Abstract
Purpose
Image guidance for minimally invasive surgery is based on spatial co-registration and fusion of 3D pre-interventional images and treatment plans with the 2D live intra-interventional images. The spatial co-registration or 3D–2D registration is the key enabling technology; however, the performance of state-of-the-art automated methods is rather unclear as they have not been assessed under the same test conditions. Herein we perform a quantitative and comparative evaluation of ten state-of-the-art methods for 3D–2D registration on a public dataset of clinical angiograms.
Methods
Image database consisted of 3D and 2D angiograms of 25 patients undergoing treatment for cerebral aneurysms or arteriovenous malformations. On each of the datasets, highly accurate “gold-standard” registrations of 3D and 2D images were established based on patient-attached fiducial markers. The database was used to rigorously evaluate ten state-of-the-art 3D–2D registration methods, namely two intensity-, two gradient-, three feature-based and three hybrid methods, both for registration of 3D pre-interventional image to monoplane or biplane 2D images.
Results
Intensity-based methods were most accurate in all tests (0.3 mm). One of the hybrid methods was most robust with 98.75% of successful registrations (SR) and capture range of 18 mm for registrations of 3D to biplane 2D angiograms. In general, registration accuracy was similar whether registration of 3D image was performed onto mono- or biplanar 2D images; however, the SR was substantially lower in case of 3D to monoplane 2D registration. Two feature-based and two hybrid methods had clinically feasible execution times in the order of a second.
Conclusions
Performance of methods seems to fall below expectations in terms of robustness in case of registration of 3D to monoplane 2D images, while translation into clinical image guidance systems seems readily feasible for methods that perform registration of the 3D pre-interventional image onto biplanar intra-interventional 2D images.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Treatment for vascular pathologies is almost exclusively in the domain of minimally invasive interventions, which are driven by increasing complexity of imaging and image processing techniques. For instance, treatment application by endovascular image-guided intervention (EIGI) [1] involves navigation of the catheter through the vasculature to the site of pathology and optimal application of treatment devices by two-dimensional (2D) fluoroscopic and angiographic image guidance. Besides the dynamic 2D+t images, recent C-arm systems enable acquisition of contrast-enhanced 3D images during EIGI, i.e., 3D digitally subtracted angiograms (3D-DSA). The 3D-DSA image is usually segmented and displayed to the clinician just before the start of EIGI so as to select the optimal 2D view(s) or working projection(s) that are then used during the EIGI to aid guidance [2]. However, the clinician still needs to mentally reconstruct the pose of interventional tools and blood flow characteristics from dynamic 2D+t images with respect to (w.r.t.) the displayed 3D vascular morphology.
A solution is to augment the pre-EIGI 3D-DSA, computed tomography or magnetic resonance angiography (CTA or MRA, respectively) with the information of the current state of EIGI present in dynamic 2D+t image(s), thus resulting in augmented 3D+t images. By incorporating augmented 3D+t images into the current EIGI workflow, the navigation and application of treatment devices could be done easier and faster. The key enabling technology for obtaining augmented 3D+t images is 3D–2D registration.
Translation of 3D–2D registration technology into clinical theater requires an extensive evaluation and rigorous validation of any newly developed and state-of-the-art methods under the same test conditions [3, 4]. In this paper, we employ an extended publicly available clinical image database [5] of 3D and 2D angiograms, all acquired during cerebral EIGI. On each dataset, accurate “gold-standard” registrations were established based on fiducial marker alignment. Quantitative and comparative evaluation of ten state-of-the-art methods for registration of 3D and 2D angiograms was performed using standardized evaluation protocols and metrics [6, 7].
State-of-the-art methods
Recently, we performed a review of state-of-the-art 3D–2D registration methods [8], wherein the methods were classified w.r.t. nature of registration basis, strategy to establish dimensional correspondences, and type of geometric transformation. Geometric transformations used are either rigid or non-rigid. The rigid methods are typically applied in cerebral EIGI, since cerebral vessels have limited non-rigid motion during EIGI. In the case of liver and abdominal EIGIs, several authors have shown that the rigid geometric transformation may not be sufficiently accurate; nevertheless, the rigid methods are still widely used to initialize the non-rigid methods [9]. Since this paper is focused on cerebral EIGI, we consider hereafter only rigid 3D–2D registration methods.
