Abstract
Purpose
Parkinson disease (PD) is a common progressive neurodegenerative disorder in our ageing society. Early-stage PD biomarkers are desired for timely clinical intervention and understanding of pathophysiology. Since one of the characteristics of PD is the progressive loss of dopaminergic neurons in the substantia nigra pars compacta, we propose a feature extraction method for analysing the differences in the substantia nigra between PD and non-PD patients.
Method
We propose a feature-extraction method for volumetric images based on a rank-1 tensor decomposition. Furthermore, we apply a feature selection method that excludes common features between PD and non-PD. We collect neuromelanin images of 263 patients: 124 PD and 139 non-PD patients and divide them into training and testing datasets for experiments. We then experimentally evaluate the classification accuracy of the substantia nigra between PD and non-PD patients using the proposed feature extraction method and linear discriminant analysis.
Results
The proposed method achieves a sensitivity of 0.72 and a specificity of 0.64 for our testing dataset of 66 non-PD and 42 PD patients. Furthermore, we visualise the important patterns in the substantia nigra by a linear combination of rank-1 tensors with selected features. The visualised patterns include the ventrolateral tier, where the severe loss of neurons can be observed in PD.
Conclusions
We develop a new feature-extraction method for the analysis of the substantia nigra towards PD diagnosis. In the experiments, even though the classification accuracy with the proposed feature extraction method and linear discriminant analysis is lower than that of expert physicians, the results suggest the potential of tensorial feature extraction.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Parkinson disease (PD) is a common progressive neurodegenerative disorder in our ageing society. Approximately 8.5 million people have been affected worldwide in 2017 [1]. The diagnosis of PD depends on the clinical features acquired from the patient’s history and neurological examination [2]. Available treatments for PD are still only for symptomatic relief and fail to stop the neurodegeneration progress. Early-stage PD biomarkers are desired for timely clinical intervention and understanding of pathophysiology [3]. Several advanced imaging markers have emerged as tools for visualising neuro-anatomic and functional processes in PD. As one of them, neuromelanin-sensitive MRI uses high-spatial-resolution T1-weighted imaging with fast spin-echo sequences at 3-Tesla MRI [4, 5]. This imaging provides a neuromelanin image with neuromelanin-sensitive contrast, and T1 high-signal-intensity areas in the midbrain represent neuromelanin-rich areas. One characteristic of PD is progressive loss of dopaminergic neurons in the substantia nigra pars compacta [6]. Furthermore, neuromelanin exists only in dopaminergic neurons of the substantia nigra pars compacta and noradrenergic neurons of locus coeruleus. Therefore, a neuromelanin image is useful for analysing the substantia nigra by capturing early PD-related changes. Figure 1 shows examples of a neuromelanin image and its annotation label indicating the substantia nigra.
To analyse the substantia nigra as volumetric data, which enables us to analyse shape and texture information simultaneously, our group tackled the automated volumetric segmentation by deep learning models [7, 8] and the statistical analysis by higher-order tensor decomposition [9]. For the segmentation, by using neuromelanin images, we achieved a more accurate segmentation [8] than T2-weighted images [7]. In the statistical analysis [9], we found that T1-weighted and neuromelanin images share similar three-dimensional patterns in a fifth-order tensor decomposition of multisequence of T1-weighted, T2-weighted and neuromelanin images. Furthermore, in the decomposition, we found that three-dimensional patterns of the substantia nigra in neuromelanin images are more compressible than T1- and T2-weighted images. These previous results indicate that we can obtain the small number of informative features of the substantia nigra from neuromelanin images. In related works, even though a deep learning model with an large dataset of T1-weighted images of the whole brain has been reported [10], the analysis of the substantia nigra on a neuromelanin image has been reported only for a small dataset of 55 patients [11]. Furthermore, this analysis manually selected a rectangular region around the substantia nigra on the axial slices of and input to a two-dimensional convolutional neural network. A volumetric data analysis for the substantia nigra is still an open problem.
