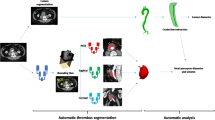
Abstract
Purpose
Abdominal aortic aneurysm (AAA) is a localized, permanent and irreversible enlargement of the artery, with the formation of thrombus into the inner wall of the aneurysm. A precise patient-specific segmentation of the thrombus is useful for both the pre-operative planning to estimate the rupture risk, and for post-operative assessment to monitor the disease evolution. This paper presents a generic approach for 3D segmentation of thrombus from patients suffering from AAA using computed tomography angiography (CTA) scans.
Methods
A fast and versatile thrombus segmentation approach has been developed. It is composed of initial centerline detection and aorta lumen segmentation, an optimized pre-processing stage and the use of a 3D deformable model. The approach has been designed to be very generic and requires minimal user interaction. The proposed method was tested on different datasets with 145 patients overall, including pre- and post-operative CTAs, abdominal aorta and iliac artery sections, different calcification degrees, aneurysm sizes and contrast enhancement qualities.
Results
The thrombus segmentation approach showed very accurate results with respect to manual delineations for all datasets (\(\hbox {Dice} = 0.86 \pm 0.06, 0.81 \pm 0.06\) and \(0.87 \pm 0.03\) for abdominal aorta sections on pre-operative CTA, iliac artery sections on pre-operative CTAs and aorta sections on post-operative CTA, respectively). Experiments on the different patient and image conditions showed that the method was highly versatile, with no significant differences in term of precision. Comparison with the level-set algorithm also demonstrated the superiority of the 3D deformable model. Average processing time was \(8.2 \pm 3.5 \hbox { s}\).
Conclusion
We presented a near-automatic and generic thrombus segmentation algorithm applicable to a large variability of patient and imaging conditions. When integrated in an endovascular planning system, our segmentation algorithm shows its compatibility with clinical routine and could be used for pre-operative planning and post-operative assessment of endovascular procedures.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The abdominal aortic aneurysm (AAA) is a localized, permanent and irreversible enlargement of the abdominal aorta to at least 1.5 times the diameter measured at renal arteries level, or exceeding the normal diameter by \({>}50\%\) [1]. Iliac artery aneurysms are often associated with AAA and are most commonly localized in the common iliac artery section (\(70\%\)). This pathology is the result of arterial walls weakening, and when blood flows against the wall, it can cause an enlargement and a loss of parallelism between its walls. The changing shape of the aorta is usually the cause of the formation of thrombus into the inner wall of the aneurysm. Even if the effect of thrombus volume on aneurysm rupture is still debated [2], accurate measurements of its maximal diameter are admitted to be a reliable estimate of aneurysm rupture risk. Post-operatively, periodical imaging is performed and monitoring the thrombus development is a key indicator for the patient recovery [3]. Complications are detected when the volume of the thrombus does not reduced, which can be due to faulty isolation from the blood flow. On the contrary, a decrease of thrombus volume indicates a suitable recovery. For those reasons, the standard follow-up procedure usually includes manual aneurysm delineation in the image. In order to reduce analysis time and to increase reproducibility, automated segmentation of thrombus would therefore be of great value for clinicians.
The preferred imaging exam for pre-operative planning of endovascular procedures is the 3D contrast computerized tomography angiography (CTA), while for post-operative assessment of AAA a CTA or an echography can be employed. The aorta lumen is relatively easy to segment, because the contrast agent injected to patient during the CTA examination highlights blood flow on the image which appears hyper-intense. On the contrary, thrombus is not highlighted because it does not receive any contrast agent. Thrombus hountzfield (HU) values on a CTA are usually between 0 and 100 HU, similar to many tissues (e.g., kidney, muscle, liver) which implies a low contrast between the thrombus and surrounding tissues (Fig. 1). Furthermore, on the contrary to organ segmentation, position and shape of the thrombus is highly heterogeneous among the population. Other challenges arise from the artifacts generated by stents, if any, the presence of calcium deposits, or the variability of the texture due to blood leaking. All of these reasons make the task of thrombus segmentation very challenging.
