Abstract
Purpose
Skull base features, including increased foramen ovale (FO) cross-sectional area, are associated with lateral skull base spontaneous cerebrospinal fluid (sCSF) leak and encephalocele. Manual measurement requires skill in interpreting imaging studies and is time consuming. The goal of this study was to develop a fully automated deep learning method for FO segmentation and to determine the predictive value in identifying patients with sCSF leak or encephalocele.
Methods
A retrospective cohort study at a tertiary care academic hospital of 34 adults with lateral skull base sCSF leak or encephalocele were compared with 815 control patients from 2013–2021. A convolutional neural network (CNN) was constructed for image segmentation of axial computed tomography (CT) studies. Predicted FO segmentations were compared to manual segmentations, and receiver operating characteristic (ROC) curves were constructed.
Results
295 CTs were used for training and validation of the CNN. A separate dataset of 554 control CTs was matched 5:1 on age and sex with the sCSF leak/encephalocele group. The mean Dice score was 0.81. The sCSF leak/encephalocele group had greater mean (SD) FO cross-sectional area compared to the control group, 29.0 (7.7) mm2 versus 24.3 (7.6) mm2 (P = .002, 95% confidence interval 0.02–0.08). The area under the ROC curve was 0.69.
Conclusion
CNNs can be used to segment the cross-sectional area of the FO accurately and efficiently. Used together with other predictors, this method could be used as part of a clinical tool to predict the risk of sCSF leak or encephalocele.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Rates of spontaneous cerebrospinal fluid (sCSF) leaks in the United States are increasing [1, 2]. sCSF leak and encephalocele of the lateral skull base are thought to be due to chronically increased intracranial pressure causing erosion of the tegmen tympani and tegmen mastoideum and has been associated with obstructive sleep apnea and idiopathic intracranial hypertension (IIH, previous known as pseudotumor cerebri or benign intracranial hypertension) [3,4,5,6]. Diagnosis of IIH by the modified Dandy criteria requires lumbar puncture to confirm elevated intracranial pressure and neuroimaging that shows no other etiology for intracranial hypertension, such as a mass or structural lesion [7,8,9,10,11]. This requires patients to undergo an invasive procedure, and it can be difficult to identify an elevated pressure if a patient is actively leaking CSF due to the temporary decrease in pressure from egress of fluid acting as a form of “auto diversion” of CSF. Patients with sCSF leak may not develop the typical signs, symptoms, and imaging findings, such as papilledema or magnetic resonance imaging (MRI) findings including posterior globe flattening, optic nerve ectasia and tortuosity, and empty sella [12,13,14,15,16,17,18,19,20] until after skull base repair, which can result in delayed or missed diagnosis [21,22,23,24,25,26].
Prior studies have found associations between skull base features identifiable on imaging studies and IIH, sCSF leak, and encephalocele, including thinning of the calvarium and skull base [27,28,29,30,31,32,33,34] and an enlarged foramen ovale (FO) [35, 36]. Manual measurement of these features requires skill in interpreting neuroimaging, is time-consuming, and is prone to inter-observer variation.
U-Net is a convolutional neural network (CNN) that is commonly used for automated image segmentation, which is the process of precisely delineating a structure on an image [37]. U-Net can perform both 2D (one slice at a time) and 3D (entire volume) predictions. 2D predictions have the benefit of being performed at full spatial resolution (~ 0.5 mm in-plane), while 3D predictions have increased accuracy due to improved spatial awareness but are performed at a lower spatial resolution (~ 1.5 mm) due to memory constraints. Previous researchers have described 2D–3D hybrid prediction techniques to get both the full resolution and accuracy benefits of 2D and 3D techniques, respectively [38, 39]. Others have also described a “cascaded” segmentation technique, first identifying a structure of interest on a whole volume, then zooming in on that structure to make additional predictions [40]. We hypothesized that a cascaded, or iterative prediction technique could provide accurate full-resolution segmentations of the FO on axial computed tomography (CT) images for area determination.
The future value of this automated FO measurement lies in the development of a predictive model of sCSF leak and encephalocele that would be non-invasive, time-efficient, and not require user expertise. This could be used to identify patients at increased risk of sCSF leak or encephalocele who may benefit from additional diagnostic testing and/or treatment. Among patients who undergo operative repair, this would also enable determining which patients are at increased risk of surgical failure (recurrent leak) or contralateral disease (new onset sCSF leak and/or encephalocele).
