Abstract
Purpose
Since pre-processing and initial segmentation steps in medical images directly affect the final segmentation results of the regions of interesting, an automatic segmentation method of a parameter-adaptive pulse-coupled neural network is proposed to integrate the above-mentioned two segmentation steps into one. This method has a low computational complexity for different kinds of medical images and has a high segmentation precision.
Methods
The method comprises four steps. Firstly, an optimal histogram threshold is used to determine the parameter \(\alpha \) for different kinds of images. Secondly, we acquire the parameter \(\beta \) according to a simplified pulse-coupled neural network (SPCNN). Thirdly, we redefine the parameter V of the SPCNN model by sub-intensity distribution range of firing pixels. Fourthly, we add an offset \(A\times S_{\mathrm{off}}\) to improve initial segmentation precision.
Results
Compared with the state-of-the-art algorithms, the new method achieves a comparable performance by the experimental results from ultrasound images of the gallbladder and gallstones, magnetic resonance images of the left ventricle, and mammogram images of the left and the right breast, presenting the overall metric UM of 0.9845, CM of 0.8142, TM of 0.0726.
Conclusion
The algorithm has a great potential to achieve the pre-processing and initial segmentation steps in various medical images. This is a premise for assisting physicians to detect and diagnose clinical cases.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Medical image segmentation, such as ultrasound image [1,2,3,4], CT image [5,6,7,8], magnetic resonance image [9,10,11], has been playing an increasingly important role in image processing field. Most fashionable segmentation algorithms in medical images invariably capture the local details of object regions to determine the segmentation results and are applied to assist physician’s diagnosis. Further, segmentation steps of these algorithms are always divided into pre-processing, initial segmentation, coarse segmentation, fine segmentation, and post-processing. Hereinto, pre-processing and initial segmentation are indispensable regardless of the properties of the images.
Medical image methods with a global threshold are usually used to segment the images into different objects [12, 13]. There are some of image threshold methods, such as static threshold methods including multi-scale 3D Otsu thresholding [14], and dynamic threshold methods including PCNN [15]. More applications of medical image threshold are obtained from the literatures of Musrrat [16] and Guo [17].
PCNN has broad applications in many aspects for image processing [18]. For examples, image segmentation [19, 20], image enhancement [21, 22] and object recognition [23, 24]. Hereinto, the PCNN has a great potential in image segmentation. Temporal similarity and spatial proximity of the output pulses of PCNN always provides an image segmentation property. One firing neuron corresponding to the pixel directly impacts its adjacent neurons and the segmentation result of the whole image. However, a majority of the prevalent PCNN algorithms require to manually or semi-automatically set parameters, and there are only several automatic segmentation algorithms containing Berg et al. [25], Ma et al. [26], and Chen et al. [27].
Although the above-mentioned methods have better segmentation effects than the basic PCNN model, it is still possible to further simplify parameters setting. What is more, physicians still need to acquire more precise segmentation results to analyze and diagnose relative clinical cases in shorter time. Therefore, we develop a novel image segmentation method based on PA-PCNN. In this method, we attempt to merge pre-processing and initial segmentation steps into one to improve segmentation precision and decrease computational complexity, and try to search a method to achieve partial segmentation for different kinds of medical images with different organ sites.
Our method contributes two new ideas. Firstly, it changes five parameters of the SPCNN model to three parameters of the PA-PCNN model. Hereinto, parameters \(\alpha _{\mathrm{f}}\) and \(\alpha _{\mathrm{e}}\) in SPCNN are combined into a parameter \(\alpha \), which represents the decay factor of the PA-PCNN. The parameter \(\beta \) is retained because of its necessity and significance. The parameter \(V_{\mathrm{E}}\) is set to V, which represents the weighing factor of the PA-PCNN. Secondly, we add a controlling parameter, which judges whether the image is over-segmentation or under-segmentation, and an offset \(S_{\mathrm{off}}\), which adjusts the value of weighing factor V, to improve segmentation accuracy rates. This image segmentation method simplifies the expression of all parameters and calculates them by optimal histogram threshold \(S'\) [28], which is appropriate for medical image segmentation.