Even if some works on thrombus segmentation were published using different image modalities such as MRI [4] or multi-spectral MRI [5], we are only interested here in segmentations from CT or CTA, adopted to be the modality of choice in endovascular procedures. Using those input images, many different techniques may be used that can be classified according to the kind of information that guides the segmentation process. First work on the subject used neural networks to learn thresholds for multilevel thresholding and a constraint-satisfaction neural network to smooth the boundaries, followed by user manual corrections [6]. In the category of level-set method, initial work was proposed by Loncaric et al. [7], who proposed a 2D framework with a level-set approach and a stopping criterion curve. Zohios et al. [8, 9] utilized geometrical methods to extract both the thrombus and the external AAA wall boundaries, with a strong assumption on the presence of calcifications. In the approach by Subasic et al. [10, 11], a sphere is initialized inside the aorta and a 2D level-set followed by 3D level-set is performed. Demerci et al. [12] used an hybrid deformable model that integrates global and local image information and combines it with shape constraints using NURBRS surface and distance functions. Some techniques are based on statistical models guided by a priori information, where information is extracted from a manually processed set of images known as training set. Using active shape or active appearance models, Bruijne et al. [13, 14] used contour obtained in the last slice for initialization, while Das et al. [15] proposed a narrow band active contour model. Dehmeshky et al. [16] proposed a grayscale and geometric appearance model. Maiora et al. [17, 18] present an active leaning image segmentation system over different image texture features such as gray-level co-occurrence matrix and local binary patterns. Macia et al. [19, 20] proposed to use a radial model with spatial constraint and prior knowledge, followed by a correction stage based on radial sectors. Using graph-cut algorithm, Lee et al. [21, 22] proposed a semi-automatic segmentation that can be interactively and easily controlled by users, and Freiman et al. [23] applied an iterative model-constrained graph-cut algorithm. Combinations of multiple algorithms were also proposed, for instance by Olabarriaga et al. [24] who employed a deformable model approach along with a nonparametric statistical gray-level appearance model, while Zhuge et al. [25] presented a level-set segmentation based on a parametric statistical model.
Even if the most recent studies succeeded in reaching reasonable good segmentation results, the majority of them is either requiring a very high degree of user interactions to be sufficiently accurate, or is showing very high computational burden (often several minutes) which is incompatible with clinical routine. Most importantly, none of these methods has been tested with on one hand a large dataset of patients, and on the other hand with different imaging and anatomical variabilities. In this paper, we will follow a 3D deformable model approach with an adequate pre-processing step that will be tuned to work with pre- and post-operative image, with/without calcifications and on different artery sections. The proposed approach does not require any training stage, requires minimal user interaction, shows very low computation time, is very accurate, and will be tested with different dataset to highlight its versatility.
Methods
Given a volume of interest from a CTA scan, we first detected the aorta centerline and segmented the lumen (“Lumen segmentation and centerline extraction” section). Based on this initial segmentation, the objective is to separate the aneurysmal thrombus from the lumen and the surrounding structures. To this end, an optimized pre-processing step is applied to the volume of interest to highlight major contours and remove the influence of the lumen (“Pre-processing” section). Then, we approach the problem of thrombus segmentation in a 3D manner using a deformable model (“3D deformable model” section) along with a mesh initialized with information about the prior lumen segmentation stage (“Mesh initialization” section). Different experiments and validations on a large datasets of patients treated with endovascular procedures will finally be conducted (“Validation” section).
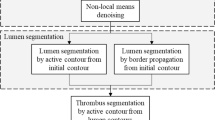
Lumen segmentation and centerline extraction
The only required user interaction is the placement of two seed points (a starting and ending point) to delimit the volume of interest where the thrombus has to be calculated (commonly the aneurysmal sac). Both seeds points are asked to be placed in the aortic lumen. After a rough segmentation of the whole aorta using binary threshold (100–600 HU) and region growing to remove small connected components, a fast-marching-based minimal path detection algorithm [26] is used to retrieve the aorta centerline. Starting from the detected centerline, the lumen is automatically segmented using a region growing algorithm that allows removing all neighboring tissues. The segmentation is then locally refined to determine the contours based on local intensity statistics. This step, which is required for endovascular planning, has already been presented in a previous paper [27] and will not be developed here. This precise segmentation of the aorta will be used for visualization purposes, but also for the pre-processing (“Pre-processing” section) and for mesh initialization (“Mesh initialization” section).
