Abstract
Purpose
Carotid artery atherosclerotic stenosis accounts for 18–25% of ischemic stroke. In the evaluation of carotid atherosclerotic lesions, the automatic, accurate and rapid segmentation of the carotid artery is a priority issue that needs to be addressed urgently. However, the carotid artery area occupies a small target in computed tomography angiography (CTA) images, which affect the segmentation accuracy.
Methods
We proposed a coarse-to-fine segmentation pipeline with the Multiplanar D-SEA UNet to achieve fully automatic carotid artery segmentation on the entire 3D CTA images, and compared with other four neural networks (3D-UNet, RA-UNet, Isensee-UNet, Multiplanar-UNet) by assessing Dice, Jaccard similarity coefficient, sensitivity, area under the curve and average hausdorff distance.
Results
Our proposed method can achieve a mean Dice score of 91.51% on the 68 neck CTA scans from Beijing Hospital, which remarkably outperforms state-of-the-art 3D image segmentation methods. And the C2F segmentation pipeline can effectively improve segmentation accuracy while avoiding resolution loss.
Conclusion
The proposed segmentation method can realize the fully automatic segmentation of the carotid artery and has robust performance with segmentation accuracy, which can be applied into plaque exfoliation and interventional surgery services. In addition, our method is easy to extend to other medical segmentation tasks with appropriate parameter settings.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Stroke is the second most common cause of mortality and the third most common cause of disability worldwide [1]. Up to 88% of strokes are ischemic in nature [2]. Extracranial atherosclerotic disease (ECAD), primarily, carotid artery atherosclerotic stenosis, accounts for 18–25% of ischemic stroke [3, 4]. Atherosclerotic plaque can encroach on the lumen and cause stenosis. Plaque disruption and thrombus formation lead to progressive narrowing of the lumen and clinical events (stroke, transient ischemic attack, etc.). Nowadays, the imaging methods used in the evaluation of carotid artery disease mainly include CTA, ultrasound, magnetic resonance imaging (MRI) and digital subtraction angiography (DSA). Among them, CTA has been widely used and as an important reference for treatment and decisions due to its high resolution, rapid scanning and 3D imaging results [5]. The localization and segmentation of the carotid artery is an important antecedent step for the evaluation of carotid atherosclerotic plaques. In clinical practice, this task mainly relies on the physician's manual annotation of the medical images, which is subjective and tedious.
In the studies of carotid artery segmentation, researchers have developed various methods, such as mathematical morphology [6], adaptive segmentation [7], region growing algorithm [8] and the level set method [9]. However, these methods are all semi-automatic, and need to select the initial point manually. With the development of machine learning technology, more and more researchers apply deep learning method to the medical image segmentation tasks, which have achieved remarkable performance. Zhou et al. [10] used CNN and UNet to segment carotid media-adventitia and lumen-intima boundary from ultrasound slices, but the method needs to select the region of interest (ROI) manually. Moreira et al. [11] used UNet to segment the longitudinal ultrasound images of carotid artery. However, the field of view (FOV) of ultrasound screening is not large, and cannot acquire the 3D images, which is still insufficient for the decision-making of clinical decisions compared with CTA. Pan et al. [12] used the cascaded optimized 3D-UNet to segment the MICCAI 2009 carotid bifurcation CTA dataset [13], but the experience-based ROI cropping methods and the unbalanced dataset (the ratio of training set and test set was 15:41) reduce segmentation accuracy, which is only 82.3%. Ziegler et al. [14] proposed an improved DeepMedic network to segment the common carotid artery, internal carotid artery and external carotid artery on the contrast-enhanced MR angiography (CE-MRA) images. But it needs to manually divide the left and right carotid artery area and perform post-processing operations with a low segmentation accuracy.
