Abstract
We present a new class of metrics for unrooted phylogenetic X-trees inspired by the Gromov–Hausdorff distance for (compact) metric spaces. These metrics can be efficiently computed by linear or quadratic programming. They are robust under NNI operations, too. The local behaviour of the metrics shows that they are different from any previously introduced metrics. The performance of the metrics is briefly analysed on random weighted and unweighted trees as well as random caterpillars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Phylogenetic metrics are often used to analyse populations of phylogenetic trees, generated by some Bayesian method or by different methods of tree reconstruction from data. Such metrics are also useful to define some empirical statistics of such populations, see, for example, Nye (2011) and Benner et al. (2014). There are already a lot of phylogenetic distances available.
The simplest one, though not the oldest one, seems to be the Robinson–Foulds distance introduced in Bourque (1978), see also Robinson and Foulds (1979) and Robinson and Foulds (1981). That one is easy and efficiently to compute in linear time (Day 1985) or even in sublinear approximation (Pattengale et al. 2007). But, it has no much power in discriminating trees, since a lot of trees with similar biological meaning have distance equal to the diameter of the unweighted tree space. Much nearer to biology seems to be a variant of the Robinson–Foulds distance, the weighted matching distance. It captures similarity of splits which entails a lot of biology and is still computable in subcubic time (Bogdanowicz and Giaro 2012; Lin et al. 2012). Another good alternative to the Robinson–Foulds metric is the quartet distance (Estabrook et al. 1985). Using the induced quartet trees instead of the induced splits, it is much more biologically plausible than the Robinson–Foulds distance and also efficiently computable (Brodal et al. 2001).
A quite natural, biology adapted way of capturing tree similarity is provided by the tree rearrangement metrics. There are different basic transformations giving rise to the NNI distance (Robinson 1971), SPR distance and TBR distance. Unfortunately, computation of those distances is NP-hard and only feasible for small trees (DasGupta et al. 1997; Allen and Steel 2001; Bonet and St. John 2010). Some fixed parameter approach to compute the (rooted) SPR distance, e.g, was done in Whidden et al. (2016). Even more at the heart of evolution is the maximum parsimony distance (Fischer and Kelk 2016; Moulton and Wu 2015). Still it is NP-hard to compute that distance, even between binary unweighted phylogenetic trees (Fischer and Kelk 2016; Kelk and Fischer 2017; Bernstein 2017).
For weighted rooted phylogenetic trees, there is the euclidean type geodesic distance on tree space introduced by Billera et al. (2001). The crucial observation was that in a natural way tree space is a space of nonpositive curvature or \(\mathrm {CAT}(0)\) space, a notion introduced by Gromov. This property implies uniqueness of geodesics. Owen and Provan (2011) provided a polynomial time algorithm for computing this metric. Although this metric was defined on rooted trees, also a version on unrooted trees is used, see, for example, Nye (2011). The \(\mathrm {CAT}(0)\) idea was used again in Gavryushkin and Drummond (2016) to develop metrics for ultrametric trees. Again, efficient computation of the geodesics is possible for at least one of the metrics. Further, different natural parametrisations may yield different geodesics.
A similar approach for weighted rooted trees, see Sokal and Rohlf (1962), uses all distances of the most recent common ancestors of pairs of taxa to the root. Recently, Kendall and Colijn (2016) returned to this idea. The authors also experimented with weighting different most recent common ancestors depending on their position in the trees.
Another way to compare phylogenetic trees is to compare the metrics they induce on the taxon set. This distance-based approach is feasible since by the work of Buneman (1971, 1974), see also Zaretskii (1965) for the unweighted case, we can identify tree-induced metrics among all metrics by the famous four-point conditions. Also, under some natural minimality assumption, the tree can be identified up to isomorphy (see Lemma 3). This approach is particularly appealing since the distance between two taxa is easy to derive, to estimate and to interpret as evolutionary distance. Another of Gromov’s ideas, Gromov’s tree, can be used to approximate arbitrary distance data by tree-induced distance data (Dress et al. 2005).
Compared to the variety of metrics reviewed above and given the popularity of distance-based methods for tree reconstruction, it is surprising that the only distance-based metrics between phylogenetic trees are pathwise difference metrics, the \(\ell ^1\) and \(\ell ^2\) versions of which are well established (Williams and Clifford 1971; Penny and Hendy 1985). The \(\ell ^\infty \) version of those metrics became of interest only recently, especially in the context of tropical geometry (Huggins et al. 2012; Bernstein and Long 2017; Coons and Rusinko 2016; Lin et al. 2017). All three metrics compare the tree-induced metrics just as real vectors. Thus, they can be computed efficiently.
In the present paper, we want to construct other distance-based phylogenetic metrics. Instead of considering the distance data as just real vectors, we are looking for metrics using the metric space properties. For (compact) metric spaces, there is the well-known Gromov–Hausdorff distance (Gromov 1981)
where the infimum is taken over all isometric embeddings of \(X,X'\) into a common metric space, and \(\rho ^H\) is the Hausdorff metric on the compacts of that space. In fact, this distance was introduced in a different way already in Edwards (1975), see Tuzhilin (2016).