Pre-processing
The pre-processing step is inspired from the work of Subasic et al. [28] and is a crucial for capturing the gradient information corresponding to the delimitation between thrombus and surrounding tissues. The overall pre-processing step is depicted on Fig. 2. First, we only want to highlight the thrombus boundaries. We therefore replaced the segmented lumen from the previous step using a constant value close to the average thrombus value (50 HU). Gradient information from the lumen/thrombus boundary is therefore removed. Then, as the nature of the noise in CT data is always non-isotropic, we applied a curvature anisotropic filtering process. This filter is an edge-preserving smoothing which preserved information on boundaries and considerably reduce noise. After smoothing, and on the contrary of other works (e.g., [28]), we do not want to use calcifications to constrain our deformable model. Adding boundary calculated from calcification information will results in an under-estimation of the total thrombus volume as the deformable shape will stop at inner boundaries. We therefore apply a passband filter from −2000 to 600 HU to remove calcifications. We then apply two different processing to keep the maximum of gradient. In the first instance, we simply apply a binary threshold (0–100 HU) on the smoothed image. In the second instance, we compute the gradient magnitude of the smoothed image which allows computing the luminance variation between neighboring voxels. To obtain gradient values that truly reflect boundaries, we apply a binary threshold on this gradient image (50–8000). Resulting images are then merged to obtain an enhanced binary image with the maximum of potential boundaries information. This enhanced binary image is then filtered to remove small foreground and background connected regions. These regions could potentially disturb the progress of the deformable model. Finally, the magnitude of the gradient is computed to be used as input of the deformable model algorithm to guide the deformation.
3D deformable model
On the contrary of lumen segmentation for endovascular sizing purposes, we are not interested here in the segmentation of collaterals, as only the diameter and volume of thrombus are of interest. We made use of a 3D deformable model [29], which is very convenient to constrain a global shape in all directions and avoid leaks when boundaries are missing. A 3D deformable model is a mesh which is deformed according to a law of motion. The mesh deformation is controlled by internal and external forces. External forces are computed from the pre-processed image, and aim to push the mesh toward areas of high image gradient which represent the thrombus boundaries. Internal forces are computed from the mesh and control the mesh deformation smoothness, and increase the mesh geometric regularity. A third force is applied to the mesh, which is an inflating force into the vertex normal direction allowing to quickly converge to boundaries by either pushing the model outward or inward. All mesh vertexes are considered as physical masses which can move according to the following law of motion:
\(\gamma \in \left[ {0,1} \right] \) is a damping factor allowing to prevent oscillations, \(P_i^t\) is the position of vertex i at time t. \(F_{\mathrm{int}}^\rightarrow \), \(F_{\mathrm{ext}}^\rightarrow \) and \(F_{\mathrm{speed}}^\rightarrow \) represent the internal, external and inflating force at vertex i. \(\alpha \), \(\beta \) and \(\kappa \) allow to respectively control the internal, external and inflating force.
Mesh initialization
Information from the centerline and the aorta lumen segmentation allow generating a tube with variable diameters (Fig. 3). The aim of this initialization tube is to be closer to the thrombus boundary than a tube with a small constant diameter. To be sure that the mesh stays inside the aneurysm sac, a margin is taken on the diameters. For each slice, 2/3 of the minimal aneurysm diameter was taken.
Validation
The proposed method was tested on four different datasets including 145 patients to reflect the versatility of the method. Mean age was \(74 \pm 5\) years, with \(61\%\) of male. The mean aorta volume in presence of AAA was 165 mL (IQR 132–218 mL), with an average maximum sac diameter of 55.9 mm (IQR 50–59.5 mm). In presence of iliac aneurysm, the maximum common iliac artery diameter was 70.8 mm (IQR 62.2–78.5 mm).