Methods
Training data gathering
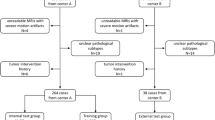
After approval from the institutional review board at the University of Nebraska Medical Center (IRB# 412–19-EX), training data was gathered from a dataset of 295 CT head studies [41]. These were performed on a variety of General Electric (GE, Boston, MA) CT scanners at our hospital and surrounding outpatient clinics. All CTs were obtained as axial/contiguous acquisitions, with 0.625 mm slice thickness and reconstructed in standard algorithm with iterative reconstruction technique. All CTs were obtained with a 512 × 512 imaging matrix, and a field-of-view of either 250 or 320 mm. Exclusion criteria were excessive motion or streak artifact obscuring measurement of the FO.
Training data labeling
CT studies were first aligned based on landmarks of the bilateral cochleas and nasal bridge approximating the anterior commissure—posterior commissure line (AC-PC line), in a technique described in a submitted, unpublished manuscript. Manual FO labels were placed for each CT image using 3D Slicer (version 4.11.20210226) [42, 43]. Labels were placed over the entire FO at the cranio-caudal center of the FO using a 3 mm spherical brush and editable intensity range of -3000–350 Houndsfield units (HU). Manual segmentation was performed by a single author (S.C.), with another author (J.C.) verifying the accuracy of the manual labels.
U-Net training
The base CT head studies and labels were augmented by a rotated version of each CT head, with up to a 30-degree rotation in the sagittal plane, and a rotated and scaled version of each CT head, rotated up to 30 degrees in the sagittal plane, and scaled by a factor of 0.5–1.0. Rotation was only done in the sagittal plane due to axial and coronal alignment performed prior to predictions. Cropped images and labels with a spacing of 0.5 × 0.5 × 0.625 and size of 128 × 128 × 64 were generated around each FO for training of a second full resolution model. Each of these 3 paired versions was then flipped. Of the 3 versions for the original and flipped images, 1 was selected from each (basically selecting each for FO once), for a total of 2 per subject and 590 total images. 90% of the images were used for training, and 10% were used for validation of the CNN.
U-Net training was performed using the Project MONAI Python toolkit [44]. Spacing for training of the full CT head images was 1.5 × 1.5 × 1.5 mm in x,y,z dimensions, resampled from 0.5 × 0.5 × 0.625. Spacing for training of the cropped images was at the original resolution of 0.5 × 0.5 × 0.625. A model for only the right FO was trained. Prediction for the left FO was accomplished by flipping the image and using the right FO model. Training was stopped when there was no improvement in mean Dice coefficient for 50 epochs.
Iterative prediction algorithm design
An iterative prediction technique was implemented. First, prediction of a FO was performed on the full head CT at 1.5 mm spacing. Then, the CT head was cropped around the FO at a size of 128 × 128 × 64 and spacing of 0.5 × 0.5 × 0.5. Prediction was then performed on that image with the separately trained cropped model. The full-resolution prediction was then transferred back onto a full resolution version of the CT head. Failures were detected by absence of prediction data, or abnormal spacing of the predicted foramina (< 35 or 65 mm apart, > 6 mm difference on z-axis, > 5 mm difference on y-axis). Figure 1 shows an example of full head and cropped sizes and spacings used for training and prediction.
Example of full head and cropped sizes and spacings used for training and prediction. Training was performed on both the full head and cropped labels. Prediction was done on the full head 1.5 mm spacing images, (a) axial and (b) coronal. Then the image was cropped around the prediction, (c) axial and (d) coronal, and prediction was done again on the cropped 0.5 mm spacing image. The full-resolution cropped prediction was then transferred back to the whole head image
Model testing
Testing of the model was performed on a separate dataset of 554 patients, including 34 patients with known sCSF leak or encephalocele confirmed at the time of surgical repair of the skull base, and 520 control patients. This dataset included CT head, CT temporal bone, CT face, and CT sinus studies. For each subject, the CT was first aligned using the same landmark-based technique. If the nasal bridge was not visible, such as on a temporal bone CT, the image was registered to a template CT head, then just the cochleas were aligned along the z- and y-axes. Predictions were then made for the right and left FO as detailed above. FO area was calculated for each predicted slice (number of voxels * voxel area), and the slice with the largest area was selected and recorded.
Processing was performed on a computer with an Intel Core i7-9700K CPU (Intel Corporation, Santa Clara, CA), and an NVIDIA GeForce GTX 1080 Ti GPU (Nvidia Corporation, Santa Clara, CA).
