Abstract
Purpose
Segmentation of liver tumours is an important part of the 3D visualisation of the liver anatomy for surgical planning. The spatial relationship between tumours and other structures inside the liver forms the basis of preoperative surgical risk assessment. However, the automatic segmentation of liver tumours from abdominal CT scans is riddled with challenges. Tumours located at the border of the liver impose a big challenge as the surrounding tissues could have similar intensities.
Methods
In this work, we introduce a fully automated liver tumour segmentation approach in contrast-enhanced CT datasets. The method is a multi-stage technique which starts with contrast enhancement of the tumours using anisotropic filtering, followed by adaptive thresholding to extract the initial mask of the tumours from an identified liver region of interest. Localised level set-based active contours are used to extend the mask to the tumour boundaries.
Results
The proposed method is validated on the IRCAD database with pathologies that offer highly variable and complex liver tumours. The results are compared quantitatively to the ground truth, which is delineated by experts. We achieved an average dice similarity coefficient of 75% over all patients with liver tumours in the database with overall absolute relative volume difference of 11%. This is comparable to other recent works, which include semiautomated methods, although they were validated on different datasets.
Conclusions
The proposed approach aims to segment tumours inside the liver envelope automatically with a level of accuracy adequate for its use as a tool for surgical planning using abdominal CT images. The approach will be validated on larger datasets in the future.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Liver tumour segmentation is an important step in the visualisation of liver anatomy for planning surgical treatment. A precise measurement of the tumour size helps to determine the required treatment options. Segmenting and defining the tumour location enable the surgeon to make an accurate assessment before deciding on the therapeutic choices. An accurate 3D visualisation also guides the surgeons calculate the size and volume of a tumour to perform an accurate resection of the liver affected. Besides removing the tumour, the surgical resection seeks to ensure a sufficient percentage of the liver remains after resection [1].
Manual segmentation of tumours based on CT scans is tedious and time-consuming. On the other hand, the automatic liver tumour segmentation is a challenging task, due to the small observable changes in the tumour and healthy tissues especially at their borders. Such a task is even more challenging for surgeons in the treatment room. The delineation of tumours has to be performed on CT scans, which are noisy and contain artefacts. Besides that, the tumours vary greatly in terms of shape, size and texture. Moreover, other surrounding organs like the gallbladder could affect the automated tumour segmentation if these organs are not excluded from the segmented liver envelope [2]. Despite these challenges which complicate tumour segmentation, the automated approach is desirable, as it is, ideally, more objective and removes dependence on human skill [3].
Segmenting of liver tumours can be applied to different image modalities like CT and MRI [4]. However, this study focuses on segmenting tumours from the portal venous phase contrast-enhanced CT scans. It introduces an automatic segmentation method to detect and segment liver tumours. The proposed method is based on an active contour deformable model combined with tumour intensity estimation, with additional post-processing steps to extract the tumours. The full automation of the tumour segmentation tasks enables surgeons to focus on more critical tasks, such as planning for resection. In addition, it removes the bias of manual segmentation. Accurate detection and segmentation of the liver tumours play an essential role in deciding on the proper treatments options and assessing the risk of liver resection.
Related works
Most liver tumour segmentation methods start by segmenting the liver envelope as a region of interest. In the literature, automated segmentation of the liver, which is another important task and affects tumour segmentation, has been applied before segmentation of liver tumours [5,6,7]. For liver tumour segmentation, a number of previous works have described methods for segmenting liver tumours. They can be broadly categorised into automatic and semiautomatic liver tumour segmentation techniques. These methods are based on different underlying approaches, such as region growing, level set contours, multi-level thresholding and graph cut techniques.
Many region growing-based tumour segmentation methods have also been proposed. Anter et al. [3] proposed a semiautomatic method of tumour segmentation using the watershed transform to extract the clusters and define the initial seeds to be used by a region growing algorithm to segment the lesions. Kumar et al. used a more precise technique based on confidence-connected region growing for liver segmentation. After extracting the liver envelope, they used alternative fuzzy C-means (AFCM) clustering for lesion segmentation. However, the FCM method suffers from noisy data or outliers [8]. Another semiautomatic region growing-based method is proposed by selecting seeds manually to update the region growing method iteratively. In their method, the region growing is constrained by the Bayesian rule and model-matching rule [9].