The 1st dataset was composed of 92 pre-operative CTAs from patients with AAA, where the section of interest was defined around the thrombus boundaries of the aneurysm (usually below the renal arteries and above the iliac bifurcation). The 2nd dataset was composed of 18 pre-operative CTAs from patients with iliac aneurysm who were also considered for endovascular procedure. The studied sections were defined around the iliac aneurysm, always within the common iliac artery portion. The majority of these patients (10 out of 18) also had an AAA. The \(3^{\mathrm{rd}}\) dataset was composed of 15 AAA patients with post-operative CTA who have been treated with endovascular aneurysm repair (EVAR). Sections of interest were defined around the aneurysm, similarly to dataset \(\#1\). Post-operative CTAs include strong streaking artifacts that can disturb the segmentation process. Lastly, and in order to test the sensitivity of the algorithm to the presence of thrombus, 20 CTAs (10 pre-operative along with 10 post-operative scans) from patients without any abdominal aneurysms were included within a \(4^{\mathrm{th}}\) dataset. All images were acquired at the Rennes University Hospital within the department of Cardiothoracic and vascular surgery, using a 64-slice scanner (General Electric Medical Systems, Milwaukee, Wisconsin, LightSpeed16). Imaging was initiated after administering 120 mL of low-osmolar iodinated contrast agent (Hexabrix, Guerbet LLC, Bloomington, Ind. Iodine concentration 320 mg/ml). Soft tissue window settings with a width of 400 HU and a center of 40HU were applied. Initial spatial resolution of CTA was on average \(0.7 \times 0.7 \times 0.8~\hbox {mm}\), while no patient had a spatial slice resolution lower than \(1~\hbox {mm}^{2}\) and an inter-slice lower than \(2 ~\hbox {mm}\). The accuracy of the thrombus segmentation was assessed against ground truth from manual segmentation using the Dice coefficient, by taking into account segmentation volumes including both the thrombus and the lumen. Jaccard index and total overlap were also computed. Manual segmentations were carefully delineated by a single expert, while a 2nd expert verified ground truths to ensure consistency.
To ensure that our proposed algorithm is not sensitive to different anatomical or image quality criteria, we divided dataset #1 (aorta sections from pre-operative CTAs) in subgroups of patients with different calcifications degrees, contrast enhancement qualities or aneurysm sizes. Each of the three criteria was decomposed into three groups of various levels with the use of the reporting standards for endovascular aortic aneurysm repair [30]. In particular, calcification degrees were classified as mild (calcification \({<} 25\%\)), moderate (\(25 \hbox { to } {<} 50 \%\)) and severe (\({>} 50\%\)). Contrast enhancement was classified as low (average voxel intensity in the lumen \({<} 200 \hbox { HU}\)), intermediate (200–400 HU) and high (\({>} 400 \hbox { HU}\)). Aneurysm size was classified as small (AAA diameter \({<} 60~\hbox {mm}\)), intermediate (\(60~\hbox {mm}\) to \(75\hbox {mm}\)) and large (\({>} 75~\hbox {mm}\)). Statistical comparisons of Dice similarity metrics were performed using the unpaired two-sample t test.
We also investigated the behavior of the 3D snake with respect to the inner parameters. This experiment is designed to look at the effect of snake parameters on the segmentation results, including the number of iterations, \(\alpha \), \(\beta \) and \(\kappa \). We varied these different parameters within their potential ranges and observed the evolution of the deformable contour over the entire gathered dataset (i.e., 145 patients).
Finally, as the level-set algorithm showed satisfactory results on the literature, we confronted this algorithm to our deformable model using the same pre-processing step to validate the choice of the deformable model. The chosen level-set approach was a fast-marching method [31], which recently showed reasonably good results [8]. Results were also confronted with the Dice coefficient over the entire gathered dataset using a paired paired-sample t test.
Results
There was high correspondence between automatic and manual measurements for the thrombus segmentation for all of the four datasets (Table 1). Dataset \(\#1\) and \(\#3\) gave good results with an average Dice similarity metric of \(0.86 \pm 0.06\) and \(0.87 \pm 0.03\), respectively. Dataset \(\#2\) gave reasonably good results with an average Dice index of \(0.81 \pm 0.06\). Not surprisingly, Dataset \(\#4\) gave the best results with an average Dice index of \(0.93 \pm 0.03\). Overall, the Dice coefficient ranged from 0.67 in the worst case to 0.94 for the best segmentation (excluding dataset \(\#4\)). Visually, the thrombus can be displayed in 3D along with the lumen segmentation (Fig. 4.) for diagnosis purposes. In 2D slices, it is, however, easier to qualitatively validate the accuracies of the segmentation (Fig. 5). The mean running time for the overall process, including lumen and thrombus segmentation, was \(8.2 \pm 3.5~\hbox {s}\) on a PC with a dual-core 3.0 GHZ processor and 8 GB of memory.