Materials
Experimental images, containing 80 ultrasound images of the gallbladder and gallstones from Gansu Provincial Hospital in China, 680 magnetic resonance images of the left ventricle from Medical Image Computing and Computer Assisted Intervention Society (MICCAI), and 322 mammogram images of left and right breast from Mammographic Image Analysis Society (MIAS) database [29], were adopted on the research. Hereinto, magnetic resonance images have three cases containing 240 images from the HF-I database, 260 images from the HF-NI database and 180 images from the HYP database. The ultrasound images, the magnetic resonance images and the mammogram images have resolution of \(512\times 512\) pixels, \(512\times 512\) pixels and \(1024\times 1024\) pixels, respectively.
The SPCNN model
Basic PCNN model
It is known that the PCNN does not require any training and only has one single layer. In contrast to other PCNN models, Chen et al.’s SPCNN model [27], derived from Zhan et al. ’s SCM model [30], has higher segmentation accuracy and lower computational complexity. Therefore, the SPCNN could be employed in this paper and is written as follows:
where
In the SPCNN model, Neuron \(N_{\mathrm{ij}}\) in position (i, j) has simplified feeding input \(F_{\mathrm{ij}}[n]\) denoted by an input stimulus \(S_{\mathrm{ij}}\), and linking input \(L_{\mathrm{ij}}[n]\) denoted by the product of a synaptic weight \(W_{\mathrm{ijkl}}\), eight neighboring outputs, and a weighing factor \(V_{\mathrm{L}}\). These inputs in internal activity \(U_{\mathrm{ij}}[n]\) are modulated by the linking strength \(\beta \). Internal activity \(U_{\mathrm{ij}}[n]\) also records its previous state by the decay factor \(e^{-\alpha _{\mathrm{f}}}\). \(\alpha _{\mathrm{f}}\) and \(\alpha _{\mathrm{e}}\) represent decay factors of internal activity \(U_{\mathrm{ij}}[n]\) and dynamic threshold \(E_{\mathrm{ij}}[n]\), respectively. Moreover, there are five adjustable parameters \(\alpha _{\mathrm{f}}\), \(\alpha _{\mathrm{e}}\), \(\beta \), \(V_{\mathrm{E}}\) and \(V_{\mathrm{L}}\), and these parameters could be set automatically
where \(S'\) and \(S_{\mathrm{max}}\) denote Otsu thresholding and the maximum intensity of the image. \(\sigma (S)\) represents standard deviation of the image. M [3] is the most significantly term of internal activity \(U_{\mathrm{ij}}[n]\) in the third iteration. In the above formulae, parameters \(\beta \), \(\alpha _{\mathrm{e}}\) and \(V_{\mathrm{E}}\) always generate the large impact for the SPCNN. Obviously, the larger the value of \(\beta \), the more strongly a neuron is influenced by its eight adjacent outputs. The larger the values of \(\alpha _{\mathrm{e}}\) or \(V_{\mathrm{E}}\), the lower segmentation accuracy rates become.
Sub-intensity ranges in the SPCNN model
In this paper, segmentation steps and sub-intensity ranges of the SPCNN are shown by two examples of the gallbladder and gallstones in ultrasound images (Figs. 1, 2). Figure 1 shows SPCNN segmentation steps which generates ineffective segmentation results at previous two iterations and effective segmentation results at subsequent iterations. Figure 2a, b shows the sub-intensity ranges of all firing neurons for Fig. 1a, d, respectively.
Parameter setting method of PA-PCNN
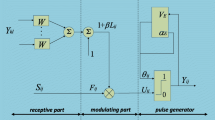
After using the SPCNN, one gray image is divided into the object and the background at the third iteration and generates further segmentation results at subsequent iterations, whereas we still need to set five parameters. Therefore, we proposed an automatic parameter setting method based on the PA-PCNN model to improve image segmentation precision and decrease computational complexity. Finally, the new method is determined at subsequent sections for detail and the general flowchart of our method is shown in Fig. 3.
Parameter \(\alpha \)
The parameters \(\alpha _{\mathrm{f}}\) and \(\alpha _{\mathrm{e}}\) in SPCNN represent decay factors of the dynamic threshold E and the internal activity U, respectively. \(\sigma \)(S) is the standard deviation of the image, whereas the parameters \(\alpha _{\mathrm{f}}\) and \(\alpha _{\mathrm{e}}\) have a large decay rates. This directly determines segmentation accuracy rates in the iteration. Therefore, by a large number of experiments, normalized optimal histogram threshold \(S'\) could be employed in our work as follows:
It is noted that the increase of the parameter \(\alpha \) is with the decrease of the threshold \(S'\). This threshold can enhance the relationship between parameters \(\alpha \) and \(\beta \).