We propose a new feature-extraction method to analyse PD-related patterns in neuromelanin images of the substantia nigra. Exploring discriminative features for classifying the substantia nigra between PD and non-PD is essential for finding PD biomarkers. As a higher-order tensor expression of volumetric images [12], we express a set of volumetric images of the extracted substantia nigra as a fourth-order tensor. Inspired by tensor-based analytical methods [13,14,15], we decompose this into a linear combination of fourth-order rank-1 tensors. By re-ordering the elements of this decomposition result, we obtain each decomposed volumetric image by identical third-order rank-1 tensors. Since decompositions of each image are based on the identical third-order rank-1 tensors, a set of weights expresses the characteristics of a volumetric image. Therefore, by selecting discriminative weights for classifying PD and non-PD as a feature vector, we achieve a feature extraction for analysing patterns of the substantia nigra. Our tensorial feature extraction offers the visualisation of discriminative patterns by reconstructing an volumetric image from selected features.
Methods
Tensor expression
In tensor algebra, the number of dimensions is referred to as the order of a tensor. We set an M-th-order tensor \(\mathcal {A} \in \mathbb {R}^{I_1 \times I_2 \times \dots \times I_M}\). An element with indices \((i_1, i_2,i_3, \dots , i_M)\) in \(\mathcal {A}\) is denoted by \(a_{i_1 i_2 \dots i_M}\). The index of a tensor is referred to as the mode of a tensor. For example, \(i_3\) is the index for mode 3.
An M-th-order tensor \(\mathcal {A}\) is a rank one if it can be expressed by M vectors by
where \(\circ \) expresses the outer product. Furthermore, we have a norm of a tensor as
Expressing data as a tensor, we can decompose it into a linear combination of rank-1 tensors and evaluate its reconstruction error with the norm.
Tensorial feature extraction
We propose a feature extraction method for classifying volumetric images between two categories. This work assumes that three-dimensional patterns of volumetric images can be decomposed into common three-dimensional patterns if volumes of interest (VOIs) are extracted and pre-registered to an appropriate coordinate system for their analysis. Setting a volumetric image to a third-order tensor \(\mathcal {X} \in \mathcal {R}^{I_1 \times I_2 \times I_3}\), we have a linear combination of R patterns such that
where a rank-1 third-order tensor \(\mathcal {Y}_r \) and scalar \(f_r\) express a decomposed three-dimensional pattern and its coefficient, respectively. Figure 2 illustrates our tensor expression of volumetric images. Since \(f_r\) indicates the importance of the r-th decomposed three-dimensional pattern \(\mathcal {Y}_r\), we can express the three-dimensional pattern of \(\mathcal {X}\) by a feature vector
For the practical computation of the above feature extraction, we introduce nonnegative tensor factorisation, an extension of canonical polyadic (CP) decomposition.
Canonical polyadic decomposition
Setting \(\mathcal {X}_{j} \in \mathbb {R}^{I_1 \times I_2 \times I_3}\) for \(j=1, 2, \dots , J\) to be volumetric images in a training dataset, we express these J volumetric images as the fourth-order tensor \(\mathcal {T} \in \mathbb {R}^{I_1 \times I_2 \times I_3 \times I_4 }\), where we set \(I_4 = J\). For \(\mathcal {T}\), we have a CP decompositionFootnote 1 [14,15,16]
by minimising a reconstruction error \(\Vert \mathcal {E} \Vert \). The number of rank-1 tensors in tensor decomposition is referred to as the CP rank of a tensor. Note that there is no orthogonal constraint for \(\varvec{u}_r^{(n)}, r=1,2,\dots , R\) for an alternative-least square solution [16,17,18] in a CP decomposition. Since a reconstructed volume image is given by
where \(u_{jr}^{(4)}\) is the j-th element of \(\varvec{u}_{r}^{(4)}\), we have a feature vector
of an image \(\mathcal {X}_j\) in a training set.
Furthermore, setting \(\check{\mathcal {X}}_{k} \in \mathbb {R}^{I_1 \times I_2 \times I_3}\) for \(k=1, 2, \dots , K\) to volumetric images in a testing dataset, we express these K volumetric images as a fourth-order tensor \(\mathcal {Q} \in \mathbb {R}^{I_1 \times I_2 \times I_3 \times \check{I}_4}\), where we set \(\check{I}_4 = K\). By computing only \(\check{\varvec{U}}^{(4)} = [\check{\varvec{u}}^{(4)}_1, \check{\varvec{u}}^{(4)}_2, \dots , \check{\varvec{u}}^{(4)}_R ]\) with the obtained matrices \(\{ \varvec{U}^{(n)} = [\varvec{u}^{(n)}_1, \varvec{u}^{(n)}_2,\) \(\dots , \varvec{u}^{(n)}_R ] \}_{n=1}^3\) in Eq. (5), we can decompose volumetric images of a testing dataset with the three-dimensional patterns in a training dataset. As a result, we have a feature vector
of an image \(\check{\mathcal {X}_k}\) in a testing set.