Statistical analysis
Demographic and clinical variables including age at time of imaging, sex, race, ethnicity, height, and weight were collected. Patient charts were reviewed for pertinent comorbidities, and the following were recorded as present or absent: diabetes mellitus (DM), osteoporosis, hypertension (HTN), chronic kidney disease (CKD), obstructive sleep apnea (OSA), idiopathic intracranial hypertension (IIH), and hydrocephalus. Patients in the sCSF leak/encephalocele group were matched 5:1 to control patients on age and sex using a nearest neighbor matching algorithm.
Categorical variables were reported as counts and percentages, and continuous variables were reported as mean and standard deviation, or median and interquartile range (IQR) if skewed. Associations between categorical variables were assessed using chi square tests, or Fisher’s exact test when expected cell sizes were low. Associations between group and continuous variables were assessed using independent samples t-tests. Receiver operating characteristic (ROC) curves were constructed to evaluate the diagnostic ability of the FO size as a binary classifier, and the area under the curve (AUC) was calculated for each ROC curve. Statistical analysis was performed in R version 4.1.2 (R Foundation for Statistical Computing, Vienna, Austria). All tests were two-tailed with statistical significance defined by an α = 0.05.
Results
Training data gathering
295 CT head studies on unique patients were selected for CNN training data creation. The minimum age was < 1 year, maximum age was 92 years, and mean (SD) age was 54.5 (19.3) years. 51% were male.
U-Net training
After augmentation by rotation, scaling, and flipping, 590 studies were semi-randomly selected for U-Net training (90%) and validation (10%) as detailed in the methods section. Mean Dice scores were 0.66 for the full head / 1.5 mm spacing right FO and 0.81 for the cropped / 0.5 mm spacing right FO.
The FO areas predicted by the CNN in this training set compared favorably with the manual ground truth measurements on the same imaging studies. The mean (SD) manually measured right FO area was 22.8 (7.1) mm2, and the left FO area was 22.7 (6.8) mm2. The mean (SD) predicted right FO area 22.4 (6.4) mm2, and the predicted left FO area was 22.2 (6.6) mm2. The difference in the sample mean foramen size between measured and predicted values was 0.4 mm2 for the right foramen (1.8% of the foramen area), and 0.5 mm2 for the left foramen (2.2% of the foramen area). At the level of individual foramina, there was a mean (SD) difference of 1.52 (2.35) mm2 between the manually measured and predicted right FO, and a mean difference of 1.73 (3.28) mm2 for the left FO, with an overall mean difference of 1.62 (2.66) mm2 (Table 1).
Model application
A separate dataset of 554 CT studies was used for evaluation of the segmentation algorithm in identifying patients with lateral skull base sCSF leak or encephalocele. The mean (SD) age of patients at the time imaging studies was 63.6 (17.9) years and ranged from 19 to 101 years. 49% were male. This included 34 imaging studies from patients with known lateral skull base sCSF leak or encephalocele that was confirmed at the time or surgical repair via a middle cranial fossa approach and 520 controls without IIH, CSF leak, or encephalocele.
Segmentation failed 9 times, including 8 times on control patients. Seven failures were due to excessive motion during image acquisition (an example is shown in Fig. 2A), and one failure was due to an uncorrectable gantry tilt issue. Segmentation failed once on a patient in the sCSF leak/encephalocele group who had numerous arachnoid granulations along the skull base obscuring the FO (Fig. 2B).
Segmentation failures: A Coronal CT image showing excess motion during image acquisition, which was responsible for the majority of segmentation prediction failures. B Failure of segmentation in patient with known idiopathic intracranial hypertension and CSF leak. Note the numerous arachnoid granulations along the central skull base obscuring the foramen ovale (arrow)
Patients in the sCSF leak/encephalocele group were matched 5:1 to controls using a nearest neighbor algorithm. Mean (SD) age was 53.3 (11.6) years in the sCSF leak/encephalocele group, and 53.2 (20.2) years in the control group. There were no significant differences in age (P = 0.95) or sex (P = 0.37) between the matched groups. Body mass index (BMI) was significantly higher in the sCSF leak/encephalocele group (median 37.0 kg/m2 vs. 24.4 kg/m2, P < 0.001). The rates of diabetes mellitus (44.1% vs. 20.6%, P = 0.004) and OSA (29.4% vs. 8.8%, P = 0.002) were higher in the sCSF leak/encephalocele group compared to the control group. The rates of osteoporosis (14.1% vs. 0%, P = 0.002) and CKD (8.8% vs. 0%, P = 0.08) were higher in the control group compared to the sCSF leak/encephalocele group. No significant differences were found for the rates of HTN (P = 0.35), IIH (P = 0.17), or hydrocephalus (no patients in either group).