From the subgroup analysis, no statistical significance was noticed between subgroups when stratifying dataset \(\#1\) with different calcification degrees, contrast enhancement qualities, or aneurysm sizes (Fig. 6). We can, however, notice a decrease of precision for patients with severe high calcification degrees on arteries. When dividing dataset \(\#1\) according to aneurysm sizes, we can also notice that errors for small aneurysm were often due to over-estimation, while errors occurring with large aneurysms would sometimes benefit from more iterations as it may be under-estimated. From the sensitivity analysis to the deformable model parameters, and when looking at each parameter independently, segmentation results using values close to the optimized set of parameters were not drastically affected. In fact, for each of the 4 studied parameter (number of iterations, \(\alpha \), \(\beta \) and \(\kappa \)), the average Dice coefficient was not significantly lower when moving in the range of \({\pm }20\%\) from the optimized value (Fig. 7). Only large differences with respect to optimized parameters (e.g., \(\beta < 0.25, \kappa > 0.75\) or number of iterations \({<} 150\)) were found to significantly decrease segmentation accuracies. Finally, the average Dice coefficient computed on the entire gathered dataset with the level-set approach was \(0.78 \pm 0.09\), which was statistically lower (\({p} < 0.001\)) than segmentations obtained with the deformable model.
Discussion
A near-automatic and generic thrombus segmentation algorithm is presented. The approach includes an optimized pre-processing step for highlighting thrombus boundaries, uses a clever mesh initialization process with prior knowledge about the aneurysm, and utilizes the power of 3D deformable model to segment the thrombus from surrounding tissues even in the presence of missing boundaries. The algorithm showed very accurate results compared to manual delineations for each of the four datasets (Table 1), and whatever the patient and image conditions (Fig. 6), showing the great versatility of the method. However, it remains some misclassification regions, where multiple tissues including the thrombus were merged without any contour information, and where even on lower or upper slices no clear information on the boundaries with respect to the thrombus were present.
The experiments on the deformable model parameters (Fig. 7.) allowed us to verify that the sensitivity of the algorithm was reasonably good, which is very interesting for reproducibility and for clinical application. In particular, the balance of parameters that has been found seems to be robust to different noises and missing contours, and has the potential to be adaptable to all anatomical morphologies of the dataset. With a gathered dataset of 145 patients, we can consider that the inter-patient variability from the population is correctly represented, and that the proposed 3D deformable model parameters are correctly optimized.
With respect to the average quality of results obtained over the entire gathered dataset, it seems compatible with values found in the literature in terms of precision. While the level-set algorithm can successfully deal with small boundary gaps, results from our comparison experiment demonstrated that it was not necessarily adequate for thrombus segmentation on the contrary to deformable model, which can prevent from large leaks. Moreover, level-set methods can be computationally expensive. Compared to statistical shape model, deformable models do not have the problem of a priori knowledge, where specific geometries may not be included into the shape model. When focusing on the global shape of the thrombus and excluding collateral arteries, 3D deformable models provide accurate results where other methods would fail due to absence of clear visible thrombus boundaries.
The main advantage of the described method is its applicability to different artery sections and to CTA scan taken at different perioperative time. The generic approach has been designed and validated on different patient conditions (with/without implanted stent, with/without thrombus), artery sections (aorta/iliac), calcification degrees, aneurysm sizes and contrast enhancement qualities. This versatility is the main originality of the proposed approach. Another advantage is that it does not require calcification segmentation. While several other methods used calcifications to constrain the deformation (e.g., [8, 9]), calcifications are masked out during our pre-processing step and do not play any role in the segmentation process, even if results from the subgroup of patients with severe calcifications showed more widespread results than with lower calcifications. It is also the first time that information from the centerline and lumen diameters are utilized to propose an initialization of the thrombus mesh to gain in precision and computation time. In terms of computation time, less than 10s for the overall process is compatible with clinical routine, whereas the majority of approaches in the literature reported computation time of more than 1min. Finally, evaluation stages of published studies are very weak, with very limited number of patients and difficulties to gather different type of CTAs. One of the other strength of this work is the evaluation stage, which includes a large number of patients suffering from aneurysms in both the aortas and iliac arteries, and which also includes post-operative CTAs. To the best of our knowledge, it is the first time that a validation process of thrombus segmentation is undertaken using a large dataset of patients.
This approach was fully integrated within the EndoSize software [27], a sizing and planning tool for endovascular surgery. It is to be noted that in the clinical workflow of the software, when the user wants to compute thrombus information, the sizing of the aneurysm is usually already performed, and the aneurysm sac is therefore already defined. No further user interaction is therefore required to compute the thrombus, making it very easy for surgeon to retrieve information about the thrombus. This segmentation will be useful for both supporting decisions including rupture risk estimation and for post-operative assessment of disease evolution. A clinical study is currently under consideration to compare pre- and post-operative thrombus volume and diameters for a large set of patients considered for EVAR using EndoSize. Further work will also include the testing of the current algorithm on thoracic aneurysms, as our current TEVAR patient database was not large enough at the time of the writing.