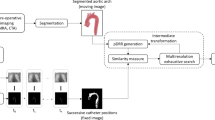
According to registration basis, 3D–2D image registration methods can be categorized as calibration-based and (extrinsic or intrinsic) image-based methods. The intrinsic image-based methods seem readily compatible with current EIGI workflow, as they rely solely on anatomical information observed in the 3D and 2D images. We will thus focus on the intrinsic image-based methods that can be further classified into intensity-, feature- and gradient-based, and hybrid methods [8]. Basic principles of these methods are illustrated in Fig. 1.
Intensity-based 3D–2D registration methods [10, 11] rely on voxel- or pixel-wise matching of 3D and 2D images by means of a similarity measure (SM). The 3D–2D dimensional correspondence can be achieved either by reconstruction of 3D image from several 2D images and 3D-to-3D image matching, or by projection of 3D image to 2D and 2D-to-2D image matching [10, 11], where the projection images are obtained by techniques like digitally reconstructed radiographs (DRRs) [10] or maximum intensity projection (MIP) [11]. Kerrien et al. [11] registered 3D-DSA and 2D-DSA cerebral images in three steps: first, initial registration was obtained by C-arm calibration, and then, the in-plane translations were determined by matching 2D-DSA and MIP images using normalized cross-correlation SM, followed by registration based on a modified optical flow method. Hipwell et al. [10] analyzed six different SMs for registration of cerebral MRA to 2D-DSA images and obtained best results using gradient difference and pattern intensity. In general, the main drawback of intensity-based methods is computationally demanding generation of DRRs/MIPs, but which may be remedied by graphical processor units (GPU)-based implementations and computational approximations.
Feature-based methods [9, 12,13,14,15,16] rely on matching corresponding features extracted from 3D and 2D images, such as vessel centerline points [12], orientations or tangent lines [12], and bifurcations [13]. To initialize the 3D–2D registration of MRA and 2D-DSA cerebral images, Feldmar et al. [12] searched for corresponding bitangent lines in 3D and 2D centerlines to initialize the registration, followed by a refinement based on modified iterative closest point (ICP) matching of 3D and 2D vessel centerline points and tangents. Kita et al. [15] found initial 2D translations by template matching of projected 3D vessel tree skeleton and the 2D-DSA image, followed by a modified ICP, in which anisotropic region akin to Voronoi regions around the projected 3D skeleton points constrained the search for corresponding 2D skeleton points. While feature-based methods achieve computational speedup through large data reduction, their registration accuracy and robustness are directly influenced by the quality of vessel segmentation.
Gradient-based methods [17,18,19] exploit the fact that the observed 2D gradient represents a component of the 3D gradient that is perpendicular to incident ray. Tomaževič et al. [17] back-projected gradients from 2D image(s) into 3D surface tangent subspace and matched the 3D gradient using weighted dot product of the two vectors. Markelj et al. [18] reconstructed a 3D gradient field by summing back-projected 2D gradients from two or more 2D images. The reconstructed 3D gradient field and the gradient field of 3D image were matched using RANSAC to estimate the initial transformation, which was subsequently refined by method of Tomaževič et al. [17]. For efficiency, gradient-based methods match only a subset of distinctive 3D and 2D gradients.
Hybrid methods [5, 20,21,22,23,24,25,26] use different registration bases in 3D and 2D and can be divided into two main subclasses. Methods from the first subclass generate the simulated 2D images by projecting a model of segmented 3D vessel, similarly as a DRR. For instance, Chan et al. [20] created a model of 3D vessel tree from MRA using a set of 3D spheres with radii equal to the radii of corresponding vessels. By projecting the 3D spheres to the imaging plane, they obtained a binary DRR, which was matched to the 2D-DSA image using sum of squared differences. The second subclass of methods first preprocess the pre-EIGI 3D image to extract features that can be efficiently projected from 3D to 2D and matched to 2D images. Jomier et al. [23] projected 3D vessel tree centerline points to 2D-DSA images and then summed the corresponding smoothed intensities, weighted by vessels’s radii. From cerebral 3D-DSA, Mitrović et al. [5] extracted 3D vessel orientations, which were projected to 2D and matched to the orientations of strong 2D intensity gradients in a local neighborhood-based SM. In general, hybrid methods employ 3D image preprocessing and omit the often difficult to perform real-time processing and/or segmentation of intra-operative 2D images.