Oliveira et al. [10] proposed another region growing method, which starts by finding initial seeds using a Gaussian mixture model (GMM) which are used in the region growing technique to perform tumour segmentation. However, the method has some limitations, especially when the tumours are located near the liver surface or when the tumour has a similar intensity to the liver tissue. Zhou et al. [11] proposed three semiautomatic tumour segmentation methods, namely a 2D knowledge-based region growing method, a 2D voxel classification method with propagation learning and 3D Bayesian region growing. However, the proposed methods suffer in segmenting tumours with blurred boundaries.
On the other hand, machine-learning techniques are utilised in tumour segmentation. Freiman et al. used an SVM to classify clusters initialised by manually selected seeds to generate another set of high-quality seeds. The high-quality seeds are then propagated based on the affinity between the pixels, controlled by an optimal propagation map. The authors tried to minimise the of user interaction; however, still there is a bias due to the expert who is selecting the initial seeds [12]. Moltz et al. [13] proposed a semiautomatic method starting with an initial coarse segmentation using adaptive thresholding, which is then followed by model-based morphological operations to remove leaks to the surrounding tissues. The method faced challenges in segmenting some types of tumours which required developing a special solutions. Li et al. [14] used a new unified level set-based algorithm that incorporates the image gradient, region competition and prior information to segment tumours from CT scans. The authors used to change the controlling parameters for different datasets to get a satisfactory results. Häme and Pollari [15] proposed another semiautomatic method. It is initialised by manual selection, followed by intensity distribution estimation to classify voxels into classes, which are passed to a hidden Markov measure aided by spherical priori shape information to segment the selected tumours. The method suffers with large tumours and high level of heterogeneity. Huang et al. proposed an automatic technique for tumour detection and semiautomatic method for tumour segmentation using kernel-based extreme learning machine (ELM). They used ELM as class learning machine to detect tumours. However, for the segmentation, they used tumour sampling in 3D space and increased the feature representation of voxels. The method suffered in segmenting tumours with voxels same like the vessels [16]. Weiwei et al. proposed a deep learning approach to segment liver tumours. A semiautomatic method combines an improved fuzzy c-mean with graph cut, in which the graph cut labelling is improved by incorporating the kernelised FCM in the energy function. The authors required a low interaction to extract the liver region of interest to reduce the computational time; the method achieved high performance with small average computational time; however, the method faces challenge in segmenting the low contrast and blurred boundaries [17].
Linguraru et al. proposed a graph cut approach constrained by shape and enhancement modes information. The method does not address the heterogeneity of some tumours, which make it struggle with unusual cases. The segmentation is further enhanced by level set active contours [18]. Wu et al. propose a graph cut method started by applying low-level processing steps to generate a tumour threshold range. They selected the seeds inside each tumour region manually, which are then used by the graph cut approach to segment the tumours [19].
The highest scoring methods are those based on an interactive technique like the graph cut technique. In addition, techniques that are based on adaptive thresholding and morphological processing achieved high scores. In this paper, we try to achieve high segmentation accuracy by proposing a fully automatic technique that reduces the bias of user interaction and the time required for segmentation.
The proposed method
Segmenting liver tumours from CT scans is as an important task in the computer-aided diagnosis systems of preoperative planning hepatic surgical treatments. The approach of segmenting the liver tumours starts by characterising them based on their intensity values. Liver tumours are classified as hypervascular and hypovascular based on the scan timing. Hypervascular tumours are arterial enhancing lesions, which are rapidly filled by contrast during the arterial phase. Most of the hypervascular lesions are benign and primary tumours. On the other hand, the hypovascular tumours are more common than the hypervascular tumours. Hypovascular tumours like malignant and metastases are the most common. The hypovascular tumours are detected as hypodense lesions in the portal venous phase [20]. This paper seeks to segment the hypovascular tumours that appear in the portal venous phase CT scans.
We propose an automatic method to segment hypovascular tumours in CT scans, which are acquired in the portal venous phase. The proposed liver tumour segmentation method is a multi-stage segmentation framework approach comprising five steps:
-
1.
Pre-processing Tumour contrast enhancement.
-
2.
Liver ROI construction Liver envelope construction.
-
3.
Initial tumour segmentation Initial tumour mask extraction.
-
4.
Localised active contour Tumour mask completion.
-
5.
Check tumour circularity Exclude erroneous segmentations.