Nonnegative tensor factorisation
We introduce nonnegative tensor factorisation (NTF) [13, 15, 16] into feature extraction. NTF is a simple extension of CP decomposition. We add nonnegative constraint \(\varvec{U}^{(n)} = [\varvec{u}^{(n)}_1, \varvec{u}^{(n)}_2, \dots , \varvec{u}^{(n)}_R ] \in \mathbb {R}_{+}^{I_n \times R}\) for \(n=1,2,3,4\), where \(\mathbb {R}_{+}\) is a set of nonnegative real values, into a decomposition in Eq. (5). In the practical computation of NTF, we iteratively update each element with a gradient-based method from initial random nonnegative values. For a decomposition of a testing dataset, we add a nonnegative constraint \(\check{U}^{(4)} \in \mathbb {R}_{+}^{\check{I}_4 \times R}\). Using the Khatori-Rao product \(\odot \), Hadamard product \(*\) and Hadamard division \(\oslash \) [15, 16, 19], Algorithms 1 and 2 summarise NTF for training and test datasets, respectively. From the results of Algorithms 1 and 2, we have \(\{ \varvec{f}_j \}_{j=1}^J\) and \(\{ \check{\varvec{f}}_k \}_{k=1}^K\).
Feature selection
We integrate the feature-selection process [20] into our method to capture discriminative features for two-category classification. For a set of extracted features \(\{ \varvec{f}_j \}_{j=1}^{J}\) of a training dataset with the condition \(R < J\), we assume each feature belongs to either the category \(\mathcal {C}_1\) or \(\mathcal {C}_2\). Therefore, features are divided into two sets, \(\{ \varvec{f}_{1i} \}_{i=1}^{N_1}\) and \(\{ \varvec{f}_{2i}^{} \}_{i=1}^{N_2}\), where \(N_1\) and \(N_2\) represent the number of images in the first and second categories, respectively. Setting \(\varvec{F}_l = [\varvec{f}_{l1}, \varvec{f}_{l2}, \dots , \varvec{f}_{l N_l} ]\) for \(l=1,2\), we have autocorrelation matrices \(\varvec{A}_1 = \frac{1}{N_1} \varvec{F}_1 \varvec{F}_1^{\top }\) and \(\varvec{A}_2 = \frac{1}{N_2} \varvec{F}_2\varvec{F}_2^{\top }\). These two matrices introduce an autocorrelation matrix of all the features by
where we set \(P(\mathcal {C})_1= \frac{N_1}{N_1+N_2}\) and \({P(\mathcal {C}_2)} = \frac{N_2}{N_1+N_2}\). For \(\varvec{A}\), we have the eigendecomposition \(\varvec{A} = \varvec{V}\varvec{\Xi }\varvec{V}^{\top }\), where \(\varvec{\Xi }= \textrm{diag}(\xi _1,\xi _2,\dots ,\xi _R)\) consists of eigenvalues \(\xi _i\) for \(i=1,2, \dots , R\) with the condition \(\xi _1\ge \xi _2 \ge \dots \ge \xi _{R} > 0\). This eigendecomposition derives a whitening matrix \(\varvec{W} = \varvec{\Xi }^{-\frac{1}{2}}\varvec{V}^{\top }\).


From the whitening matrix \(\varvec{W}\) and Eq. (9), we have
where \(\varvec{I}\) is an identity matrix. The solutions of the eigenvalue problems \( \tilde{\varvec{A}}_l \varvec{\phi }_{l,i} = \lambda _{l,i} \varvec{\phi }_{l,i}\) and Eq. (10) derive relations \(\tilde{\varvec{A}}_2 \varvec{\phi }_{2,i} = ( \varvec{I} - \tilde{\varvec{A}}_1 ) \varvec{\phi }_{2,i}\) and \(\tilde{\varvec{A}_1} \varvec{\phi }_{2,i} =(1 - \lambda _{2,i} ) \varvec{\phi }_{2,i} \) for \(l =1,2\). These lead to
and
for \(i=1,2, \dots , R\) and \(l =1,2\). These relations imply that the eigenvectors corresponding to the large eigenvalues of \(\tilde{\varvec{A}}_1\) contribute to expressing \(\mathcal {C}_1\), although they make a minor contribution to expressing \(\mathcal {C}_2\).