Methods in comparative evaluation
Ten state-of-the-art 3D–2D registration methods were implemented; namely two intensity-, two gradient-, three feature-based and three hybrid methods. The intensity-based methods were based on maximum intensity projection (MIP) of 3D-DSA and maximized mutual information (MIP-MI) and gradient correlation (MIP-GC) SMs. Gradient-matching methods were the back-projection gradient-based [17] (BGB) and robust gradient reconstruction-based [18] (RGRB). Of the feature-based methods ICP with distance transform of 2D vessel tree skeleton [16] (DT-ICP), ICP with region-based correspondence search [15] (RCS-ICP) and segmentation-driven ICP registration [14] (SEG-ICP) were implemented. The three hybrid methods were based on matching of 2D-DSA to binary DRR of projected spheres of the 3D vessel tree model [20] (MBDRR), matching by summing the smoothed 2D-DSA intensities at positions of projected 3D vessel tree centerline points (MSSI) and matching of geometric primitives [5] (i.e., vessels’ centerline points, orientations and radii) (MGP). Table 1 gives the list of tested methods and their classification.
In all tested methods, the rigid-body parameters were optimized simultaneously by Powell’s directional set method [27]. The methods were mainly implemented and executed in MATLAB (The MathWorks, Inc., USA) on Intel(R) Core(TM) i7 CPU 860 @ 2.80 GHz machine with 8 GB memory, with the exception of MIP generation, which was written in CUDA and executed on NVIDIA GeForce GTS 450 graphics card.
Clinical image datasets
For evaluation, we employed 25 clinical image datasets, each containing 3D and 2D images of patients that underwent cerebral EIGI. Two patients underwent arteriovenous malformation (AVM) and 23 patients underwent aneurysm treatment. All images were acquired on Siemens Axiom Artis dBA biplane flat detector angiography system. Image sets consisted of enhanced and non-enhanced cone-beam computed tomography (CBCT) images, which were used to generate 3D-DSA image. All 3D images had voxel size of \(0.46 \times 0.46 \times 0.46\,\hbox {mm}^{3}\) and image dimension up to \(512 \times 512 \times 391\). Prior to registration, the 3D images were resampled to 0.75 mm isotropic resolution using cubic spline interpolation to reduce size and speed up the registration process.
For each patient, a set of 2D-DSA and contrast-enhanced 2D fluoroscopic images (2D-MAX) were acquired during EIGI (Fig. 2). The 2D images had pixel sizes either \(0.154 \times 0154\,\hbox {mm}^{2}\) or \(0.184 \times 0.184\,\hbox {mm}^{2}\) with dimensions up to \(2480 \times 1920\). The 2D images were acquired in lateral (LAT) and anterior–posterior (AP) gantry orientation so as to best capture cerebral vasculature of interest for EIGI.
“Gold-standard” registration
Establishing an accurate “gold-standard” registration of 3D and 2D images was based on retrospective calibration of the C-arm system and finding the optimal rigid transformation, using fiducial markers visible both in 3D and 2D images. Procedure to obtain “gold-standard” 3D–2D registration was detailed in a previous paper [5], and an automated “gold-standard” creation framework was presented in [28].
Experiments and results
The clinical image datasets were used to quantitatively evaluate the performance of ten state-of-the-art 3D–2D registration methods (Table 1). The experiments consisted of registrations of a 3D-DSA image to either one or two 2D views, referred to as monoplane or biplane registration, respectively. The two intensity-based, three hybrid, BGB and DT-ICP methods were capable of both mono- and biplane registration. RCS-ICP and SEG-ICP methods were developed specifically for monoplane registration, while RGRB required at least two 2D views or biplane registration context; therefore, these methods were tested accordingly in mono- or biplane registration context. Furthermore, two subsets of experiments were performed, one by using 2D-DSA and the other by using 2D-MAX images.
Evaluation protocol
Standardized methodologies as proposed by van de Kraats et al. [7] and Markelj et al. [6] were used to evaluate the performance of the methods across the 25 clinical image datasets. Registration error was measured by mean reprojection distance (mRPD) and mean target registration error (mTRE) for monoplane and biplane registration, respectively. Figure 3 illustrates the two measures of registration error.