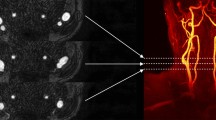
For the entire 3D CTA image, it is usually hard to be fed into the segmentation network directly due to the limited computing resources. If use the entire CTA image with down-sampling fed into the segmentation network, it will cause another resolution loss problem, which will reduce the segmentation accuracy. Moreover, if a random patch sampling strategy is adopted, due to the serious imbalance between the objective regions and the background as shown in Fig. 1, most patches will only contain the background, which will bring additional calculation costs without improvement of segmentation accuracy. Therefore, there are two major problems in the 3D carotid artery CTA image segmentation, one is automatically carotid artery segmentation without selecting the ROI manually, another is the data imbalance, which the object area is much less than the background. In order to address these challenges, this paper proposed a C2F segmentation pipeline with the Multiplanar D-SEA UNet to segment the carotid artery from the entire 3D CTA image automatically. The experiment results show that the C2F segmentation pipeline can avoid resolution loss and improve segmentation accuracy. At the same time, Multiplanar D-SEA UNet has robust performance in various CTA cases.
The main contribution of our paper includes:
-
1.
We introduced a coarse-to-fine segmentation pipeline, developed a coarse-to-fine carotid artery segmentation pipeline based on CTA images, which can realize the automatic segmentation of the full resolution 3D images.
-
2.
We proposed a segmentation network named Multiplanar D-SEA UNet by extending Multiplanar-UNet combined with the deep-supervision mechanism, squeeze and excitation block, and attention block for better feature extraction.
-
3.
Experiments on the clinical carotid artery datasets from Beijing Hospital demonstrate the effectiveness of proposed method in both automatic segmentation and segmentation accuracy and can be extended to other medical images.
Materials and methods
Dataset
68 neck CTA scans from Beijing Hospital were enrolled in this study. The CT scanners used to acquire the samples come from three manufacturers (Toshiba, Siemens and General Electric Company). Scan parameters include the average resolution 512 × 512 × 425, and axial scan interval 0.5–1 mm. The basic characteristics of the patients include the average age of 68.3 (standard deviation (SD) ± 12.8), gender distribution (78% male, n = 53; 22% female, n = 15) and time range of image inspection (2015–2020). In this study, 50 and 18 cases were randomly used as the training and test set.
Two neuroradiologists with 7 and 12 years of dedicated experience annotated the carotid artery masks as ground truth. The annotation task was performed using 3D Slicer (version 4.10.2, www.slicer.org) with the spherical paint. If they are inconsistent, a senior expert will make the final judgment.
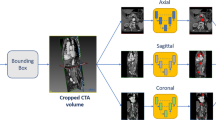
Coarse-to-fine segmentation pipeline (C2F)
The C2F segmentation pipeline contains coarse and fine segmentation modules, and each module employs a similar deep learning segmentation network. The coarse segmentation module automatically detects the coarse region of the carotid artery from the entire CTA image. Based on the candidate regions acquired from the coarse module, the aim of the fine segmentation module is to achieve the accurate carotid artery boundary segmentation results, as shown in Fig. 2.
In coarse segmentation process, the original image \(I_{r}\) is down-sampled, which will be sent to the trained segmentation network to obtain the coarse segmentation mask \(I_{m}\). Then, the region of interest (ROI) of the carotid artery in \(I_{m}\) can be calculated. According to the ROI, the carotid region \(I_{{{\text{cr}}}}\) can be obtained by cropping the original image \(I_{r}^{^{\prime}}\). The purpose of coarse segmentation is to get the coarse region of carotid, so the down-sampling operation will not affect the final results.
In fine segmentation process, the carotid artery region \(I_{{{\text{cr}}}}\) predicted from the coarse segmentation is sent to the trained segmentation network to obtain the fine carotid segmentation \(I_{f}\). Then padding \(I_{f}\) with the area which is cut off in the cropping step, the final result of carotid artery segmentation \(I_{p}\) can be obtained.
Multiplanar D-SEA UNet
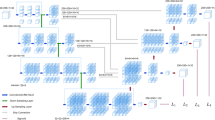
In this work, we proposed a new segmentation network named Multiplanar D-SEA UNet as C2F pipeline’s segmentation network. It includes the recently proposed multiplanar 3D image preprocessing method [15] and our proposed D-SEA UNet network, which is extended from UNet [16] with the deep-supervision mechanism [17], squeeze and excitation block [18] and attention block [19].