Conclusion
We proposed a fast and generic approach for near-automatic segmentation of thrombus from CTA scans for endovascular procedures. The proposed approach is composed of an initial centerline detection and aorta lumen segmentation, an optimized pre-processing stage and the use of a 3D deformable model. Our experimental results showed that, with minimal interactions, the approach is accurate (\(\hbox {Dice} \,{>}0.85\) and \({>} 0.80\) for abdominal aorta and iliac artery sections, respectively), fast (\(\hbox {processing time}\, {<} 10\hbox { s}\)), and most importantly robust to varying aneurysm sizes, calcification degrees, contrast enhancement qualities, aneurysm locations and presence of implanted stents. A precise patient-specific segmentation of the thrombus is useful for both the pre-operative planning to estimate the aneurysm rupture risk, and for post-operative assessment to monitor the disease evolution. The large dataset employed (145 patients) here allow us to take conclusion about the clinical utilization of the proposed approach, which has already been implemented into an endovascular planning software.
References
Sakalihasan N, Limet R, Defawe O (2005) Abdominal aortic aneurysm. Lancet 365:1577–1589
Wever JJ, Blankensteijn JD, Mali WTM, Eikelboom BC (2000) Maximal aneurysm diameter follow-up is inadequate after endovascular abdominal aortic aneurysm repair. Eur J Vasc Endovasc Surg 20:177–182
Walker GT, Kalva SP, Yeddula K, Wicky S, Kundu S, Dresher P, d’Othee J, Rose SC, Cardella JF (2010) Clinical practice guidelines for endovascular abdominal aortic aneurysm repair: written by the standards of practice committee for the society of interventional radiology and endorsed by the cardiovascular and interventional radiological society of Europe and the Canadian interventional radiology association. J Vasc Interv Radiol 21:1632–1655
Martinez-Muñoz S, Ruiz-Fernandez D, Galiana-Merino JJ (2016) Automatic abdominal aortic aneurysm segmentation in MR images. Exp Syst Appl 54:78–87
Rodriguez-Vila B, Tarjuelo-Gutierrez J, Sánchez-González P, Verbrugghe P, Fourneau I, Maleux G, Herijgers P, Gomez EJ (2015) Automated delineation of vessel wall and thrombus boundaries of abdominal aortic aneurysms using multispectral MR images. Comput Math Methods Med 2015:e202539
Shiffman S, Rubin GD, Napel S (1996) Semiautomated editing of computed tomography sections for visualization of vasculature. Presented at the April 15
Loncaric S, Subasic M, Sorantin, E (2000) 3-D deformable model for aortic aneurysm segmentation from CT images. In: Proceedings of the 22nd annual international conference of the IEEE engineering in medicine and biology society, 2000, vol 1, pp 398–401
Zohios C, Kossioris G, Papaharilaou Y (2012) Geometrical methods for level set based abdominal aortic aneurysm thrombus and outer wall 2D image segmentation. Comput Methods Programs Biomed 107:202–217
Kossioris G, Papaharilaou Y, Zohios C (2008) Detection of lumen, thrombus and outer wall boundaries of an abdominal aortic aneurysm From 2D medical images using level set methods. In: Proceedings of ASME Summer Bioengineering Conference, Marco Island, 25–29 June 2008. pp 889–890
Subasić M, Loncarić S, Sorantin E (2005) Model-based quantitative AAA image analysis using a priori knowledge. Comput Methods Programs Biomed 80:103–114
Subasic M, Loncaric S, Sorantin E (2003) Region-based deformable model for aortic wall segmentation. In: Proceedings of the 3rd international symposium on image and signal processing and analysis, 2003, vol 2. ISPA 2003. pp 731–735
Demirci S, Lejeune G, Navab N (2009) Hybrid deformable model for aneurysm segmentation. In: IEEE international symposium on biomedical imaging: from nano to macro, 2009. ISBI ’09, pp 33–36
de Bruijne M, van Ginneken B, Viergever MA, Niessen WJ (2004) Interactive segmentation of abdominal aortic aneurysms in CTA images. Med Image Anal 8:127–138
de Bruijne M, van Ginneken B, Viergever MA, Niessen WJ (2003) Adapting active shape models for 3D segmentation of tubular structures in medical images. Inf Process Med Imaging Proc Conf 18:136–147