Example registration results as a color-coded overlay of the 3D-to-2D projected DSA onto 2D DSA image and corresponding mTRE values. Note that initial displacement was a maximum of 20 mm mTRE and a failure threshold for the final mRPD or mTRE (mRPD \(\le \) mTRE) was set to 2 mm. The 2 mm threshold is approximately the radius of large cerebral vessels, which are typically in focus of cerebral endovascular interventions
Each method was evaluated across multiple registration tests, in which 3D images were initially displaced from the “gold-standard” position by a known, randomly generated six degrees of freedom (3 rotations, 3 translations) rigid transformation and then registered by the automated method. Initial displacements were in the range of 0–20 mm of mTRE, with 20 registrations per 1 mm subinterval. Although our previous study [29] suggests that machine-based initial registration on C-arm systems may lead to initial mTRE of up to 37 mm, to limit the amount of computations, the 20-mm interval as suggested by two of the standardized validation protocols [6, 7] was chosen in this study. An example of the degree of 3D–2D alignment error with 20-mm initial mTRE is shown in Fig. 4. Clearly, the 20-mm mTRE alignment can be easily achieved by manual co-registration prior to running the automated 3D–2D image registration.
Registration was considered successful if the final registration error (mRPD or mTRE) was less than 2 mm. The 2 mm threshold is also application motivated since it represents approximately the radius of large cerebral vessels, which are typically in focus of cerebral endovascular interventions, and such registration accuracy thus provides good enough localization for navigation and guidance during the intervention. This threshold may be reasonable also for other vascular anatomies, since the fixed size of the catheter will limit the scope of endovascular intervention to vessels of certain minimal radius. Similar thresholds were previously used by other authors in the other application contexts of 3D/2D registration. Exemplar registrations with various mTRE values are shown in Fig. 4.
Registration accuracy was defined as mean ± STD of mRPD or mTRE of the successful registrations, while the capture range (CR) was defined as the lowest mTRE subinterval with less than 95% of successful registrations. The 95% percentile was chosen to eliminate the impact of outliers in the results. Success rate (SR) was defined as the percentage of successful registrations. To visualize registration error, the range of initial mTRE was divided into 20 accumulative subintervals: 0–1, 0–2, ..., 0–20 mm, and for each subinterval the 95th percentile of mRPD or mTRE registration error was computed. Registration times were also measured.
Registration results
Registration results are presented in Tables 2 and 3 for monoplane and biplane registration, respectively, while Fig. 5 shows accumulative mTRE subintervals for all tested methods.
Registration accuracy
For registrations of 3D-DSA to either mono- or biplane 2D-DSA views, all methods achieved subvoxel registration accuracy (< 0.75 mm, which is the voxel size of the 3D image). The intensity-based methods [10] performed best, with mRPDs and mTREs less than 0.4 and 0.3 mm for mono- and biplane 2D-DSA registration, respectively. In biplane registration, the gradient-based RGRB [18] method achieved comparable accuracy (Table 3). Surprisingly, the feature-based DT-ICP [16] and SEG-ICP [14] methods were quite accurate even though they relied on error-prone 3D and 2D centerline extraction. The accuracy of BGB [17] was comparable to DT-ICP, while the hybrid methods were the least accurate.
When registering 3D-DSA to the 2D-MAX images, the accuracy of all methods decreased, but was still far below the failure threshold of 2 mm. The MIP-MI, MBDRR [20] and MSSI [23] methods were most affected with approximately twice higher registration error. Registration accuracy of gradient-based methods (MIP-GC, BGB [17], RGRB [18] and MGP [5]) remained comparable to that of 3D- to 2D-DSA registrations. The DT-ICP and SEG-ICP [14] methods used the segmentations and centerlines extracted from the 2D-DSA, assuming that these can be similarly extracted from 2D-MAX.
Registration error in terms of mRPD (left) and mTRE (right) of tested state-of-the-art methods for mono- and biplane registration, respectively, for each of the 1 mm subintervals of the initial mTRE (horizontal axes). The error was computed across the 25 image datasets. A robust method exhibits small registration error across a wide range of initial mTRE. See Tables 2 and 3 for corresponding methods’ symbols
Robustness
The ability of 3D–2D registration method to maintain its performance in the presence of disruptive factors, such as image artifacts, interventional tools, bony structures, and occlusions, is referred to as robustness. In this study, robustness was expressed by SR and CR and by comparing performances on 2D-DSA and 2D-MAX images.