Parameter \(\beta \)
According to (8) and (10), since \(S_{\max }\approx 1\) in most medical images, we can reset the parameter \(\beta \) as
Parameter V
In the SPCNN model, \(\beta V_{\mathrm{L}}\) is always a factor as a whole. What is more, according to (10), the value of the parameter \(V_{\mathrm{L}}\) is equal to 1. Therefore, we can remove the parameter \(V_{\mathrm{L}}\) and the expression of new linking input is given as
Subsequently, we will redefine parameter \(V_{\mathrm{E}}\) as V and the formula in [18] can be adopted as
According to (16), dynamic threshold \(E[1+l-1]\) in the decay step \(1+l-1\) would be larger than that of the internal activity \(U[1+l]\) in the decay step \(1+l\), while the minimum value of dynamic threshold \(E[1+l]\) in the decay step \(1+l\) would be less than that of internal activity \(U[1+l+1]\) in the decay step \(1+l+1\). This indicates that there is a sub-intensity range of firing pixels in the decay step \(1+l+1\). Besides, if one medical image generates the first segment in the second iteration, we could determine parameter l as 0 and the firing neurons can satisfy the firing conditions
U[0] and E[0] are always set to 0 and U[1] is more than 0 under normal circumstances. This obeys the first firing condition in (17). Thus, to acquire the first image segment in the second iteration, we can redefine the firing condition
According to (3), the minimum intensity of firing pixels in U[2] is expressed as
and according to (5), \(E[1]=V_{E}=V\). The formula (18) is rewritten as
In addition, Otsu thresholding \(S'\), which always generates a larger value than \(S_{\mathrm{low}}\), can be substituted into (20) and the above formula is redefined as
Subsequently, we obtain the unique value of V in light of the highest limiting in (21)
To further improve image segmentation precision, we could add an offset \(S_{\mathrm{off}}\) and the controlling parameter A, and the new formula is given as
where the parameter \(S_{\mathrm{off}}\) denotes a small offset. A denotes a parameter to judge whether the testing result is over-segmentation or under-segmentation as shown in (24)
According to (24), the formula with \(A=1\) denotes over-segmentation of images without the offset \(S_{\mathrm{off}}\). At this time, the value of V would be added by the offset \(S_{\mathrm{off}}\) to reduce the number of firing pixels and obtain reasonable segmentation results. What is more, the formula with \(A=-1\) shows an opposite situation. Finally, the steps of the whole algorithm are shown in Algorithm 1.

In the Algorithm 1, \(F_{\mathrm{x}}\) and \(F_{\mathrm{y}}\) denote the average gradient value of the whole image in the x direction and y direction, respectively. \(F_{\mathrm{mean}}\) is expressed as
and the parameter B is shown as
In (26), the parameter C is a weighing factor (here, \(C=100\)). In summary, the parameters of the PA-PCNN are not independent but interact with each other by optimum histogram threshold \(S'\). The PA-PCNN model is shown in Fig. 4, and its formulae are described as follows:
where
Experiments and analysis
To compare our algorithm with the state-of-the-art algorithms, five competitive methods including SPCNN [27], NSCM [31], Otsu [28], Kittler [32], ISPCNN [15], and three metrics including uniformity measurement (UM) [33], contrast measurement (CM) [34], runtime measurement (TM) are employed on our research. Moreover, 80 ultrasound images of the gallbladder and gallstones, 680 magnetic resonance images of the left ventricle, and 322 mammogram images of left and right breast are used as experimental images. All tests are run on MATLAB 7.11.0 with Intel(R) Core(TM) i3 M 350 at 2.27GHz from Satellite L600 of Toshiba.
Segmentation evaluation criteria
For the evaluation of ultrasound images, magnetic resonance images and mammogram images, three metrics analyzing the segmentation precision and computational complexity of the object and the background can be used in this paper. Hereinto, the metric UM denotes the uniformity of the segmented region and is written as
where C denotes the maximum normalized intensities of the whole image. For a segmented image, \(R_{\mathrm{j}}\) and A denote the \(j\hbox {th}\) region and its area, respectively. n denotes the number of regions. i denotes the number of gray-level (i.e., i is set to 2 in the binary image). Further, the larger the value of UM, the better segmentation effect becomes. Subsequently, the metric CM denotes the contrast of pixel intensities
where \(f_\mathrm{o}\) and \(f_\mathrm{b}\) denote the mean value of the object and the background, respectively. The larger the value of CM, the higher segmentation accuracy rates of the image becomes.