Das B, Mallya Y, Srikanth S, Malladi R (2006) Aortic thrombus segmentation using narrow band active contour model. In: Conference proceedings of the annual international conference of the IEEE engineering in medicine and biology society IEEE engineering in medicine biology society annual conference, vol 1, pp 408–411
Dehmeshki J, Amin H, Ebadian-dehkordi M, Jouannic A, Qanadli S (2009) Automatic detection, segmentation and quantification of abdominal aortic aneurysm using computed tomography angiography. In: Proceedings of the medical image understanding and analysis, pp 32–36
Maiora J, Papakostas GA, Kaburlasos VG, Grana M (2014) A proposal of texture features for interactive CTA segmentation by active learning. Stud Health Technol Inform 207:311–320
Maiora J, Graña M (2012) Abdominal CTA image analysis through active learning and decision random forests: Application to AAA segmentation. In: The 2012 international joint conference on neural networks (IJCNN), pp 1–7
Macía I, Arenas M, Legarreta JH, Robin I, Graña M, Rajasekharan S (2012) Fast AAA thrombus segmentation from CTA images for endovascular repair follow-up. In: Medical image computing and computer-assisted intervention–MICCAI 2012 15th international conference, vol 7510. Springer, Berlin, pp 72–79
Macía I, Graña M, Maiora J, Paloc C, de Blas M (2011) Detection of type II endoleaks in abdominal aortic aneurysms after endovascular repair. Comput Biol Med 41:871–880
Lee K, Johnson RK, Yin Y, Wahle A, Olszewski ME, Scholz TD, Sonka M (2010) Three-dimensional thrombus segmentation in abdominal aortic aneurysms using graph search based on a triangular mesh. Comput Biol Med 40:271–278
Lee K, Yin Y, Wahle A, Olszewski ME, Sonka M (2008) 3-D segmentation and quantitative analysis of inner and outer walls of thrombotic abdominal aortic aneurysms. In: Hu XP, Clough AV (eds) Medical imaging 2008: physiology, function, and structure from medical images, vol 6916, SPIE Proceedings, Bellingham, WA, pp 691626.1–691626-9. doi:10.1117/12.773394
Freiman M, Esses SJ, Joskowicz L, Sosna J (2010) AN iterative model-constrained graph-cut algorithm for abdominal aortic aneurysm thrombus segmentation. In: 2010 IEEE international symposium on biomedical imaging: from nano to macro, pp 672–675
Olabarriaga SD, Rouet J-M, Fradkin M, Breeuwer M, Niessen WJ (2005) Segmentation of thrombus in abdominal aortic aneurysms from CTA with nonparametric statistical grey level appearance modeling. IEEE Trans Med Imaging 24:477–485
Zhuge F, Rubin GD, Sun S, Napel S (2006) An abdominal aortic aneurysm segmentation method: level set with region and statistical information. Med Phys 33:1440–1453
Insight Journal (ISSN 2327-770X)—Fast marching minimal path extraction in ITK, http://www.insight-journal.org/browse/publication/213
Kaladji A, Lucas A, Kervio G, Haigron P, Cardon A (2010) Sizing for endovascular aneurysm repair: clinical evaluation of a new automated three-dimensional software. Ann Vasc Surg 24:912–920
Subasic M, Loncaric S, Sorantin E (2000) 3-D image analysis of abdominal aortic aneurysm. Stud Health Technol Inform 77:1195–1200
Kass M, Witkin A, Terzopoulos D (1988) Snakes: active contour models. Int J Comput Vis 1:321–331
Chaikof EL, Blankensteijn JD, Harris PL, White GH, Zarins CK, Bernhard VM, Matsumura JS, May J, Veith FJ, Fillinger MF, Rutherford RB, Kent KC (2002) Ad hoc committee for standardized reporting practices in vascular surgery of the society for vascular surgery/American association for vascular surgery: reporting standards for endovascular aortic aneurysm repair. J Vasc Surg 35:1048–1060
Sethian J (1999) Fast marching methods. SIAM Rev 41:199–235
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Lalys, F., Yan, V., Kaladji, A. et al. Generic thrombus segmentation from pre- and post-operative CTA. Int J CARS 12, 1501–1510 (2017). https://doi.org/10.1007/s11548-017-1591-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-017-1591-8