In this work, setting \(\varvec{\Phi } = [\varvec{\phi }_{1,1}, \varvec{\phi }_{1,2}, \dots , \varvec{\phi }_{1,d}]\), we project extracted feature vectors by
to select \(\mathcal {C}_1\)-related features.
Classification and its visual interpretation
We classify a feature-selected vector and reconstruct its three-dimensional pattern for visual interpretation. For the classification of \(\varvec{g} \in \{ \varvec{g}_j \}_{j=1}^J \cup \{ \check{\varvec{g}}_k \}_{k=1}^K\), we have a discriminant function
where \(\varvec{w}=[w_1, w_2, \dots , w_d]^{\top }\) and \(w_0\) are a weight vector and thresholding criterion, respectively. We compute \(\varvec{w}\) by Fisher’s linear discriminant analysis (LDA) [21] from the feature-selected vectors of a training dataset. Setting means \(\mu _1\) and \(\mu _2\) of projected feature vectors in \(\mathcal {C}_1\) and \(\mathcal {C}_2\), respectively, in a projected space given by \(\varvec{w}\), we define \(w_0 = \lambda \mu _1 + (1 - \lambda ) \mu _2 \) with a hyperparameter \(\lambda \in [0, 1]\). Using Eq. (15), we classify \(\varvec{g}\) as follows:
We set \(\tilde{g}_p\) with an index \(p=1, 2, \dots , d\) to the pth element of \( \tilde{\varvec{g}}=\varvec{w} \odot \varvec{g}\). Since positive elements of \(\tilde{\varvec{g}}\) show the selected features’ relevance to \(\mathcal {C}_1\), we have \(\varvec{g}^{+} = (g^{+}_p)\) by filtering
Using an element \(f_r^{+}\) of \(\varvec{f}^{+}=\varvec{\Phi }\varvec{g}^{+}\), we can reconstruct \(\mathcal {C}_1\)-related three-dimensional patterns in \(\mathcal {X} \in \{ \mathcal {X}_j \}_{j=1}^J \cup \{ \check{\mathcal {X}}_k \}_{k=1}^K\) from its feature-selected vector \(\varvec{g}\) by
where ReLU replaces negative values in an input tensor with zero.
Experiments
Datasets
Patients diagnosed with PD based on the UK PD Society Brain Bank clinical criteria and healthy subjects without any neurological diseases [22], who visited Juntendo Hospital between 2019 and 2022, were recruited with IRB approval. Using a 3T scanner (MAGNETOM Prisma, Siemens Healthcare), we collected neuromelanin images of 263 patients: 124 PD and 139 non-PD patients with additional spectral presaturation inversion-recovery pulses [2]. General scan parameters were as follows: 600/12 ms repetition time/echo time; echo train length of 14; 2.5 mm slice thickness; 0.5 mm slice gap; 3.0 mm spacing between slices; 512 \(\times \) 359 acquisition matrix; 220 \(\times \) 220 mm field of view (0.43 \(\times \)0.43 mm pixel size); 175 Hz/pixel bandwidth, three-averages; 7:15 min of total scan time.
In this collection, first, we obtained T1-weighted images by setting AC-PC lines to be parallel against axial slices in each patient. The size of a T1-weighted image is \(224 \times 300 \times 320\) voxels, each of which is 0.80 mm \(\times \) 0.80 mm \(\times \) 0.80 mm. Second, we obtained neuromelanin images for all the patients. The size of a neuromelanin image is \(512 \times 512 \times 14\) voxels, each of which is 0.43 mm \(\times \) 0.43 mm \(\times \) 3.00 mm. Third, for each patient, we registered a neuromelanin image to a T1-weighted image by rigid transform, where we used the FMRIB software library’s linear registration tool [23].