For registration of 3D-DSA to single 2D-DSA image, MIP-MI and MPG [5] methods demonstrated the highest robustness over all evaluated methods (Fig. 5). The RCS-ICP [15] performed quite robust with SR of 78.15% and CR of 9 mm, owing to good estimation of in-plane translations. In registrations of 3D-DSA to biplane 2D-DSAs, MGP [5] method outperformed all the other evaluated methods.
Feature-based DT-ICP method and SEG-ICP [14] performed poorly because of non-corresponding centerlines extracted from 3D and 2D images. SEG-ICP was especially sensitive to overlapping of the vessels on 2D images, since equal values of topological timestamps in 3D and 2D vessel tree skeletons did not correspond to the correct 3D–2D point pair. Lastly, the MIP-GC, BGB [17] and RGRB [18] methods had constantly lower SR in comparison with other methods as these are based on matching locally sensitive gradient information.
Although the methods based on intensity information, i.e., MIP-MI, MBDRR [20] and MSSI [23], performed quite well on 2D-DSA images, in 2D-MAX images, they were affected by lower vessel-to-background contrast and the presence of bony structures and interventional tools, which caused, on average, a decrease in SR of 39.6%. On the other hand, methods using gradient information, i.e., MIP-GC, BGB [17], RGRB [18] and MGP [5], produced comparable results using either 2D-DSA or 2D-MAX images. (Average SR difference was 4.3%.)
Execution time
The obtained results show significant advantage of one feature-based DT-ICP [16] and two hybrid methods, i.e., MSSI [23] and MGP [5], with mean registration time in the order of seconds achieved in all registration tests. As expected, the slowest methods were those using simulated projection, i.e., MIP-GC, MIP-MI [10] and MBDRR [20], with mean registration times at least a factor of two longer than all other methods.
Discussion
Translating the 3D–2D registration technology into EIGI requires extensive evaluation and rigorous validation of the state-of-the-art methods. This paper makes two important contributions in this respect: (1) a clinical image database of 25 patients with corresponding reference or “gold-standard” registrations was created, and (2) ten state-of-the-art 3D–2D registration methods were objectively and comparatively evaluated.
Is the 3D–2D registration technology ready for application in EIGI? The results of methods’ evaluation indicate that there does not exist a method clearly superior in all registration attributes (registration accuracy, robustness and execution time). However, there are methods clearly superior according to certain registration attributes. As expected, the intensity- and gradient-based methods were the most accurate, while the feature-based and hybrid methods were somewhat less accurate owing to errors in 3D and/or 2D segmentation. It should be noted that intensity-based methods are perfectly suited for the case of monomodal registration as performed herein. Nevertheless, all of the tested methods achieved registration accuracy far below failure threshold of 2 mm. Intensity-based methods were highly sensitive to low vessel-to-background contrast, bony structures and interventional tools visible in 2D-MAX images. Feature-based methods are also sensitive to these artifacts due to the need for 2D image segmentation; however, in this study the segmentations were performed on 2D-DSA images. On the other hand, the gradient-matching methods were more robust. Feature-based and hybrid methods had execution times feasible for application in EIGI. Overall, the hybrid MGP [5] method demonstrated the highest potential among all methods (Fig. 5). The answer to the posed question is “Yes” in the case of biplane, and “Yes, provided a good initial alignment” in case of monoplane registration.
The above answers are also supported by the capabilities and operation of a commercial application SmartBrush Angio (www.brainlab.com), which enables the fusion of pre-interventional 3D and intra-interventional 2D images using an automated registration method. Albeit the specifics of the employed method are not given, they state that “aligning the images manually aids the fusion algorithm and improves [registration] results.” Before proceeding to fusion, the algorithm requires visual verification of the registration similarly as in Fig. 4 and lets the user manipulate the scale and rotations of the 3D vessel layer, to better match the underlying 2D image data. This process clearly aims to reduce the initial registration error to within the capture range of the method being used.
The choice of the optimizer may have a large impact on the success rate and capture range. The probability to get stuck in a local optimum greatly affects the capture range and is a function of the optimizer and the smoothness of the similarity measure in the search space. Our choice of the Powell optimizer was a trade-off between the sensitivity to local optima and efficiency. Other authors have used simple gradient descent and quasi-Newton-based methods, which could be more efficient, but also more susceptible to local optima. On the other hand, stochastic optimizers like the covariance matrix adaptation evolution strategy [30] and simulated annealing could be less sensitive to local optima, but they require orders of magnitude more evaluations of the cost function.