Pre-processing: tumour contrast enhancement
In this work, our main concern is the hypovascular tumours, which appear darker in the portal venous phase scanning. The CT scans for evaluation are acquired in the portal venous phase. The tumours do not contain the same as contrast as liver parenchyma. In the portal venous phase, the liver is loaded with the contrast by the portal vein to detect the hypovascular tumours. Despite that, tumours and the liver tissue could have similar density values, especially where they border each other. In this step, the aim is to increase the contrast between the liver tissue and the tumour to aid in the extraction of the tumour from the target CT scan. This step should make the tumour appear darker compared to the liver tissue.
To enhance the tumour contrast in the liver parenchyma, an edge-enhancing diffusion filtering technique (EED) is used. We use an anisotropic diffusion filtering technique developed by Mendrik et al. [21]. In anisotropic enhancing diffusion filters, instead of using a scalar diffusion, they used a diffusion tensor to adapt the diffusion along image structures. Thus, diffusion filtering describes the image structure using a structure tensor, which is based on the use of a structure description such as structural features or local coherence of structure, as a diffusion tensor to steer the diffusion [22].
The EED filter is designed to enhance the contrast, filters the noise in the homogeneous regions and preserves the boundaries of the shape [21]. EED filtering enhances the contrast of tumours by enhancing the homogeneities inside the liver and tumour tissue regions. In addition, it preserves the boundaries between tumours and liver tissue. Figure 1 shows an example of a CT scan before and after being enhanced using EED filtering. The intensity of the liver parenchyma is enhanced and appears brighter than the tumour regions, while the tumours appear darker compared to the liver tissue.
Liver ROI construction
In most of the previously proposed tumour segmentation techniques, the method starts by having a pre-segmented liver envelope as a region of interest within which the liver tumours are segmented. In liver segmentation, tumours that are located inside the liver and surrounded by the liver tissue can are segmented by extending an initial mask or using region filling. However, in some datasets, the tumour could exist near the liver surface. When this type of tumour is segmented, it will result in a segmented liver envelope with a concave on the surface of the segmented liver.
In this step of our proposed approach, the aim is to construct a liver ROI that includes both tumours located inside the liver tissue and the tumours located near the liver surface. For this purpose, the results generated in our previous work [23] are used. This liver ROI will be used in the next step to extract the initial masks of the liver tumours.
In this step, a localised mean shape model is utilised to create an enhanced liver ROI. The localised mean shape is sought to be located within the liver region and covers most of the liver organ. In order to enhance the construction of the liver ROI, the localised shape model is improved upon and deformed to better represent the liver envelope.
For this purpose, an intensity-based registration using the mutual information metric is used. The localised shape model is registered to a previously segmented liver mask. As a result, the liver shape model is deformed to match the surface of the liver in the CT image without being affected by tumour concaves that may appear on the liver surface. Figure 2 shows an example of the constructed liver ROI and demonstrates how the registered mean shape successfully includes the tumorous regions in the resulting segmented liver.
Initial tumour segmentation
In this step, the aim is to find the range of tumour intensities inside the liver region. After the enhancement step (“Pre-processing: tumour contrast enhancement” section), the contrast between liver tissue and tumours is improved. Based on that, the liver ROI is assumed to include three classes of tissues, which can be distinguished by their intensities. The first class is liver tissue, which represents the majority liver volume. The second class is higher intensity tissue, which includes vessels or parts from surrounding organs that are erroneously included in the ROI. These exist because the focus of this paper is to segment hypovascular tumours and hence slight errors in the constructed liver ROI are tolerated. The third class of tissue is those with lower intensities, which include the tumour tissue. Again, due to the slightly relaxed accuracy sought for the constructed liver ROI, this third class of tissue has voxels from surrounding anatomy such as muscles and other organs such as parts of the gallbladder, which have intensity values smaller than the tumour intensities.
Adaptive local thresholding is applied to automatically perform intensity analysis in order to identify the main intensity ranges for the three classes of tissue inside the constructed liver ROI. The diagram in Fig. 3 represents the process of the adaptive thresholding carried on the liver ROI. The Otsu algorithm is used to implement the multiple local thresholding, to find the estimated thresholds. With the range of intensities in the ROI represented as e to d, the thresholding steps are as follows:
-
1.
A threshold f is found to divide the ROI intensities (\(e-d)\) into two classes.
-
2.
A second threshold g divides the range \(f-d\). The range \(g-d\) is designated as the higher intensity class (vessels, other anatomy).
-
3.
A third threshold h, divides the range \(e-g\). The range \(h-g\) is designated as liver parenchyma.