For these neuromelanin images, a board-certified radiologist with ten years of experience specialising in neuroradiology annotated regions of the substantia nigra by using MRIcron [24]. Based on the annotations, we cropped the VOIs of the substantia nigra from the neuromelanin images. Setting the centre of a VOI to be the centre of gravity in a region of the substantia nigra, we expressed a VOI of each sequence as third-order tensors of \(64 \times 64 \times 64\) voxels. In each third-order tensor, after setting elements of the outer region of substantia nigra to be zero, we normalised all the elements of each third-order tensor in the range of [0, 1]. We randomly divided these images into training and testing datasets, as summarised in Table 1. Figure 3 summarises the age distributions in our datasets.
Evaluation of substantia nigra classification
To evaluate the proposed feature-extraction method, we classify extracted feature vectors by a linear function \(g(\varvec{f}) = \varvec{w}^{\top }\varvec{f} +w_0\), where \(\varvec{f} \in \{ \varvec{f}_j \}_{j=1}^J \cup \{ \check{\varvec{f}}_k \}_{k=1}^K\). We extract feature vectors from training and testing datasets by Algorithms 1 and 2, respectively, for \(R=64, 155, 250, 500, 1000,2000, 4000\). Figure 4a shows CP-rank and mean relative reconstruction error \(\mathbb {E}(\Vert \mathcal {X} - \sum _{r=1}^R f_r \mathcal {Y}_r \Vert / \Vert \mathcal {X} \Vert )\) for each CP rank. Using extracted features for the training dataset, we computed \(\varvec{w}\) and \(w_0\) in the same manner described in Sect. Methods. As the evaluation metric of classification accuracy, we set sensitivity and specificity to the ratios of correctly classified images in PD and non-PD categories, respectively. Figure 5 summarises classification accuracy for training and testing datasets for different \(\lambda \).
To evaluate the validity of our feature selection, we computed classification accuracy for the selected features with the procedure described in Sect. Methods. Figure 6 shows the classification accuracy with feature selection for \(R=64\) and \(d=7, 15, 18\). Figure 5b illustrates the eigenvalue distribution of the normalised autocorrelation matrix for the PD category. Figure 7 presents examples of reconstructed volumetric images before and after feature selection. For the reconstruction from selected features, we used Eq. (18).
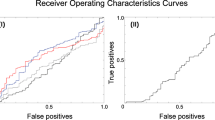
For a comparative evaluation, we computed the classification accuracy of a support vector machine (SVM) [25]. We trained a linear SVM model using the extracted feature vectors of the training dataset without the feature selection. For training a linear SVM model, we set the weight of the regularisation to one and used the balanced weight \((N_1+N_2)/2N_l\) for the lth category. To enable SVM to output estimated probability for each category, we adopted Platt’s likelihood [26]. If an SVM’s output probability for the PD category exceeds a criterion, we classify input as \(\mathcal {C}_1\). Using the criterion from 0.00 to 1.00 with the step size of 0.05, we plotted the ROC curves of training and testing datasets for the linear-SVM model. For the computation of SVM, we used scikit-learn. Figure 8 summarises the comparative evaluation between LDA with feature selection (ours) and SVM without feature selection.
To further explore our approach, we performed fivefold cross-validation by splitting all the data into training and testing datasets. In training steps in each fold, we selected features and computed \(\varvec{w}\) for \(d=\)8, 12, 16, 20, 24, 28, 32, 48, 64. Figure 9 shows the results of the fivefold cross-validation.
Discussion
Figure 5 indicates that increasing the CP rank results in the extraction of indiscriminative features for unseen images in the substantia nigra classification, even though Fig. 4a shows that increasing the CP rank contributes to decreasing the reconstruction error. For the given 155 images in the training dataset, Fig. 5 shows that CP ranks from 64 to 500 are acceptable for the classification. For these CP ranks, Fig. 5b shows insufficient sensitivities and specificities of approximately 0.60. NTF was originally designed only for analysis, not for classification. Therefore, these results imply the necessity of feature selection to capture category-specific discriminative features.
Figure 4b shows that fewer than 18 eigenvectors express essential patterns for the PD category. As shown in Fig. 6, our feature selection using the appropriate eigenvectors improved the sensitivity and specificity of the testing dataset. Figure 8 illustrates the higher generalisation ability of our approach than the linear SVM. Even though there is a gap in classification accuracy between training and testing datasets for the linear SVM, our proposed method achieved the small one. The proposed method achieved a sensitivity of 0.72 and a specificity of 0.64 for training and testing datasets.