Most methods in the evaluation were originally devised for establishing correspondence of the vascular information between 3D and 2D images. Obtaining the vascular information, however, generally requires that the 3D and 2D angiograms are acquired through the injection of potentially toxic iodine (or gadolinium in case of MRA) contrast agent into the patient’s blood stream, with the exception of time-of-flight MRA technique. For this reason, the reduction in contrast agent usage is a relevant clinical application, but which should be approached with different 3D–2D registration methods. For instance, registration could exploit proxy structures like the bony landmarks, which is a promising avenue for research in the context of cerebral angiograms, since cerebral vessels generally do not move with respect to the bony structures and provided that the vascular anatomy does not change as a result of pathologic process or treatment between the acquisition of 3D and 2D images.
Using MRA techniques to acquire the pre-interventional 3D image also helps to reduce the overall patient’s dose exposure. However, multimodality 3D–2D registration is then required, which might prove more challenging than monomodal registration considered in this study. This is also supported by the observed results for intensity-based methods like MIP-MI, using which the 3D- to 2D-DSA registrations were far more successful than the 3D-DSA to 2D-MAX registrations, a precursor to performance characteristics in true multimodality 3D–2D registration. Using monoplane 3D–2D registration, which is most frequently the case during EIGI, but also more challenging as the results show, is also preferred over the biplane setting because of reduced patient dose.
In conclusion, translation to EIGI seems feasible, especially for situations, in which the 3D image is registered to biplane 2D images, while for registration to monoplane 2D image a good initial alignment may be required. Generally, most methods seem to fall below expectations in terms of robustness when it comes to monoplane registrations (Table 2); thus, improvements should focus on increasing their SR and CR. Besides, reliable automatic identification of registration failure may be needed for real applications of 3D–2D registration in EIGI and thus remain a challenge for future work. To encourage the further development in the field, the entire clinical image database will be made available upon request from the authors (http://lit.fe.uni-lj.si/tools).
References
Rudin S, Bednarek DR, Hoffmann KR (2008) Endovascular image-guided interventions (EIGIs). Med Phys 35(1):301
Ruijters D, Homan R, Mielekamp P, van de Haar P, Babic D (2011) Validation of 3D multimodality roadmapping in interventional neuroradiology. Phys Med Biol 56(16):5335–5354
Jannin P, Krupinski E, Warfield S (2006) Validation in medical image processing. IEEE Trans Med Imaging 25(11):1405–1409
Jannin P, Grova C, Maurer CR (2006) Model for defining and reporting reference-based validation protocols in medical image processing. Int J Comput Assist Radiol Surg 1(2):63–73
Mitrović U, Špiclin Ž, Likar B, Pernus F (2013) 3D–2D registration of cerebral angiograms: a method and evaluation on clinical images. IEEE Trans Med Imag 32(8):1550–1563
Markelj P, Likar B, Pernuš F (2010) Standardized evaluation methodology for 3D/2D registration based on the visible human data set. Med Phys 37(9):4643–4647
van de Kraats E, Penney G, Tomaževič D, van Walsum T, Niessen W (2005) Standardized evaluation methodology for 2-D–3-D registration. IEEE Trans Med Imaging 24(9):1177–1189
Markelj P, Tomaževič D, Likar B, Pernuš F (2012) A review of 3D/2D registration methods for image-guided interventions. Med Image Anal 16(3):642–661
Groher M, Zikic D, Navab N (2009) Deformable 2D–3D registration of vascular structures in a one view scenario. IEEE Trans Med Imaging 28(6):847–860
Hipwell JH et al (2003) Intensity-based 2-D–3-D registration of cerebral angiograms. IEEE Trans Med Imaging 22(11):1417–1426
Kerrien E, Berger M-O, Maurincomme E, Launay L, Vaillant R, Picard L (1999) Fully automatic 3D/2D subtracted angiography registration. In: Medical image computing and computer-assisted intervention—MICCAI 1999. Springer, London, pp 664–671