The mean (SD) FO area was significantly higher in the sCSF leak/encephalocele group: 25.4 (6.1) mm2 versus 22.2 (6.2) mm2 (P = 0.008) (Table 2). There was a larger difference between left and right FO among patients in the sCSF leak/encephalocele group, with a mean (SD) difference of 7.2 (5.3) mm2 between the 2 sides, compared to the control group, which had a mean (SD) difference of 4.2 (4.7) mm2, but this did not reach statistical significance (P = 0.56). Comparison of the larger of the 2 foramina for each patient yielded an even greater difference between the 2 groups: 29.0 (7.7) mm2 versus 24.3 (7.6) mm2 (P = 0.002) (Fig. 3). A binomial test of the patients with measurable foramina showed that in neither the sCSF leak/encephalocele group (n = 33, P = 0.49, 95% CI 0.39–0.75) nor the control group (n = 179, P = 0.18, 95% CI 0.48–0.63) was the left or right foramen significantly larger than compared to the contralateral side. The laterality of the larger FO was ipsilateral to the laterality of the CSF leak or encephalocele in only 17 (50%) patients, which was not significantly different from chance (P = 0.52, 95% CI 0.34–0.69).
Comparison of mean areas of the foramen ovale for control patients versus patients with sCSF leak or encephalocele. Significant differences in the means were found between the two groups for the left FO, right FO, mean (mean value comparing each patient’s left and right side) FO, and the larger (the larger of the two comparing each patient’s left versus right side) FO. Error bars are the standard error of the mean
ROC curves were constructed to assess the diagnostic ability of the FO area as a binary classifier to identify patients with sCSF leak/encephalocele. Figure 4 shows ROC curves created to show the predictive value in identifying patients in the sCSF leak/encephalocele group compared to age- and sex- matched controls using (A) the mean FO size (mean of left and right foramen) for each patient, and (B) using the larger FO (left or right) for each patient. The area under the ROC curve (AUC) was 0.65 when using the area of the average (mean of left and right) FO for each patient. This increased to an AUC of 0.69 when using the area of the larger of the 2 foramina (left or right) for each patient in both the sCSF leak/encephalocele and control groups. Using a cutoff value of 30 mm2 to diagnose an enlarged FO, as described by Butros et al. in patients with IIH,19 gives a sensitivity of 30% and a specificity of 79% for the sCSF leak/encephalocele group.
ROC curves were constructed to show the predictive value in identifying patients in the sCSF leak/encephalocele group compared to age- and sex- matched controls using (A) the mean foramen ovale size (mean of left and right foramen) for each patient, and (B) using the larger foramen ovale (left or right) for each patient
Discussion
Our data show the utility of using a CNN with an iterative technique to produce accurate full-resolution 3D segmentation of the FO utilizing U-Net. This allows the segmentation and analysis of smaller detailed structures such as the FO, circumventing memory restrictions previously encountered with full-resolution prediction. It also continues to benefit from the spatial awareness of 3D as opposed to 2D prediction, further increasing accuracy. While the increased accuracy of segmentation at full resolution is intuitive, the Dice score increase from 0.66 to 0.81 also objectively demonstrates an improvement. The model was also quite robust, only failing to segment the FO on CT studies for clear reasons (e.g., motion artifact, unidentifiable FO). Finally, no extra manual effort was required for this implementation, as the cropped images and labels were automatically generated. Differences in measurements between the manual segmentations and the predicted (deep learning algorithm) measurements were < 2 mm2 on average and are not simply due to errors of the CNN but are a function of the inherent challenge in the interpretation of voxels at the edge of foramen, with intermediate opacity between bone and soft tissue, which is a challenge that is always present in manual measurements as well.
The clinical utility of this technique lies in its ability to predict the risk of patients developing a lateral skull base sCSF leak or encephalocele, using data (CT images) that are already readily available for many patients. There is currently no agreement on screening or management of sCSF leak in patients with IIH or other pathologies associated with elevated intracranial pressure [45]. While reported rates of surgical repair are high, there is room for improvement in identifying patients at high risk of sCSF leak or encephalocele, which could potentially be prevented through targeted interventions, and among patients who undergo surgical repair, identifying patients at risk of surgical failure due to increased intracranial pressure that was not identified prior to repair due to a CSF fistula [46,47,48,49].