Unfortunately, computing the Gromov–Hausdorff distance between finite metric spaces is NP-hard (Mémoli 2007), even considering the metric spaces induced by metric trees (Agarwal et al. 2015). There is recent work on approximation algorithms (Agarwal et al. 2015) and relaxations of the metric or related optimisation problems (Mémoli 2007; Villar et al. 2016).
Applied to tree-induced metrics, this definition induces a semimetric on the space of all weighted trees. But, we cannot distinguish trees with permuted labels. To give a short argument, consider the two potential phylogenies in Fig. 1. The left one is common believe in the evolutionary history of human, bear, dog and sheep. The right one is completely unacceptable. This means that any meaningful distance between those two trees must be positive. Unfortunately, Gromov–Hausdorff distance doesn’t have this property since permutation of the leaf labels induces an isomorphism of metric spaces. Since we want to compare the whole trees, not just tree shapes, we have to modify the metric (1). That makes the definition more complicated (see Sect. 2) since we have to match the leaf labels, but the idea of joint embeddings remains. Fortunately, our metric becomes efficiently computable this way. Since there are several reasonable candidates to substitute the Hausdorff metric in (1), we derive three different metrics. In all three cases, the value of the metric is the solution of a linear or quadratic program of polynomial size. Much of the mathematics presented in Sect. 4 aims at reducing the size of those programs to get solutions as fast as possible.
For mathematical reasons, it is very convenient to include also semimetrics on the taxon set in the definition. This situation may occur in phylogenetics if we do not resolve the topology by all singleton splits, see, for instance, Robinson and Foulds (1981).
Summarisingly, we are looking for metrics on the space of weighted phylogenetic trees which are both computable in polynomial time and able to capture some biological similarity. We show in Sect. 2 how to apply Gromov’s idea of joint embeddings to define metrics on the space of semimetrics. Section 3 defines their counterparts on the spaces of weighted X-trees, unweighted phylogenetic trees and unweighted binary phylogenetic trees. Then, Sect. 4 demonstrates how to compute these metrics efficiently. We compare the metrics with the pathwise difference metrics and the NNI distance in Sect. 5. Some special computations in Sect. 6 show how our metrics behave under the change of one or two edge lengths. A small simulation study is done in Sect. 7. Finally, Sect. 8 discusses several extensions and open questions.
2 Distances Between Semimetrics
For a finite set X denote by M(X) the set of all semimetrics on X, i.e. all \(\rho :X\times X\rightarrow \mathbb {R}_{\ge 0}\) such that for all \(x,y,z\in X\) \(\rho (x,x)=0\), \(\rho (x,y)=\rho (y,x)\) and \(\rho (x,y)\le \rho (x,z)+\rho (z,y)\). Frequently, we describe such a semimetrics in an equivalent fashion by \(\rho :\left( {\begin{array}{c}X\\ 2\end{array}}\right) \rightarrow \mathbb {R}_{\ge 0}\) where \(\left( {\begin{array}{c}X\\ 2\end{array}}\right) =\left\{ \left\{ x,y\right\} :x,y\in X,x\ne y\right\} \). Let \(\mathcal M=\left\{ (Y,\rho ):\#Y<\infty ,\rho \in M(Y)\right\} \) denote the set of all finite semimetric spaces. Isometries \(\varphi :(X,\rho )\rightarrow (Y,\rho ')\) preserve the semimetrics, i.e. for all \(x,y\in X\) \( \rho (x,y)=\rho '(\varphi (x),\varphi (y))\), but need not be injective.
Definition 1
Let X be a finite set. Then, we define three functions \(\tilde{D}_1,\tilde{D}_2,\tilde{D}_\infty \) on \(M(X)\times M(X) \) by
where the infimum is taken over all \((Y,\bar{d})\in \mathcal M\) and all isometries \(\varphi :(X,\rho )\rightarrow (Y,\bar{d})\), \(\psi :(X,\rho ')\rightarrow (Y,\bar{d})\).
Note that \(\tilde{D}_\infty \) is nearest to the Gromov–Hausdorff distance, which we should implement via
It is not complicated to deduce (see, for example, Mémoli 2007) that then the optimum is achieved by matching the points of \(\varphi (X)\) with points of \(\psi (Y)\) and find a matching with all distances as small as possible. In difference to this, all \(D_i\) match \(\varphi (x)\) with \(\psi (x)\) to compute the distance. This implies that just permuting labels to define \(\rho '\) yields nonnull distances, see the discussion around Fig. 1. Another consequence is that we compute upper bounds for the Gromov–Hausdorff distance, but this is not needed later.