Feldmar J, Ayache N, Betting F (1997) 3D–2D projective registration of free-form curves and surfaces. Comput Vis Image Und 65(3):403–424
Groher M, Jakobs TF, Padoy N, Navab N (2007) Planning and intraoperative visualization of liver catheterizations: new CTA protocol and 2D–3D registration method. Acad Radiol 14(11):1325–1340
Groher M, Bender F, Hoffmann R-T, Navab N (2007) Segmentation-driven 2D–3D registration for abdominal catheter interventions. In: Medical image computing and computer-assisted intervention—MICCAI, 2007, vol 10. Springer, Berlin, pp 527–535
Kita Y, Wilson DL, Noble A (1998) Real-time registration of 3D cerebral vessels to X-ray angiograms. In: Medical image computing and computer-assisted interventation—MICCAI 1998, vol 1496. Springer, Berlin, pp 1125–1133
Rivest-Hénault D, Sundar H, Cheriet M (2012) Nonrigid 2D/3D registration of coronary artery models with live fluoroscopy for guidance of cardiac interventions. IEEE Trans Med Imaging 31(8):1557–1572
Tomaževič D, Likar B, Slivnik T, Pernuš F (2003) 3-D/2-D registration of CT and MR to X-ray images. IEEE Trans Med Imaging 22(11):1407–1416
Markelj P, Tomaževič D, Pernuš F, Likar B (2008) Robust gradient-based 3-D/2-D registration of CT and MR to X-ray images. IEEE Trans Med Imaging 27(12):1704–1714
Livyatan H, Yaniv Z, Joskowicz L (2003) Gradient-based 2-D/3-D rigid registration of fluoroscopic X-ray to CT. IEEE Trans Med Imaging 22(11):1395–1406
Chan HM, Chung ACS, Yu SCH, Wells WM III (2004) 2D–3D vascular registration between digital subtraction angiographic (DSA) and magnetic resonance angiographic (MRA) images. Presented at the IEEE international symposium on biomedical imaging: nano to macro, vol 1, pp 708–711
Turgeon G-A, Lehmann G, Guiraudon G, Drangova M, Holdsworth D, Peters T (2005) 2D–3D registration of coronary angiograms for cardiac procedure planning and guidance. Med Phys 32(12):3737–3749
Vermandel M, Betrouni N, Gauvrit J-Y, Pasquier D, Vasseur C, Rousseau J (2006) Intrinsic 2D/3D registration based on a hybrid approach: use in the radiosurgical imaging process. Cell Mol Biol 52(6):44–53
Jomier J, Bullitt E, Van Horn M, Pathak C, Aylward SR (2006) 3D, 2D model-to-image registration applied to TIPS surgery. In: Medical image computing and computer-assisted intervention—MICCAI 2006, vol 9. Springer, Berlin, pp 662–669
Ruijters D, ter Haar Romeny BM, Suetens P (2009) Vesselness-based 2D–3D registration of the coronary arteries. Int J Comput Assist Radiol Surg 4(4):391–397
Metz C et al (2013) Registration of 3D+t coronary CTA and monoplane 2D+t X-ray angiography. IEEE Trans Med Imaging 32(5):919–931
Copeland AD, Mangoubi RS, Desai MN, Mitter SK, Malek AM (2010) Spatio-temporal data fusion for 3D+T image reconstruction in cerebral angiography. IEEE Trans Med Imaging 29(6):1238–1251
Powell MJD (1964) An efficient method for finding the minimum of a function of several variables without calculating derivatives. Comput J 7(2):155–162
Madan H, Pernuš F, Likar B, Špiclin Ž (2017) A framework for automatic creation of gold-standard rigid 3D–2D registration datasets. Int J CARS 12(2):263–275
Mitrović U, Pernuš F, Likar B, Špiclin Ž (2015) Simultaneous 3D–2D image registration and C-arm calibration: application to endovascular image-guided interventions. Med Phys 42(11):6433–6447
Hansen N (2012) The CMA evolution strategy: a comparing review. In: Lozano JA, Larrañaga P, Inza I, Bengoetxea E, (eds) Springer, Berlin pp 75–102
Funding
This research was supported by the Slovenian Research Agency (Grants Nos. P2-0232, J2-5473, J7-6781, J2-7211 and J2-8173).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest related to this research work.
Ethical standard
For this type of study, formal consent is not required.
Rights and permissions
About this article
Cite this article
Mitrović, U., Likar, B., Pernuš, F. et al. 3D–2D registration in endovascular image-guided surgery: evaluation of state-of-the-art methods on cerebral angiograms. Int J CARS 13, 193–202 (2018). https://doi.org/10.1007/s11548-017-1678-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-017-1678-2