Prior studies have shown the application of deep learning algorithms for automated detection of intracranial hemorrhage, calvarial fracture, midline shift, and mass effect in patients with head trauma or stroke symptoms using datasets of head CT scans [50,51,52]. A deep learning framework has also been used for differentiating patients with normal cognition, mild cognitive impairment, Alzheimer’s disease (AD), and non-AD dementias using MRI brain imaging together with demographic and clinical data [53]. Similarly, beyond the usefulness of a non-invasive imaging-based diagnostic tool to identify patients at risk of sCSF leak or encephalocele, the methods used in the present study could be used together with other demographic and clinical variables as part of a larger model to stratify and direct patients to undergo further diagnostic workup with lumbar puncture to confirm IIH or to suggest treatment with weight loss interventions, acetazolamide, or ventriculoperitoneal shunting to reduce intracranial pressure [54, 55].
The present study has several limitations. First, alignment of non-head CT studies (e.g., CT sinus, CT temporal bone) could be somewhat inconsistent if all landmarks were not present for alignment in the axial plane, which could slightly alter the area measurement for that study. Registration to an aligned template was meant to mitigate that inconsistency. Also, the precise areas reported in this study are specific to the training labels and model utilized for prediction. Labeling was done with a -3000–350 HU editable intensity range. As the interface between bone and soft tissue is somewhat indistinct, a higher range would have led to larger FO areas. Precise reproduction of study results would need to adhere to the above editable intensity range for labeling. Finally, generalizability of the findings may be reduced since the training dataset was derived from a single CT scanner vendor (GE), though from a variety of scanner models. Wider use of the prediction model would likely require additional training data from more vendors.
Conclusion
An iterative prediction technique allows for accurate and full resolution 3D segmentation of medical images. We described the use of an automated method of measuring the FO and the application of this technique for predicting patients with sCSF leak or encephalocele. Future applications combining these measurements with other clinically relevant data may further increase the predictive power and clinical utility.
References
Nelson RF, Gantz BJ, Hansen MR (2015) The rising incidence of spontaneous cerebrospinal fluid leaks in the United States and the association with obesity and obstructive sleep apnea. Otol Neurotol 36:476–480. https://doi.org/10.1097/MAO.0000000000000535
Totten DJ, Schueth E, Saltagi MZ, Rabbani C, Harris AH, Tressman D, Hohmann SF, Nelson RF (2022) Trends in spontaneous cerebrospinal fluid leak repairs in the United States, 2009–2018. Otol Neurotol Open 2:e021. https://doi.org/10.1097/ONO.0000000000000021
Kutz JW, Tolisano AM (2019) Diagnosis and management of spontaneous cerebrospinal fluid fistula and encephaloceles. Curr Opin Otolaryngol Head Neck Surg 27:369–375. https://doi.org/10.1097/MOO.0000000000000568
Rabbani CC, Saltagi MZ, Nelson RF (2019) The role of obesity, sleep apnea, and elevated intracranial pressure in spontaneous cerebrospinal fluid leaks. Curr Opin Otolaryngol Head Neck Surg 27:349–355. https://doi.org/10.1097/MOO.0000000000000562
Bakhsheshian J, Hwang MS, Friedman M (2015) Association between obstructive sleep apnea and spontaneous cerebrospinal fluid leaks: a systematic review and meta-analysis. JAMA Otolaryngol Head Neck Surg 141:733–738. https://doi.org/10.1001/jamaoto.2015.1128
Rabbani C, Saltagi MZ, Ye MJ, Ye MJ, Patel JM, Manchanda S, Nelson RF (2018) Association of obstructive sleep apnea with calvarial and skull base thinning. JAMA Otolaryngol Head Neck Surg 144:513–518. https://doi.org/10.1001/jamaoto.2018.0347
Binder DK, Horton JC, Lawton MT, McDermott MW (2004) Idiopathic intracranial hypertension. Neurosurgery 54:538–552. https://doi.org/10.1227/01.NEU.0000109042.87246.3C