The three-dimensional graph of the final segmentation results: Data in the first, second and third row denote the evaluation results of the metrics UM, CM and TM, respectively; data in the first, second and third columns denote the evaluation results of the our method, the SPCNN method, the NSCM method, the Otsu method, the Kittler method and the ISPCNN method, respectively
The segmentation results of the ultrasound and the mammogram images: Images in the column (1) denote original images; images in the columns (2), (3), (4), (5), (6) and (7) denote segmentation results of our method, the SPCNN method, the NSCM method, the Otsu method, the Kittler method and the ISPCNN method, respectively; images in the rows a and b denote segmentation results of mammogram images; images in the rows c and d denote segmentation results of ultrasound images
Experiment analysis and discussion
In our experiments, all images are divided into five groups including 80 ultrasound images in the first group, 240 magnetic resonance images of the HF-I database in the second group, 260 magnetic resonance images of the HF-NI database in the third group, 180 magnetic resonance images of the HYP database in the third group and 322 mammogram images in the fourth group. Moreover, we compare our method with other methods to obtain ultimate evaluation results in Table 1 and their three-dimensional graph is shown in Fig. 5. We also select one image in each group as the example to show experimental results (Figs. 6, 7).
The segmentation results of the MR images: Images in the column (1) denote original images; images in the columns (2), (3) and (4) denote segmentation results of the our method, the SPCNN method, the SCM method, the Otsu method, the Kittler method and the ISPCNN method, respectively; images in the rows a denote segmentation results for the HF-I dataset; images in the rows b denote segmentation results for the HF-NI dataset; images in the rows c denote segmentation results for the HYP dataset
According to the above Table 1 and Fig. 5, compared with other state-of-the-art algorithms, our algorithm has a good performance for the evaluations of the metrics.
Discussion
In this paper, we present an image segmentation method based on PA-PCNN to further simplify computations and improve initial segmentation accuracy for various medical images. In contrast to other methods of the PCNN, our method only set three parameters \(\alpha \), \(\beta \), V, which are associated with each other by the Otsu thresholding \(S'\), and retain main properties of the basic PCNN model. Our method, which determines rapidly and effectively the initial segmentation result in the pre-processing step and initial segmentation step for various medical images, obviously reduces the runtime and improve segmentation accuracy rates of the whole method including the coarse-fine segmentation and the post-processing. So, the whole segmentation method including our method is more easily to generate a good final segmentation result, which brings a great help to diagnose clinical cases for physicians. For example, for gallbladder and gallstone regions segmentation, the computing program of the whole segmentation method including PA-PCNN is copied to the chip controlled by ultrasonic diagnostic apparatus. When physicians use detectors to seek out lesion regions, the monitor will show rapidly segmentation results in most cases.
Our method for medicine has three main significances: (1) It obtains segmentation results more rapidly and accurately for physicians. (2) It avoids the otherness from subjective experience and human knowledge. (3) It improves the reliability and accuracy of clinical diagnosis for physicians.
By a large number of experiments employing three metrics, five comparative methods and 1082 medical images containing 80 ultrasound images of the gallbladder and gallstones, 680 magnetic resonance images of the left ventricle and 322 mammogram images of left and right breast, we demonstrate that the image segmentation method has low computational complexity and high initial segmentation accuracy rates, presenting the overall metric UM of 0.9845, CM of 0.8142, TM of 0.0726.
In the future, we firstly continue to add other types of medical images with other organ sites into our dataset. Secondly, for several types of medical images, we will combine coarse segmentation steps into modified PA-PCNN for further adding the applicable range of the method. Thirdly, we will use the whole segmentation method including PA-PCNN to validate the effectiveness and robustness of our method for gallbladder and gallstones in clinical diagnosis.
Conclusion
This paper indicates that the proposed method has better testing performance than other comparative methods because all calculated parameters are automatically acquired by self-adaptive ways. The method has a great potential to achieve the pre-processing and initial segmentation for various medical images. Although the segmentation ways of other clinical cases are being discussed, the new method is still a promising method for medical image segmentation.