-
4.
A final threshold i, divides the range \(e-h\). The range \(i-h\) is designated as tumour tissue.
As shown in Fig. 3, only intensities between i to h are assumed to be of tumour tissue. Lower intensity voxels are assumed to be the surrounding muscles and other anatomy.
Figure 4 shows the initial segmentation of the tumours. Their intensities are located within the estimated range of tumours intensities (\(i-h)\), from the constructed liver ROI. Figure 4 demonstrates that the tumour tissue is differentiated from other tissue inside the ROI.
Localised active contour
The previous step extracts an initial tumour mask from the constructed liver ROI. However, this initial mask might only include part of the tumour due to incorrect liver ROI construction, as shown in Fig. 5, which is especially true for tumours located at the liver surface.
In this step, a level set-based active contour method is used to extend the initial tumour mask to the boundary of the tumours in the CT image. In this work, in order to avoid the problems of the global region-based active contour method [17], a localised region-based active contour method is used, as proposed by Lankton et al. [24]. Instead of modelling the whole image, the contour is modelled by many neighbourhood local regions, and each local region is considered separately. The input for the localised active contour step is the initial tumour mask. The localised active contour aims to deform the tumours masks to the tumour regions that have intensity values located within the estimated intensity range of the tumours.
The process of mask deformation is applied to each tumour separately to avoid overlap with other tumours. Figure 5 shows the tumour segmentation after applying the localised active contour.
Tumour circularity check
Due to the relaxed accuracy of the constructed liver ROI in the second step above, tissue which does not belong to the liver is included in the ROI. These erroneously included tissues may have intensity values within the estimated tumour intensity range. Consequently, the initial mask of the tumours may include parts from the surrounding tissue. Those non-tumorous parts would also be deformed in the active contour step and appear in the final tumour segmentation result. In this step, the aim is to remove the erroneous segmented tumours, by discriminating the non-tumour structures out from true tumour tissue. For this purpose, we proposed a knowledge-based discriminative metric based on the clinical feature that the hypovascular tumours have a spherical shape [25]. A circularity metric is used to check each segmented tumour. If the segmented tumour adheres to this circularity metric, it will be considered a true tumour.
The tumour discrimination metric is based on fitting a minimum circle to enclose each part of the tumour that appears in each slice as shown in Fig. 6. The area of the fitted circle is computed for each slice. This step is repeated for all other tumour parts. Finally, the fitted areas for all tumour’s parts are summed up to find the minimum volume that represents the tumour. The object is considered a tumour if the ratio between the actual tumour volume, which we examined, to the calculated fitted area is above a degree of circularity. In this paper, we experimentally measured the accepted degree of circularity, which is 0.42. The discrimination metric is calculated based on Eq. 1 shown below;
Results and discussion
The proposed method is validated on the challenging 3D IRCAD dataset. These datasets offer highly variable and complex liver tumours. The proposed method is validated against datasets with pathology cases, which have 111 tumour cases residing inside the liver envelope. The method outputs are compared to the manual ground truth tumours, which are delineated by experts.
Segmentation results
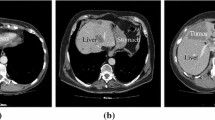
Figure 7 shows a visualisation of the results of the proposed tumour segmentation method for three different patients, with the CT images shown in an intensity window of 0 to 150 Hounsfield values, in which the tumour and liver tissue appear clearly. The first two columns (a and b) in Fig. 7 show two different axial slices of the segmented tumours for each dataset. The result of the proposed automatic segmentation method is shown as a blue boundary, while the ground truth segmentation is marked as a red boundary. It can be seen that the proposed method successfully extracts the tumour boundaries and is close to the ground truth delineation in the CT images.
Column (c) in Fig. 7 shows the 3D rendering of the segmented tumours inside a translucent liver envelope of the selected datasets. The 3D rendering of the segmented tumours visualises the location and size of the tumours with respect to the liver. This visualisation would enable surgeons to understand the geometry and location of the cancerous lesion. In addition, the tumours could be labelled with different colours to enable surgeons to interact with each tumour separately during treatment.