As shown in Fig. 3, the 3D image input of size \(w \times h \times d\) is sampled into multiple slice sequences using trilinear and nearest neighbor interpolation along multiple direction axes. For each slice sequence, the D-SEA UNet segmentation network is used to predict a full 3D candidate segmentation result \(\hat{v} \in {\mathbb{R}}^{w \times h \times d \times C}\), where \(C\) is the total number of classes. At last, map all 3D candidate segmentation results to a final segmentation \(\hat{P} \in {\mathbb{R}}^{w \times h \times d \times C}\) by the fusion model.
The fusion model is a simple linear regression model, which calculates \(\hat{P}\) by a weighted sum of \(\hat{v}\). It is defined as follows:
where \(M = 6\) is the number of 3D candidate segmentation results. \(\hat{v}_{m,k}\) denotes the probability of class \(k\) in the \(m{\text{th}}\) candidate segmentation result \(\hat{v}_{m}\). \(W_{m,k} \in {\mathbb{R}}^{w \times h \times d}\) weights the class \(k\) as predicted from \(\hat{v}_{m}\). \(\beta_{k}\) is bias parameter. The number of classes \(C\) is set to 2 in this study, including carotid artery and background. The parameters of fusion model are obtained through training.
In the proposed D-SEA UNet segmentation network, the basic architecture UNet consists of an encoder and a decoder which symmetrically distributes on the both sides. And the features of different levels in the encoder are merged into the decoder by skip connection. To extract deeper features, this paper sets the base filter to 90 and the network depth to 5.
The deep-supervision mechanism performs \(1 \times 1\) convolution and up-sampling operations on the output of each layer in decoder, then superimposes them on the final network output, as shown in the dashed branches on the right of D-SEA UNet in Fig. 3.
The squeeze and excitation block (SE) is added after the convolution operation in decoder, which is used to emphasize important channels and ignore less important ones. The detailed structure of SE is shown in Fig. 4a. For the input feature map \(U \in {\mathbb{R}}^{w \times h \times c}\), use a global average pooling layer to produce the vector \(Z \in {\mathbb{R}}^{1 \times 1 \times c}\), then the channel-specific descriptor is defined as:
where \(w_{1} \in {\mathbb{R}}^{{c \times \frac{c}{r}}} \) and \(w_{2} \in {\mathbb{R}}^{{\frac{c}{r} \times c}}\) are weights of two fully connect layers. The parameter \(r = 4\) is the channel squeeze factor. \(\delta \left( \cdot \right)\) is ReLU activation and \(\sigma \left( \cdot \right)\) is sigmoid layer. The output of SE block is defined as \(\hat{U} = \left[ {\hat{z}_{1} u_{1} , \hat{z}_{2} u_{2} , \ldots , \hat{z}_{c} u_{c} } \right]\), where \(\hat{z}_{i}\) indicates the importance of the \(i{\text{th}}\) channel in \(U\), which is either scaled up or down.
In addition, a simplified attention block was added to the skip connection. It takes the encoder features and the corresponding decoder features as input. While retaining the original feature information of encoder, the decoder features focus more attention on the carotid artery. Figure 4b shows the detailed structure. It is worth mentioning that the attention block was only used in fine segmentation.
The loss function of D-SEA UNet is based on the categorical cross-entropy, which is defined as follows:
where \( N\) is the number of pixels in a batch during training. \(y_{n}^{k}\) is the binary label of pixel \(n\) to the class \(k\) in ground truth \(y\). \(\hat{y}_{n}^{k}\) is the probability of pixel \(n\) belonging to class \(k\), which is the output of the softmax layer in D-SEA UNet.
Implementation details
In the image preprocessing, the outlier-robust scaling is applied for data normalization, which scales intensity values according to the median and interquartile range. For each sampled image in a batch, this paper applied the Random Elastic Deformations augmentation strategy [20] with a probability of 1/3. The elasticity constants and deformation intensity multipliers are sampled uniformly from (20, 30) and (100, 500).
The iteration of the segmentation network is 150, and the batch size is 8. Use the Adam optimizer to update the gradient with an initial learning rate of 0.0005, the momentum β1 = 0.9 and β2 = 0.999. If the network stabilizes after 10 epochs, the learning rate will reduce to half. The experiment was deployed under the Keras framework and trained with NVIDIA Titan GPU.