References
Hareendranathan A, Mabee M, Punithakumar K, Noga M, Jaremko J (2016) A technique for semiautomatic segmentation of echogenic structures in 3D ultrasound, applied to infant hip dysplasia. Int J Comput Assist Radiol Surg 11(1):31–42. doi:10.1007/s11548-015-1239-5
Nobel J, Boukerroui D (2006) Ultrasound image segmentation: a survey. IEEE Trans Med Imaging 25(8):987–1010. doi:10.1109/TMI.2006.877092
Gupta J, Gosain B, Kaushal S (2010) A comparison of two algorithms for automated stone detection in clinical B-mode ultrasound images of the abdomen. Int J Clin Monit Comput 24(5):341–362. doi:10.1007/s10877-010-9254-0
Lian J, Ma Y, Ma Y, Shi B, Liu J, Yang Z, Guo Y (2017) Automatic gallbladder and gallstone regions segmentation in ultrasound image. Int J Comput Assist Radiol Surg. doi:10.1007/s11548-016-1515-z
Yang X, Ye X, Slabaugh G (2015) Multilabel region classification and semantic linking for colon segmentation in CT colonography. IEEE Trans B Biomed Eng 62(3):948–959. doi:10.1109/TBME.2014.2374355
Zou X, Li Z (2016) TV-based correction for beam hardening in computed tomography. J Med Imaging Heal Inf 6(7):1701–1707. doi:10.1166/jmihi.2016.1875
Dandin O, Teomete U, Osman O, Tulum G, Ergin T, Sabuncuoglu M (2016) Automated segmentation of the injured spleen. Int J Comput Assist Radiol Surg 11(3):351–368. doi:10.1007/s11548-015-1288-9
Hanaoka S, Masutani Y, Nenoto M, Nomura Y, Miki S, Yoshikawa T, Hayashi N, Ohtomo K, Shimizu A (2017) Landmark-guided diffeomorphic demons algorithm and its application to automatic segmentation of the whole spine and pelvis in CT images. Int J Comput Assist Radiol Surg 12(3):413–430. doi:10.1007/s11548-016-1507-z
Wang Z, Zhang X, Dou W, Zhang M, Chen H, Lu M, Li S (2016) Best Window Width Determination and Glioma Analysis Application of Dynamic Brain Network Measure on Resting-State Functional Magnetic Resonance Imaging. J Med Imaging Heal Inf 6(7):1735–1740. doi:10.1166/jmihi.2016.1881
Ma Y, Wang L, Ma Y, Dong M, Du S, Sun S (2016) Novel automatic segmentation of left ventricle in cardiac cine MR images. Int J Comput Assist Radiol Surg 11(11):1951–1964. doi:10.1007/s11548-016-1429-9
Faghih Roohi S, Aghaeizadeh Zoroofi R (2013) 4D statistical shape modeling of the left ventricle in cardiac MR images. Int J Comput Assist Radiol Surg 8(3):335–351. doi:10.1007/s11548-012-0787-1
Sezgin M, Sankur B (2004) Survey over image thresholding techniques and quantitative performance evaluation. J Electron Imaging 13(1):146–165. doi:10.1117/1.1631315
Lee S, Chung S, Park R (1990) A comparative performance study of several global thresholding techniques for segmentation. Comput Vis Graph Image Process 52(2):171–190. doi:10.1016/0734-189X(90)90053-X
Feng Y, Zhao H, Li X, Zhang X, Li H (2016) A multi-scale 3D Otsu thresholding algorithm for medical image segmentation. Digit Signal Process 60:186–199. doi:10.1016/j.dsp.2016.08.003
Yang Z, Dong M, Guo Y, Gao X, Wang K, Shi B, Ma Y (2016) A new method of micro-calcifications detection in digitized mammograms based on improved simplified PCNN. Neurocomputing. doi:10.1016/j.neucom.2016.08.068
Musrrat A, Ch W, Pant M (2013) Multi-level image thresholding by synergetic differential evolution. Appl Soft Comput 17(3):1–11. doi:10.1016/j.asoc.2013.11.018
Guo Y, Dong M, Yang Z, Gao X, Wang K, Luo C, Ma Y, Zhang J (2016) A new method of detecting micro-calcification clusters in mammograms using contourlet transform and non-linking simplified PCNN. Comput Methods Progam Biomed 130:31–45. doi:10.1016/j.cmpb.2016.02.019