It is clear from the three examples in the first two columns in Fig. 7 that the difference between the proposed method and the corresponding ground truth occurs on the tumour surface, which indicates that the method under-segments the tumours in some cases. However, the method segmentation contour (blue) follows closely the tumour boundaries, which appear very clear in the image, compared to the manual delineation of the experts (red) which extend beyond the visible boundary. Usually, for surgical purposes, the experts add a safety margin to their delineations to include those tumorous tissues on the borders between clear tumours and healthy tissues. These tumorous tissues have very similar density values to the healthy liver tissue. Experts add the safety margin to ensure resecting all tumorous tissue during surgical treatments to reduce the possibility of tumours regeneration. This could make our method to under segment the tumours compared to the manually segmented tumours, which would affect the results in the quantitative assessment as presented in the next section.
Quantitative evaluation
As mentioned above, the tumour segmentation is performed on the 3D IRCAD dataset which contains a large number of tumours. The results of the proposed method are evaluated using a group of performance measures against the manually segmented gold standards. These measures are the Absolute Relative Volume Difference (ARVD), Jaccard Index (JI) and Dice Coefficient (DICE).
Table 1 provides the quantitative evaluation of the results of the proposed method. In addition, the table shows the number of tumours that exist in each patient. A total of 111 tumours have been segmented in this study across all patient. The average ARVD value is 11.43%, the average JI is 60.16%, and the average DICE is 74.96%. As explained in the previous section and shown by Fig. 7, the proposed method results in contours that follow the tumours’ boundaries, compared to the ground truth delineation which appears to slightly over segment the visible tumours’ boundaries.
The lack of reference datasets makes the quantitative comparison between the different liver tumour segmentation methods in the literature difficult. Many proposed methods used their private datasets. On the other hand, the datasets from MICCAI 2008 liver tumour segmentation challenge are not publicly available. Taking this into account, we attempt to present the results of our proposed method along with other techniques proposed to solve the problem of liver tumour segmentation in Table 2.
As shown in Table 2, the presented methods have been validated against a different number of tumours from different sources of data. We also present the amount of user interaction for the methods shown in Table 2. The amount of user interaction plays an important role in enhancing the segmentation output. As the level of interaction increases, the accuracy of the segmentation results may improve as shown by some semiautomatic methods. We note that our results are comparable to another automated approach [13] in the DSC value. However, we produce a lower ARVD. The dataset used in our evaluation (3D IRCAD) also has more tumours than that used in [13].
Conclusion
In this paper, we described an automatic segmentation method of liver tumours without the need for user interaction. The proposed framework combines multiple steps, starting with anisotropic filtering, followed by the construction of an initial tumours mask using adaptive thresholding which is improved upon by using localised active contours to deform the tumour mask to their boundaries. The proposed method is validated against a large number of tumours (111), with different size, shape and number of tumours in each patient dataset. The 3D visualisation and quantitative evaluation using different measures show high accuracies, demonstrating the effectiveness of the proposed method. We propose that the method can be used in larger computer-aided systems for liver surgical planning, and we aim to evaluate it against larger datasets in the future.
References
Blum HE (2005) Hepatocellular carcinoma: therapy and prevention. World J Gastroenterol 11:7391–400
Göçeri EA (2013) Comparative evaluation for liver segmentation from spir images and a novel level set method using signed pressure force function. Dissertation, İzmir Institute of Technology
Anter AM, Azar AT, Hassanien AE, El-Bendary N, Elsoud MA (2013) Automatic computer aided segmentation for liver and hepatic lesions using hybrid segmentations techniques. In: Federated conference on computer science and information systems (FedCSIS), 2013, pp 193–198
Göçeri E, Gürcan MN, Dicle O (2014) Fully automated liver segmentation from SPIR image series. Comput Biol Med 53:265–278