Results and analysis
Evaluation metrics
Five evaluation metrics used to evaluate the performance of the segmentation network in this study, including Dice coefficients (Dice), Jaccard similarity coefficient (Jaccard), Sensitivity (Sen), Area Under the Curve (AUC) and Average Hausdorff Distance (AVD) [21]. Among them, Dice is the most used metric in validating medical volume segmentations, which measure the similarity between the ground truth and segmentation result, which is defined as:
where TP, FP, and FN represent the amount of true positive, false positive and false negative, respectively. Jaccard is given by the expression:
Sensitivity measures the portion of positive voxels in the ground truth that is also identified as positive in the segmentation result, and it is defined as follows:
AUC is the area under the ROC curve (Receiver Operating Characteristic). For the case where a segmentation result is compared to a ground truth, we consider a definition of the AUC according to [22], namely the area of the trapezoid, which is given by:
High values indicate good segmentation results in these four metrics.
AVD mainly calculates the distance between two finite point sets. The definition of AVD is as follows:
where \(A\) and B are the ground truth and segmentation result voxel sets respectively. N is the number of points in voxel set \(A\) and \(||a - b||\) is the Euclidean distance. A lower value means that the segmentation result is closer to the ground truth.
Comparison with different networks
As there is no published prior work for comparison, in order to show the performance of our proposed method for 3D carotid segmentation, we compared it with the 3D image segmentation classic networks such as 3D-UNet [23], RA- UNet [24] and Isensee-UNet [17], and the state-of-the-art architecture Multiplanar-UNet [15]. Among them, 3D-UNet is a simple extension of UNet. RA-UNet is an improved 3D UNet network with adding residual module [25] and attention mechanism. Isensee-UNet adds deep-supervision to 3D-UNet and uses an equally weighted Dice coefficient.
Firstly, in order to evaluate the performance of the designed C2F segmentation pipeline, we compared the segmentation result of each network before and after using the C2F pipeline as shown in Table 1. It can be observed that the segmentation result of different networks under the C2F pipeline is improved with a 9.81–16.11% increase in Dice score, indicating that the C2F pipeline can effectively improve the segmentation performance and avoid resolution loss.
Then, compared with other networks, our proposed Multiplanar D-SEA UNet under the C2F pipeline achieved the best Dice 91.51%, the best Jaccard 84.46%, the best Sen 89.62%, the best AUC 0.948 and the best AVD 0.21 mm, which is closest to the ground truth. In addition, the metrics of Multiplanar-UNet and Multiplanar D-SEA UNet are significantly better than other 3D-UNet networks. It means that in the 3D carotid artery segmentation task, the multiplanar 2D segmentation network is better than the 3D segmentation network, which can capture the details of features from different views.
Figure 5 is the box plot of different networks’ evaluation metrics under the C2F pipeline. The box plot is a statistical chart to display the data distribution. It is often used to observe shape information such as the median, outlier and distribution interval of the data. It can be seen the results of 3D-UNet, RA-UNet and Isensee-UNet have a wide range of distribution, which indicates that their test results are greatly affected by the test samples. By contrast, the results of Multiplanar-UNet are more concentrated, and Multiplanar D-SEA UNet has the best and densest data distribution results, showing that Multiplanar D-SEA UNet has the better robustness.
The 3D display of the carotid artery segmentation results under the C2F pipeline is shown in Fig. 6. In case01–03, it can be seen that (a) 3D-UNet, (b) RA-UNet are difficult to ensure the continuity of the carotid artery. (c) Isensee-UNet performs well in case 03, but there are ruptures in case01 and case02. (d) Multiplanar-UNet appears a rupture in the lower part of the right carotid artery in Case01. Only (e) Multiplanar D-SEA Unet retains the integrity and continuity of blood vessels to the greatest extent, showing the best performance. In Case02, the four networks (a), (b), (c) and (d) are all susceptible to the influence of the background tissues, which can cause over- segmentation. (e) Multiplanar DSA-UNet exhibits the slightest over-segmentation and is closer to the ground truth, which is stably in various CTA cases.