We also presented the reconstruction of the selected features in Fig. 7. Generally, the pars compacta of the substantia nigra is divided into ventral and dorsal tiers, and each tier is further subdivided into medial to lateral regions. In PD, the ventrolateral tier of the substantia nigra loses first, and then the ventromedial tier also loses. Typically, the cells of 70–90% in the ventrolateral tier have been lost by the time a PD patient dies [27]. In Fig. 7, our feature selection captures three-dimensional patterns related to the ventrolateral tiers.
In Fig. 9, the three of fivefold show the feature selection’s improvement of sensitivity and specificity. These indicate the validity of the proposed method. However, the optimal number of the feature selection for those folds differs. Furthermore, for the other folds, the feature selection does not work. These results imply that more data collection is needed. In the case of a sufficiently large dataset, after randomly removing 10–25% of the dataset, we can have approximately the same distribution shape as the original dataset. If we randomly remove 10–25% of a small dataset, we might have a different distribution shape from the original dataset. To achieve expert-level precise classification [28, 29] of the substantia nigra between PD and non-PD, we are required to capture more discriminative features from a large dataset.
Conclusions
This paper proposed a tensorial feature-extraction method for substantia nigra analysis towards PD diagnosis. Since category-specific features should be discriminative, we evaluated the accuracy of classifying the substantia nigra between PD and non-PD cases. The experimental evaluations demonstrated the method’s validity, where the classification accuracy is acceptable as a preliminary study. Unlike deep learning approaches, our proposed method directly reconstructs important patterns to interpret classification results visually.
Notes
This decomposition is also called as CANDECOMP, PARAFAC or CANDECOMP/PARAFAC (CP).
References
James SL, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, Abbastabar H, Abd-Allah F, Abdela J, Abdelalim A, Abdollahpour I, Abdulkader RS, Abebe Z, Abera SF, Abil OZ, Abraha HN, Abu-Raddad LJ, Abu-Rmeileh NME, Accrombessi MMK, Acharya D et al (2018) Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the global burden of disease study 2017. The Lancet 392(10159):1789–1858
Berre AL, Kamagata K, Otsuka Y, Andica C, Hatano T, Saccenti L, Ogawa T, Takeshige-Amano H, Wada A, Suzuki M, Hagiwara A, Irie R, Hori M, Oyama G, Shimo Y, Umemura A, Hattori N, Aoki S (2019) Convolutional neural network-based segmentation can help in assessing the substantia nigra in neuromelanin MRI. Neuroradiology 61:1387–1395
He N, Chen Y, LeWitt PA, Yan F, Haacke EM (2023) Application of neuromelanin MR imaging in Parkinson disease. J Magn Resonance Imaging 57:337–352
Sasaki M, Shibata E, Tohyama K (2006) Neuromelanin magnetic resonance imaging of locus ceruleus and substantia nigra in Parkinson’s disease. Neuro Rep 17(11):1215–1218
Kashihara K, Shinya T, Higaki F (2011) Neuromelanin magnetic resonance imaging of nigral volume loss in patients with Parkinson’s disease. J Clin Neurosci 18(8):1093–1096
Drui G, Carnicella S, Carcenac C, Favier M, Bertrand A, Boulet S, Savasta M (2014) Loss of dopaminergic nigrostriatal neurons accounts for the motivational and affective deficits in Parkinsons disease. Mol Psychiatry 19:358–367
Hu T, Itoh H, Oda M, Hayashi Y, Lu Z, Saiki S, Hattori N, Kamagata K, Aoki S, Kumamaru KK, Akashi T, Mori K (2022) Enhancing model generalization for substantia nigra segmentation using a test-time normalization-based method. In: Proceedings of the 25th international conference on medical image computing and computer assisted intervention, LNCS 13437: 736–744
Hu T, Itoh H, Oda M, Saiki S, Hattori N, Kamagata K, Aoki S, Mori K (2023) Priority attention network with Bayesian learning for fully automatic segmentation of substantia Nigra from neuromelanin MRI. In: Proceedings of the SPIE medical imaging 2023: image processing SPIE 12464: 124643G
Itoh H, Hu T, Oda M, Saiki S, Kamagata K, Hattori N, Aoki S, Mori K (2022) Pattern analysis of substantia Nigra in Parkinson disease by fifth-order tensor decomposition and multi-sequence MRI. In: Proceedings of the 3rd international workshop on multiscale multimodal medical imaging LNCS 13594:63–75