Priyadarsini S, Selvathi D (2012) Survey on segmentation of liver from CT images. In: Proceedings of 2012 IEEE international conference on advanced communication, control and computing technologies (ICACCCT), 2012, pp 234–238
Pamulapati V, Venkatesan A, Wood BJ, Linguraru MG (2012) Liver segmental anatomy and analysis from vessel and tumor segmentation via optimized graph cuts. In: Yoshida H, Sakas G, Linguraru MG (eds) Abdominal imaging. Computational and Clinical Applications. ABD-MICCAI 2011. Lecture Notes in Computer Science, vol 7029. Springer, Berlin, Heidelberg, pp 189–197
Göçeri E (2016) Fully automated liver segmentation using Sobolev gradient-based level set evolution. Int J Numer Methods Biomed Eng 32:e02765
Kumar SS, Moni RS, Rajeesh J (2013) Automatic liver and lesion segmentation: a primary step in diagnosis of liver diseases. Signal Image Video Process 7:163–172
Yingyi Q, Wei X, Wee KL, Qi T, Jiayin Z, Jiang L, Thazin H, Sudhakar KV, Shih-chang W (2008) Semi-automatic segmentation of liver tumors from CT scans using Bayesian rule-based 3D region growing. In: MICCAI workshop, vol 41, pp 1–10
Oliveira DA, Feitosa RQ, Correia MM (2011) Segmentation of liver, its vessels and lesions from CT images for surgical planning. Biomed Eng Online 10:30
Zhou JY, Wong DW, Ding F, Venkatesh SK, Tian Q, Qi YY, Xiong W, Liu JJ, Leow WK (2010) Liver tumour segmentation using contrast-enhanced multi-detector CT data: performance benchmarking of three semiautomated methods. Eur Radiol 20:1738–1748
Freiman M, Cooper O, Lischinski D, Joskowicz L (2011) Liver tumors segmentation from CTA images using voxels classification and affinity constraint propagation. Int J Comput Assist Radiol Surg 6:247–255
Moltz JH, Bornemann L, Dicken V, Peitgen HO (2008) Segmentation of liver metastases in CT scans by adaptive thresholding and morphological processing. In: MICCAI workshop, vol 41, p 195
Li BN, Chui CK, Chang S, Ong SH (2012) A new unified level set method for semi-automatic liver tumor segmentation on contrast-enhanced CT images. Expert Syst Appl 39:9661–9668
Häme Y, Pollari M (2012) Semi-automatic liver tumor segmentation with hidden Markov measure field model and non-parametric distribution estimation. Med Image Anal 16:140–149
Huang W, Li N, Lin Z, Huang GB, Zong W, Zhou J, Duan Y (2013) Liver tumor detection and segmentation using Kernel-based extreme learning machine. In: 35th annual international conference of the IEEE EMBS Osaka, Japan, pp 3662–3665
Wu W, Wu S, Zhou Z, Zhang R, Zhang Y (2017) 3D liver tumor segmentation in CT images using improved fuzzy C-means and graph cuts. Biomed Res Int 2017:1–11
Linguraru MG, Richbourg WJ, Liu J, Watt JM, Pamulapati V, Wang S, Summers RM (2012) Tumor burden analysis on computed tomography by automated liver and tumor segmentation. IEEE Trans Med Imaging 31:1965–1976
Wu W, Wu S, Zhang R, Zhou Z (2016) Fast graph cuts based liver and tumor segmentation on olumetric CT images. In: Joint international conference on service science, management and engineering and international conference on information science and technology, pp 3–7
Brancatelli G, Baron RL, Peterson MS, Marsh W (2003) Helical CT screening for hepatocellular carcinoma in patients with cirrhosis: frequency and causes of false-positive interpretation. Am J Roentgenol 180:1007–1014
Mendrik AM, Vonken EJ, Rutten A, Viergever MA, Van Ginneken B (2009) Noise reduction in computed tomography scans using 3-D anisotropic hybrid diffusion with continuous switch. IEEE Trans Med Imaging 28:1585–1594
Weickert J (1998) Anisotropic diffusion in image processing. Image Rochester NY 256:170
Irr OIA, Rahni AAA (2015) Automatic volumetric localization of the liver in abdominal CT scans using low level processing and shape priors. In: IEEE international conference on signal and image processing applications (ICSIPA), 2015, pp 434–438
Lankton S, Member S, Tannenbaum A (2008) Localizing region-based active contours. IEEE Trans Image Process 17(11):2029–2039
Kim KB, Kim CW, Kim GH (2008) Area extraction of the liver and hepatocellular carcinoma in CT scans. J Digit Imaging 21(Suppl 1):S89–103
Vorontsov E, Abi-Jaoudeh N, Kadoury S (2014) Metastatic liver tumor segmentation using texture-based omni-directional deformable surface models. Springer, Cham, pp 74–83
Funding
This research is supported by the Malaysian Ministry of Higher Education and Universiti Kebangsaan Malaysia (Grant Number GUP-2014-066).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. For this type of study, formal consent is not required.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Alirr, O.I., Rahni, A.A.A. & Golkar, E. An automated liver tumour segmentation from abdominal CT scans for hepatic surgical planning. Int J CARS 13, 1169–1176 (2018). https://doi.org/10.1007/s11548-018-1801-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-018-1801-z