Friedman DI, Jacobson DM (2002) Diagnostic criteria for idiopathic intracranial hypertension. Neurology 59:1492–1495. https://doi.org/10.1212/01.WNL.0000029570.69134.1B
Friedman DI, Liu GT, Digre KB (2013) Revised diagnostic criteria for the pseudotumor cerebri syndrome in adults and children. Neurology 81:1159–1165. https://doi.org/10.1212/WNL.0b013e3182a55f17
Bidot S, Levy JM, Saindane AM, Oyesiku NM, Newman NJ, Biousse V (2019) Do most patients with a spontaneous cerebrospinal fluid leak have idiopathic intracranial hypertension? J Neuroophthalmol 39:487–495. https://doi.org/10.1097/WNO.0000000000000761
Scoffings DJ (2021) Imaging of acquired skull base cerebrospinal fluid leaks. Neuroimaging Clin N Am 31:509–522. https://doi.org/10.1016/j.nic.2021.05.009
Bidot S, Saindane AM, Peragallo JH, Bruce BB, Newman NJ, Biousse V (2015) Brain imaging in idiopathic intracranial hypertension. J Neuroophthalmol 35:400–411. https://doi.org/10.1097/WNO.0000000000000303
Kwee RM, Kwee TC (2019) Systematic review and meta-analysis of MRI signs for diagnosis of idiopathic intracranial hypertension. Eur J Radiol 116:106–115. https://doi.org/10.1016/j.ejrad.2019.04.023
Agid R, Farb RI, Willinsky RA, Mikulis DJ, Tomlinson G (2006) Idiopathic intracranial hypertension: the validity of cross-sectional neuroimaging signs. Neuroradiology 48:521–527. https://doi.org/10.1007/s00234-006-0095-y
Maralani PJ, Hassanlou M, Torres C, Chakraborty S, Kingstone M, Patel V, Zackon D, Bussière M (2012) Accuracy of brain imaging in the diagnosis of idiopathic intracranial hypertension. Clin Radiol 67:656–663. https://doi.org/10.1016/j.crad.2011.12.002
Goddard JC, Meyer T, Nguyen S, Lambert PR (2010) New considerations in the cause of spontaneous cerebrospinal fluid otorrhea. Otol Neurotol 31:940–945. https://doi.org/10.1097/MAO.0b013e3181e8f36c
Quatre R, Attye A, Righini C et al (2017) Spontaneous cerebrospinal fluid rhinorrhea: association with body weight and imaging data. J Neurol Surg Part B Skull Base 78:419–424. https://doi.org/10.1055/s-0037-1603731
Allen KP, Perez CL, Kutz JW, Reyt E, Giai J, Schmerber S, Karkas A (2014) Elevated intracranial pressure in patients with spontaneous cerebrospinal fluid otorrhea. Laryngoscope 124:251–254. https://doi.org/10.1002/lary.24251
Stevens SM, Rizk HG, Golnik K, Andaluz N, Samy RN, Meyer TA, Lambert PR (2018) Idiopathic intracranial hypertension: contemporary review and implications for the otolaryngologist. Laryngoscope 128:248–256. https://doi.org/10.1002/lary.26581
Kuzan BN, Ilgın C, Kuzan TY, Dericioğlu V, Kahraman-Koytak P, Uluç K, Çimşit NÇ (2022) Accuracy and reliability of magnetic resonance imaging in the diagnosis of idiopathic intracranial hypertension. Eur J Radiol 155:110491. https://doi.org/10.1016/j.ejrad.2022.110491
Brainard L, Chen DA, Aziz KM, Hillman TA (2012) Association of benign intracranial hypertension and spontaneous encephalocele with cerebrospinal fluid leak. Otol Neurotol 33:1621–1624. https://doi.org/10.1097/MAO.0b013e318271c312
Alonso RC, De La Peña MJ, Caicoya AG, Rodriguez MR, Moreno EA, de Vega Fernandez VM (2013) Spontaneous skull base meningoencephaloceles and cerebrospinal fluid fistulas. Radiographics 33:553–570. https://doi.org/10.1148/rg.332125028
Aaron G, Doyle J, Vaphiades MS, Riley KO, Woodworth BA (2014) Increased intracranial pressure in spontaneous CSF leak patients is not associated with papilledema. Otolaryngol Head Neck Surg 151:1061–1066. https://doi.org/10.1177/0194599814551122
Davidson B, Nassiri F, Mansouri A, Badhiwala JH, Witiw CD, Shamji MF, Peng PW, Farb RI, Bernstein M (2017) Spontaneous intracranial hypotension: a review and introduction of an algorithm for management. World Neurosurg 101:343–349. https://doi.org/10.1016/j.wneu.2017.01.123
Naples JG, Shah RR, Ruckenstein MJ (2019) The evolution of presenting signs and symptoms of lateral skull base cerebrospinal fluid leaks. Curr Opin Otolaryngol Head Neck Surg 27:344–348. https://doi.org/10.1097/MOO.0000000000000561
Schlosser RJ, Wilensky EM, Grady MS, Bolger WE (2003) Elevated intracranial pressures in spontaneous cerebrospinal fluid leaks. Am J Rhinol 17:191–195