Camacho M, Wilms M, Mouches P, Almgren H, Souza R, Camicioli R, Ismail Z, Monchi O, Forkert ND (2023) Explainable classification of Parkinson’s disease using deep learning trained on a large multi-center database of T1-weighted MRI datasets,. Neuro Image Clin 38:103405
Shinde S, Prasad S, Saboo Y, Kaushick R, Saini J, Pal PK, Ingalhalikar M (2019) Predictive markers for Parkinson’s disease using deep neural nets on neuromelanin sensitive MRI. NeuroImage: Clin 22:101748
Itoh H, Imiya A, Sakai T (2016) Pattern recognition in multilinear space and its applications: mathematics, computational algorithms and numerical validations. Mach Vis Appl 27:1259–1273
Smilde A, Bro R, Geladi P (2008) Multi-way analysis: applications in the chemical sciences, 1st edn. Wiley, New York
Kroonenberg PM (2008) Applied multiway data analysis, 1st edn. Wiley-Interscience, New York
Cichocki A, Zdunek R, Phan AH, Amari S (2009) Nonnegative matrix and tensor factorizations: applications to exploratory multi-way data analysis and blind source separation. Wiley, New York
Kolda TG, Bader BW (2009) Tensor decompositions and applications. SIAM Rev 51(3):455–500
Carroll J, Chang J-J (1970) Analysis of individual differences in multidimensional scaling via an n-way generalization of Eckart-Young decomposition. Psychometrika 35(3):283–319
Harshman RA (1970) Foundations of the PARAFAC procedure: Models and conditions for an "explanatory" multi-model factor analysis. UCLA Working Papers in Phonetics 16:1–84
Shashua A, Hazan T (2005) Non-negative tensor factorization with applications to statistics and computer vision. In: Proceedings of the international conference on machine learning, pp 792–799
Fukunaga K, Koontz WLG (1970) Application of the Karhunen-Loéve expansion to feature selection and ordering. IEEE Trans Comput c–19(4):311–318
Duda RO, Hart PE, Stork DH (2000) Pattern classification, 2nd edn. Wiley Interscience, New York
Hughes AJ, Susan ED, Linda K, Andrew JL (1992) Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg 55(3):181–184
Jenkinson M, Bannister P, Brady JM, Smith SM (2002) Improved optimisation for the robust and accurate linear registration and motion correction of brain images. NeuroImage 17(2):825–841. https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FLIRT
Chris R, Matthew B (2001) Stereotaxic display of brain lesions. Behav Neurol 12:192–200. https://www.nitrc.org/projects/mricron/
Vapnik V, Lerner A (1963) Pattern recognition using generalized portrait method. Autom Remote Control 24:774–780
Platt JC (1999) Probabilistic outputs for support vector machinesand comparisons to regularized likelihood methods. Adv Large Margin Classifier: 61-74
Ellison E, Chimelli L., Harding B, Love S, Lowe J, Roberts G, Vinters H (1998) Neuropathology. Mosby-Year Book
Rizzo G, Copetti M, Arcuti S, Martino D, Fontana A, Logroscino G (2016) Accuracy of clinical diagnosis of Parkinson disease. Neurology 86:566–576
Cho SJ, Bae YJ, Kim JM, Balik SH, Sunwoo L, Choi BS, Kim JH (2021) Diagnostic performance of neuromelanin-sensitive magnetic resonance imaging for patients with Parkinson’s disease and factor analysis for its heterogeneity: a systematic review and meta-analysis. Eur Radiol 31:1268–1280
Acknowledgements
This study was funded by grants from AMED (22dm0307101h0004), MEXT/JSPS KAKENHI (21K19898, 23K16900).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Mori K is supported by Cybernet Systems and Olympus (research grant) and by NTT outside the submitted work. The other authors have no conflict of interest.
Ethical approval
All the procedures performed in studies involving human participants were in accordance with the ethical committees of Nagoya University (No. 382) and Juntendo University (H19-179) and the 1964 Helsinki Declaration and subsequent amendments or comparable ethical standards. Informed consent was obtained by an opt-out procedure from all individual participants in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Itoh, H., Oda, M., Saiki, S. et al. Preliminary study of substantia nigra analysis by tensorial feature extraction. Int J CARS (2024). https://doi.org/10.1007/s11548-024-03175-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11548-024-03175-2