Ablation experiments
In order to evaluate the contributions of the added components in the proposed C2F Multiplanar D-SEA UNet, we conducted a series of ablation experiments.
As shown in Table 2, the first row represents the results that use the proposed method and the remaining rows correspond to the results after removing a component.
According to the first two rows, the designed C2F pipeline makes a great contribution to the segmentation accuracy as mentioned in the previous section, indicating that the C2F pipeline can be used as an effective processing technology for segmentation when the objective regions are far less than the background. Then, the first and third rows reveal that the deep-supervision mechanism can increase the mean value of each metric by combining the deep and shallow features in decoder. As shown in the first and fourth rows, the SE block can also improve the network’s performance, which aims to emphasize the important channels after the convolution operation. Finally, we observed that the attention block related to the first and fifth rows is shown to contribute to the segmentation accuracy.
Conclusion
In this paper, we proposed a C2F pipeline with the Multiplanar D-SEA UNet to segment the carotid artery from the entire 3D CTA images automatically. The experiment results of the clinical datasets indicate that our approach is robust, and it remarkably outperforms state-of-the-art 3D image segmentation methods used in carotid artery dataset. We also confirmed by ablation experiments that the C2F pipeline, deep-supervision mechanism, SE block and attention block are considerably useful techniques for learning discriminative features and improving segmentation performance. This methodology can also be applied to the different CTA images from different clinical centers with appropriate parameter settings, which does not need the physicians to intervene. All the segmentation process is automatic and can be applied into the clinical practice.
Abbreviations
- CTA:
-
Computed tomography angiography
- CNN:
-
Convolutional neural network
- DSA:
-
Digital subtraction angiography
- ECAD:
-
Extracranial atherosclerotic disease
- C2F:
-
Coarse-to-fine
- Sen:
-
Sensitivity
- AUC:
-
Area under the curve
- AVD:
-
Average Hausdorff distance
References
Collaborators G2 (2018) Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet (London, England) 392:1859–1922
Brott T, Halperin J, Abbara S, Bacharach J, Barr J, Bush R, Cates CU, Creager M, Fowler SB, Friday G, Hertzberg V, McIff EB, Moore W, Panagos P, Riles T, Rosenwasser R, Taylor A (2011) 2011 ASA/ACCF/AHA/AANN/AANS/ACR/ASNR/CNS/SAIP/SCAI/SIR/SNIS/SVM/SVS guideline on the management of patients with extracranial carotid and vertebral artery disease: executive summary. Stroke 42(8):e420–e463
Ooi Y, Gonzalez N (2015) Management of extracranial carotid artery disease. Cardiol Clin 33(1):1–35
Saba L, Saam T, Jäger H, Yuan C, Wintermark M (2019) Imaging biomarkers of vulnerable carotid plaques for stroke risk prediction and their potential clinical implications. Lancet Neurol 18:559–572
Saxena A, Ng E, Lim S (2019) Imaging modalities to diagnose carotid artery stenosis: progress and prospect. BioMed Eng Online 18:1–23
Beare R, Chong W, Ren M, Das G, Srikanth V, Phan T (2010) Segmentation of carotid arteries in CTA images. Int Conf Digital Image Comput Tech Appl 2010:69–74
Santos FL, Joutsen A, Terada M, Salenius J, Eskola H (2014) A semi-automatic segmentation method for the structural analysis of carotid atherosclerotic plaques by computed tomography angiography. J Atheroscler Thromb 21(9):930–940
Bozkurt F, Köse C, Sarı A (2018) An inverse approach for automatic segmentation of carotid and vertebral arteries in CTA. Expert Syst Appl 93:358–375
Tang H, Walsum TV, Hameeteman R, Shahzad R, Vliet LV, Niessen W (2013) Lumen segmentation and stenosis quantification of atherosclerotic carotid arteries in CTA utilizing a centerline intensity prior. Med Phys 40(5):051721
Zhou R, Fenster A, Xia Y, Spence J, Ding M (2019) Deep learning-based carotid media-adventitia and lumen-intima boundary segmentation from three-dimensional ultrasound images. Med Phys 46:3180–3193