Nelson RF, Hansen KR, Gantz BJ, Hansen MR (2015) Calvarium thinning in patients with spontaneous cerebrospinal fluid leak. Otol Neurotol 36:481–485. https://doi.org/10.1097/MAO.0000000000000552
Stevens SM, Lambert PR, Rizk H, McIlwain WR, Nguyen SA, Meyer TA (2015) Novel radiographic measurement algorithm demonstrating a link between obesity and lateral skull base attenuation. Otolaryngol Neck Surg 152:172–179. https://doi.org/10.1177/0194599814557470
Rizk HG, Hatch JL, Stevens SM, Lambert PR, Meyer TA (2016) Lateral skull base attenuation in superior semicircular canal dehiscence and spontaneous cerebrospinal fluid otorrhea. Otolaryngol Neck Surg 155:641–648. https://doi.org/10.1177/0194599816651261
Hatch JL, Schopper H, Boersma IM, Rizk HG, Nguyen SA, Lambert PR, McRackan TR, Meyer TA (2018) The bone mineral density of the lateral skull base and its relation to obesity and spontaneous cerebrospinal fluid leaks. Otol Neurotol 39:e831–e836. https://doi.org/10.1097/MAO.0000000000001969
Yancey KL, Manzoor NF, Kelly PD, Yawn RJ, O’Malley M, Rivas A, Haynes DS, Bennett ML (2020) Impact of obesity and obstructive sleep apnea in lateral skull base cerebrospinal fluid leak repair. Laryngoscope 130:2234–2240. https://doi.org/10.1002/lary.28421
Sharma D, Tucker BJ, Millay DS, Rubel KE, Saltagi M, Nelson RF, Illing EA, Ting JY (2021) Calvarium thinning in patients with spontaneous cerebrospinal fluid leaks of the anterior skull base. Laryngoscope 131:1271–1276. https://doi.org/10.1002/lary.29130
Yancey KL, Manzoor NF, Yawn RJ, O’Malley M, Rivas A, Bennett ML, Haynes DS (2021) Cerebrospinal fluid leaks of the posterior fossa: patient characteristics and imaging features. J Neurol Surg Part B Skull Base 82:345–350. https://doi.org/10.1055/s-0039-1697976
Barke M, Castro HM, Adesina OO, Chuang AZ, Do T, Patel RP, Richani K (2022) Thinning of the skull base and calvarial thickness in patients with idiopathic intracranial hypertension. J Neuro-Ophthalmol 42(2):192–198. https://doi.org/10.1097/WNO.0000000000001504
Butros SR, Goncalves LF, Thompson D, Agarwal A, Lee HK (2012) Imaging features of idiopathic intracranial hypertension, including a new finding: widening of the foramen ovale. Acta Radiol 53:682–688. https://doi.org/10.1258/ar.2012.110705
Curry SD, Granger KW, Richman EH, Liu C, Moore GF, Barnes CA, Surdell DL, Hatch JL (2022) Expansion of the foramen ovale in patients with cerebrospinal fluid leak or encephalocele. Otol Neurotol 43:845–851. https://doi.org/10.1097/MAO.0000000000003583
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, 1st edn. Springer, Cham, pp 234–241
Chang PD, Kuoy E, Grinband J, Weinberg BD, Thompson M, Homo R, Chen J, Abcede H, Shafie M, Sugrue L, Filippi CG, Su MY, Yu W, Hess C, Chow D (2018) Hybrid 3D/2D convolutional neural network for hemorrhage evaluation on head CT. Am J Neuroradiol 39:1609–1616. https://doi.org/10.3174/ajnr.A5742
Song G, Nie Y, Zhang J, Chen G (2020) Research on the fusion method of 2D and 3D UNet in pulmonary nodules segmentation task. 2020 International Conference on Computer Science and Management Technology (ICCSMT). IEEE, Shanghai, China, pp 44–47
Wang G, Li W, Ourselin S, Vercauteren T (2018) Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In: Crimi A, Bakas S, Kuijf H et al (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 178–190
Huff TJ, Ludwig PE, Salazar D, Cramer JA (2019) Fully automated intracranial ventricle segmentation on CT with 2D regional convolutional neural network to estimate ventricular volume. Int J Comput Assist Radiol Surg 14:1923–1932. https://doi.org/10.1007/s11548-019-02038-5
Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R (2012) 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30:1323–1341. https://doi.org/10.1016/j.mri.2012.05.001
Kikinis R, Pieper SD, Vosburgh KG (2014) 3D Slicer: a platform for subject-specific image analysis, visualization, and clinical support. In: Jolesz FA (ed) Intraoperative Imaging and Image-Guided Therapy. Springer, New York, New York, NY, pp 277–289
The MONAI Consortium. (2021) Project MONAI. https://github.com/Project-MONAI. Accessed November 14. https://github.com/Project-MONAI.