Zhan K, Shi J, Wang H, Xie Y, Li Q (2016) Computational mechanisms of pulse-coupled neural networks: a comprehensive review. Arch Comput Methods Eng. doi:10.1007/s11831-016-9182-3
Ma H, Cheng X (2014) Automatic image segmentation with PCNN algorithm based on grayscale correlation. Int J Signal Process 7(5):249–258. doi:10.14257/ijsip.2014.7.5.22
Zhuang H, Low K, Yau W (2012) Multichannel Pulse-Coupled-Neural-Network-Based Color Image Segmentation for Object Detection. IEEE Trans Ind Electron 59(8):3299–3308. doi:10.1109/TIE.2011.2165451
Zheng W, Pu T, Chen J, Zeng H (2012) Image contrast enhancement by contour let transform and PCNN. In: Audio lang image process (ICALIP) international conference, pp 735–739. doi:10.1109/ICALIP.2012.6376711
Xu G, Li C, Zhao J, Lei B (2014) Multiplicative decomposition based image contrast enhancement method using PCNN factoring model. In : Intelligent control and automation (WCICA), pp 1511–1566. doi:10.1109/WCICA.2014.7052943
Yu B, Zhang L (2004) Pulse-coupled neural networks for contour and motion matchings. IEEE Trans Neural Netw 15(5):1186–1201. doi:10.1109/TNN.2004.832830
Chen Y, Ma Y, Park S (2015) Region-based object recognition by color segmentation using a simplified PCNN. IEEE Trans Neural Netw Learn Syst 26(8):1682–1697. doi:10.1109/TNNLS.2014.2351418
Berg H, Olsson R, Lindblad T, Chilo J (2008) Automatic design of pulse coupled neurons for image segmentation. Neurocomputing 71(10):1980–1993. doi:10.1016/j.neucom.2007.10.018
Ma Y, Qi C (2006) Study of automated PCNN system based on genetic algorithm. J Syst Simul 18(3):722–725
Chen Y, Park S, Ma Y, Ala R (2011) A new automatic parameter setting method of a simplified PCNN for image segmentation. IEEE Trans Neural Netw 22(6):880–892. doi:10.1109/TNN.2011.2128880
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9(1):62–66. doi:10.1109/TSMC.1979.4310076
Suckling J, Parker J, Dance D, Astley S, Hutt I, Boggis C, Ricketts I, Stamatakis E, Cerneaz N, Kok S (1994) The mammographic image analysis society digital mammogram database. In :Excerpta medica international congress series, pp 375–378
Zhan K, Zhang H, Ma Y (2009) New spiking cortical model for invariant texture retrieval and image processing. IEEE Trans Neural Netw 20(12):1980–1986. doi:10.1109/TNN.2009.2030585
Zhan K, Shi J, Li Q, Teng J (2015) Image segmentation using fast linking SCM. Int Jt Confere Neural Netw (IJCNN). doi:10.1109/IJCNN.2015.7280579
Kittler J, Illingworth J (1986) Minimum error thresholding. Pattern Recognit 19(1):41–47. doi:10.1016/0031-3203(86)90030-0
Sahoo P, Soltani S, Wong A (1988) A survey of thresholding techniques. Comput Graph Vis Image Process 41(2):233–260. doi:10.1016/0734-189X(88)90022-9
Levine M, Naxif A (1985) Dynamic measurement of computer generated image segmentation. IEEE Transactions Pattern Anal Mach Intell 7(2):155–164. doi:10.1109/TPAMI.1985.4767640
Acknowledgements
The authors thank all the reviewers for their valuable comments, which further improved the quality of the paper. This study was funded National Natural Science Foundation of China (Grant Numbers 61175012 & 61201422), Natural Science Foundation of Gansu Province of China (Grant Number 148RJZA044) and Youth Foundation of Lanzhou Jiaotong University of China (Grant Numbers 2013004 & 2014005).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Lian, J., Shi, B., Li, M. et al. An automatic segmentation method of a parameter-adaptive PCNN for medical images. Int J CARS 12, 1511–1519 (2017). https://doi.org/10.1007/s11548-017-1597-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-017-1597-2