Moreira AS (2019) Carotid lumen segmentation using a neural network approach. Dissertation, University of Porto
Pan XY (2019) Carotid bifurcation segmentation based on the deep neural network UNet. Dissertation, Zhejiang University
Hameeteman R, Zuluaga MA, Freiman M, Joskowicz L, Cuisenaire O, Florez-Valencia L, Gülsün M, Krissian K, Mille J, Wong WC, Orkisz M, Tek H, Hoyos MH, Benmansour F, Chung AC, Rozie S, Gils MV, Borne LV, Sosna J, Berman P, Cohen N, Douek P, Sánchez I, Aissat M, Schaap M, Metz C, Krestin G, Lugt A, Niessen W, Walsum TV (2011) Evaluation framework for carotid bifurcation lumen segmentation and stenosis grading. Med Image Anal 15(4):477–488
Ziegler M, Alfraeus J, Bustamante M, Good E, Engvall J, de Muinck E, Dyverfeldt P (2021) Automated segmentation of the individual branches of the carotid arteries in contrast-enhanced MR angiography using DeepMedic. BMC Med Imag 21(1):1–10
Perslev M, Dam E, Pai A, Igel C (2019) One network to segment them all: a general, lightweight system for accurate 3D medical image segmentation. In: MICCAI, pp 30–38
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A (eds) Medical image computing and computer-assisted intervention – MICCAI 2015. MICCAI 2015. Lecture notes in computer science, vol 9351, Springer, pp 234–241
Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH (2018) Brain tumor segmentation and radiomics survival prediction: contribution to the BRATS 2017 challenge. In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M (eds) Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. BrainLes 2017. Lecture notes in computer science, vol 10670, Springer, pp 287–297
Roy AG, Navab N, Wachinger C (2019) Recalibrating fully convolutional networks with spatial and channel “squeeze and excitation” blocks. IEEE Trans Med Imag 38:540–549
Oktay O, Schlemper J, Folgoc LL, Lee MJ, Heinrich M, Misawa K, Mori K, McDonagh SG, Hammerla N, Kainz B, Glocker B, Rueckert D (2018). Attention U-Net: learning where to look for the pancreas. http://arxiv.org/abs/1804.03999.
Castro E, Cardoso JS, Pereira JC (2018) Elastic deformations for data augmentation in breast cancer mass detection. IEEE EMBS Int Conf Biomed Health Inform (BHI) 2018:230–234
Taha AA, Hanbury A (2015) Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imag 15(1):29
Powers D (2011) Evaluation: from precision, recall and F-factor to ROC, informedness, markedness correlation. Mach Learn Technol 2:37–63
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W (eds) Medical image computing and computer-assisted intervention – MICCAI 2016. MICCAI 2016. Lecture notes in computer science, vol 9901, Springer, pp 424–432
Jin Q, Meng Z, Sun C, Wei L, Su R (2020) RA-UNet: a hybrid deep attention-aware network to extract liver and tumor in CT scans. Front Bioeng Biotechnol 8:1471
Kou C, Li W, Liang W, Yu Z, Hao J (2019) Microaneurysms segmentation with a U-Net based on recurrent residual convolutional neural network. J Med Imag 6:025008–025008
Funding
This work was funded by the Capital’s Funds for Health Improvement and Research (2020-4-4053), the National Natural Science Foundation of China (KKA309004533), the Enterprise funded projects from Fudan University (20275), the Independent Research fund of Key Laboratory of Industrial Dust Prevention and Control & Occupational Health and Safety, Ministry of Education (Anhui University of Science and Technology) (EK20201003) and the Shandong Key Laboratory of Intelligent Buildings Technology (SDIBT202006).
Author information
Authors and Affiliations
Contributions
JW collected and labeled the medical images. YY and RY conducted the experiments, wrote the draft. HW and DG participated in data and material review. JL and ZY participated in experiment design, reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Consent to participate
Agree.
Consent for publication
Agree
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
For the retrospective studies formal consent is not required.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, J., Yu, Y., Yan, R. et al. Coarse-to-fine multiplanar D-SEA UNet for automatic 3D carotid segmentation in CTA images. Int J CARS 16, 1727–1736 (2021). https://doi.org/10.1007/s11548-021-02471-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02471-5