Tam EK, Gilbert AL (2019) Spontaneous cerebrospinal fluid leak and idiopathic intracranial hypertension. Curr Opin Ophthalmol 30:467–471. https://doi.org/10.1097/ICU.0000000000000603
Lobo BC, Baumanis MM, Nelson RF (2017) Surgical repair of spontaneous cerebrospinal fluid (CSF) leaks: a systematic review. Laryngoscope Investig Otolaryngol 2:215–224. https://doi.org/10.1002/lio2.75
Chaaban MR, Illing E, Riley KO, Woodworth BA (2014) Spontaneous cerebrospinal fluid leak repair: a 5-year prospective evaluation. Laryngoscope 124:70–75. https://doi.org/10.1002/lary.24160
Vivas EX, Mccall A, Raz Y, Fernandez-Miranda JC, Gardner P, Hirsch BE (2014) ICP, BMI, surgical repair, and CSF diversion in patients presenting with spontaneous CSF otorrhea. Otol Neurotol 35:344–347. https://doi.org/10.1097/MAO.0b013e3182a473cf
Teachey W, Grayson J, Cho D-Y, Riley KO, Woodworth BA (2017) Intervention for elevated intracranial pressure improves success rate after repair of spontaneous cerebrospinal fluid leaks. Laryngoscope 127:2011–2016. https://doi.org/10.1002/lary.26612
Chilamkurthy S, Ghosh R, Tanamala S, Biviji M, Campeau NG, Venugopal VK, Mahajan V, Rao P, Warier P (2018) Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. The Lancet 392:2388–2396. https://doi.org/10.1016/S0140-6736(18)31645-3
Wang X, Shen T, Yang S, Lan J, Xu Y, Wang M, Zhang J, Han X (2021) A deep learning algorithm for automatic detection and classification of acute intracranial hemorrhages in head CT scans. NeuroImage Clin 32:102785. https://doi.org/10.1016/j.nicl.2021.102785
Lee JY, Kim JS, Kim TY, Kim YS (2020) Detection and classification of intracranial haemorrhage on CT images using a novel deep-learning algorithm. Sci Rep 10:20546. https://doi.org/10.1038/s41598-020-77441-z
Qiu S, Miller MI, Joshi PS, Lee JC, Xue C, Ni Y, Wang Y, De Anda-Duran I, Hwang PH, Cramer JA, Dwyer BC, Hao H, Kaku MC, Kedar S, Lee PH, Mian AZ, Murman DL, O’Shea S, Paul AB, Saint-Hilaire MH, Alton Sartor E, Saxena AR, Shih LC, Small JE, Smith MJ, Swaminathan A, Takahashi CE, Taraschenko O, You H, Yuan J, Zhou Y, Zhu S, Alosco ML, Mez J, Stein TD, Poston KL, Au R, Kolachalama VB (2022) Multimodal deep learning for Alzheimer’s disease dementia assessment. Nat Commun 13:3404. https://doi.org/10.1038/s41467-022-31037-5
Supuran CT (2015) Acetazolamide for the treatment of idiopathic intracranial hypertension. Expert Rev Neurother 15:851–856. https://doi.org/10.1586/14737175.2015.1066675
Stevens SM, Smith CJ, Lawton M (2019) Postoperative management of patients with spontaneous cerebrospinal fluid leak. Curr Opin Otolaryngol Head Neck Surg 27:361–368. https://doi.org/10.1097/MOO.0000000000000559
Funding
No funding was received for conducting this study.
Author information
Authors and Affiliations
Contributions
Steven D. Curry: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing; Kieran S. Boochoon: Data curation, Investigation, Writing – review & editing; Geoffrey C. Casazza: Methodology, Investigation, Resources, Writing – review & editing; Daniel L. Surdell: Methodology, Investigation, Resources, Writing – review & editing; Justin A. Cramer: Conceptualization, Data curation, Investigation, Methodology, Resources, Software, Writing – review & editing.
Corresponding author
Ethics declarations
Conflict of interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of the University of Nebraska Medical Center (June 5, 2019/IRB# 412–19-EX).
Informed consent
Informed consent was waived by the IRB due to the retrospective study design without publication of individually identifiable data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Curry, S.D., Boochoon, K.S., Casazza, G.C. et al. Deep learning to predict risk of lateral skull base cerebrospinal fluid leak or encephalocele. Int J CARS (2024). https://doi.org/10.1007/s11548-024-03259-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11548-024-03259-z