects on illumina NGS data analysis. PLoS One 8, e85024.4
Jiang RH, de Bruijn I, Haas BJ, Belmonte R, Lobach L, Christie J, van den Ackerveken G, Bottin A, Bulone V, Diaz-Moreno SM, Dumas B, Fan L, Gaulin E, Govers F, Grenville-Briggs LJ, Horner NR, Levin JZ, Mammella M, Meijer HJ, Morris P, Nusbaum C, Oome S, Phillips AJ, van Rooyen D, Rzeszutek E, Saraiva M, Secombes CJ, Seidl MF, Snel B, Stassen JH, Sykes S, Tripathy S, van den Berg H, Vega-Arreguin JC, Wawra S, Young SK, Zeng Q, Dieguez-Uribeondo J, Russ C, Tyler BM, van West P (2013) Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica. PLoS Genet 9, e1003272
Kelley DR, Schatz MC, Salzberg SL (2010) Quake: quality-aware detection and correction of sequencing errors. Genome Biol 11:R116
Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, Robert-Seilaniantz A, Bailey K, Holub E, Studholme DJ, Maclean D, Jones JD (2011) Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol 9, e1001094
Laurie JD, Ali S, Linning R, Mannhaupt G, Wong P, Guldener U, Munsterkotter M, Moore R, Kahmann R, Bakkeren G, Schirawski J (2012) Genome comparison of barley and maize smut fungi reveals targeted loss of RNA silencing components and species-specific presence of transposable elements. Plant Cell 24:1733–1745
Levesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, Raffaele S, Robideau GP, Thines M, Win J, Zerillo MM, Beakes GW, Boore JL, Busam D, Dumas B, Ferriera S, Fuerstenberg SI, Gachon CM, Gaulin E, Govers F, Grenville-Briggs L, Horner N, Hostetler J, Jiang RH, Johnson J, Krajaejun T, Lin H, Meijer HJ, Moore B, Morris P, Phuntmart V, Puiu D, Shetty J, Stajich JE, Tripathy S, Wawra S, van West P, Whitty BR, Coutinho PM, Henrissat B, Martin F, Thomas PD, Tyler BM, De Vries RP, Kamoun S, Yandell M, Tisserat N, Buell CR (2010) Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol 11:R73
Lim EC, Muller J, Hagmann J, Henz SR, Kim ST, Weigel D (2014) Trowel: a fast and accurate error correction module for Illumina sequencing reads. Bioinformatics 30:3264–3265
Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B (2012) RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40:W622–W627
Martin JA, Wang Z (2011) Next-generation transcriptome assembly. Nat Rev Genet 12:671–682
Meiser A, Balint M, Schmitt I (2013) Meta-analysis of deep-sequenced fungal communities indicates limited taxon sharing between studies and the presence of biogeographic patterns. New Phytol. doi:10.1111/nph.12532
Metzker ML (2010) Sequencing technologies - the next generation. Nat Rev Genet 11:31–46
Ozsolak F, Milos PM (2011) RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12:87–98
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7, e30619
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Meta HITC, Bork P, Ehrlich SD, Wang J (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65
Quinn L, O’Neill PA, Harrison J, Paskiewicz KH, McCracken AR, Cooke LR, Grant MR, Studholme DJ (2013) Genome-wide sequencing of Phytophthora lateralis reveals genetic variation among isolates from Lawson cypress (Chamaecyparis lawsoniana) in Northern Ireland. FEMS Microbiol Lett 344:179–185
Raffaele S, Kamoun S (2012) Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol 10:417–430
Schatz MC, Delcher AL, Salzberg SL (2010) Assembly of large genomes using second-generation sequencing. Genome Res 20:1165–1173
Schmidt PA, Balint M, Greshake B, Bandow C, Rombke J, Schmitt I (2013) Illumina metabarcoding of a soil fungal community. Soil Biol Biochem 65:128–132
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864
Sharma R, Mishra B, Runge F, Thines M (2014) Gene loss rather than gene gain is associated with a host jump from monocots to dicots in the smut fungus Melanopsichium pennsylvanicum. Genome Biol Evol 6:2034–2049
Sharma R, Gassel S, Steiger S, Xia X, Bauer R, Sandmann G, Thines M (2015) The genome of the basal agaricomycete Xanthophyllomyces dendrorhous provides insights into the organization of its acetyl-CoA derived pathways and the evolution of Agaricomycotina. BMC Genomics 16:233
Stratton M (2008) Genome resequencing and genetic variation. Nat Biotechnol 26:65–66
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Wang L, Wang S, Li W (2012) RSeQC: quality control of RNA-seq experiments. Bioinformatics 28(16):2184–2185. doi:10.1093/bioinformatics/bts356
Yang X, Liu D, Liu F, Wu J, Zou J, Xiao X, Zhao F, Zhu B (2013) HTQC: a fast quality control toolkit for Illumina sequencing data. BMC Bioinformat 14:33
Yoshida K, Schünemann VJ, Cano LM, Pais M, Mishra B, Sharma R, Lanz C, Martin FN, Kamoun S, Krause J, Thines M, Weigel D, Burbano HA (2013) The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. eLife 2, e00731
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829
Acknowledgments
We thank Claus Weiland for support with respect to cluster access. This work was supported by the research funding programme LOEWE “Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz” of Hesse’s Ministry of Higher Education, Research, and the Arts in the framework of IPF. We are grateful to Bagdevi Mishra and Deepak Kumar Gupta for testing of the programme and for providing important feedback and comments.
Author contributions
MT and RS designed the study. MT and RS conceived analyses and provided ideas. MT and RS developed the algorithms and RS wrote the scripts. RS and MT wrote the manuscript.
Competing interest statement
MT is a section editor of Mycological Progress.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure S1
Percentage of reads left after applying different read processing parameters for the library of 250 bp insert size. (PPTX 116 kb)
Supplementary Figure S2
Data coverage left after applying different read processing parameters for the library of 250 bp insert size. (PPTX 119 kb)
Supplementary Figure S3
Percentage of reads left after applying different read processing parameters for the library of 8 kb insert size. (PPTX 121 kb)
Supplementary Figure S4
Data coverage left after applying different read processing parameters for the library of 8 kb insert size. (PPTX 127 kb)
ESM 5
(TXT 14 kb)
ESM 6
(TXT 4 kb)
ESM 7
(TXT 3 kb)
Table S1
Comparison of various tools available for the filtering of short reads from high throughput sequencing platforms. (DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Sharma, R., Thines, M. FastQFS – A tool for evaluating and filtering paired-end sequencing data generated from high throughput sequencing. Mycol Progress 14, 60 (2015). https://doi.org/10.1007/s11557-015-1077-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-015-1077-4