We will see now that it is enough to have just one model space Y. This simplifies the optimisation problems in Definition 1 considerably. Frequently we need identical copies of our taxon set X. Using a slightly informal notation, we denote them \(X'=\left\{ x':x\in X\right\} \) and \(X''=\left\{ x'':x\in X\right\} \). To \(\rho ,\rho '\in M(X)\), we associate now the set \(E(\rho ,\rho ')\) of their extensions
So every extension reproduces the distances from \(\rho \) on X and the distances from \(\rho '\) on \(X'\). Just the distances \(\bar{d}(x,y')\) and \(\bar{d}(x',y)\), \(x,y\in X\) between the two images of X are not fully determined, but only constrained through the metric properties of \(\bar{d}\). It is important and easy to see that \(E(\rho ,\rho ')\), considered as a subset of \(\mathbb {R}^{(X\cup X')\times ( X\cup X')}\), is convex.
Let \(\left||\cdot \right||_i\) denote the usual \(\ell ^i\)-norm on \(\mathbb {R}^X\). We obtain a simple reformulation of Definition 1:
Lemma 1
For \(i=1,2,\infty \)
Thus, to compute the distances \(\tilde{D}_i(\rho ,\rho ')\), just one convex function over the convex set \(E(\rho ,\rho ')\) has to be minimised. Compactness of the sublevel sets of the norms \(\left||\cdot \right||_i\) gives directly
Lemma 2
For \(i=1,2,\infty \), there exists a \(d^*_i\in E(\rho ,\rho ')\) such that
Now we are ready to prove that we defined metrics.
Theorem 1
\(\tilde{D}_i\), \(i=1,2,\infty \), are metrics on M(X).
Proof
Symmetry is clear.
If \(\tilde{D}_i(\rho ,\rho ')=0\), choose \(d_i^*\in E(\rho ,\rho ') \) according to the previous lemma. Obviously, we obtain \(d_i^*(x,x')=0\) for all \(x\in X\). The triangle inequality implies for all \(x,y\in X\)
such that \(\rho =\rho '\).
Now fix \(\rho ,\rho ',\rho ''\in M(X)\). Using again Lemma 2, we choose \(d_1\in M(X\cup X')\) extending \(\rho ,\rho '\) and \(d_2\in M(X'\cup X'')\) extending \(\rho ',\rho ''\) such that
Following Cristina (2008) or Lemma 7 in “Appendix”, we find some \(d\in M(X\cup X'\cup X'')\) extending both \(d_1,d_2\):
We see now from monotonicity of the \(\ell ^i\)-norms on \(\mathbb {R}_{\ge 0}^X\) and their triangle inequalities that
\(\square \)
3 Distances Between X-Trees
We are mainly interested in metrics on tree space. To get a metric space from a tree (or a graph), we metrise trees by the lengths of shortest paths. To start, let us introduce some phylogenetic and graph theoretic notions. For more details, see Semple and Steel (2003).
Let \(G=(V,E,q)\) be a weighted connected graph, i.e. \(E\subseteq \left( {\begin{array}{c}V\\ 2\end{array}}\right) \) and \(q:E\rightarrow \mathbb {R}_{\ge 0}\). Then, we define the induced semimetric on V by
where
is the length of the path \(x_0x_1\dots x_m\) from \(x_0\) to \(x_m\). For unweighted graphs (V, E), we assume \(q(\left\{ x,y\right\} )=1\) for all \(\left\{ x,y\right\} \in E\).
We only consider unrooted trees, i.e. connected acyclic graphs (V, E). A weighted X-tree is a quadruple \((V,E,q,\mu )\), where (V, E) is a tree, \(\mu :X\rightarrow V\) is a (not necessarily injective) map and \(q:E\rightarrow \mathbb {R}_{>0}\) is a weight function. Additionally, it is required that all vertices \(v\in V\) of degree \(\le 2\) are included in \(\mu (X)\). Identifying isomorphic variants, let the tree space T(X) be the set of all weighted X-trees. Unweighted phylogenetic X-trees are characterised by all edges having weight 1 and by \(\mu \) being an injective map onto the set of all vertices \(v\in V\) of degree 1, which implies absence of vertices of degree 2. The corresponding subspace of T(X) will be denoted \(T_1(X)\). In a binary (bifurcating) phylogenetic X-tree, all vertices have degree 1 or 3. We denote the set of binary phylogenetic X-trees by \(T_1^2(X)\).
For an X-tree, \(\tau =(V,E,q,\mu )\) denote the induced semimetric on X by \(\rho _\tau \):
This means that \(\rho _\tau (x,y)\) is the length of the shortest path between the leaves (labelled) x and y. Then, we define for two weighted X-trees \(\tau ,\tau '\in T(X)\) and \(i=1,2,\infty \)
Again, all three \(D_i\) are metrics on tree space. This can be seen from the following characterisation of tree-induced semimetrics, provided in essence by Buneman (1971).
Lemma 3
For \(\rho \in M(X)\), there exists a weighted X-tree \(\tau \) with \(\rho =\rho _\tau \) if and only if for all \(x,y,z,w\in X\) the four-point condition
is fulfilled.
Given such a \(\rho \in M(X)\), all X-trees \(\tau \) with \(\rho =\rho _\tau \) are isomorphic.
Proof
Necessity of (4) is proved in the same way as for metrics \(\rho \), see Lemma 7.1.7 of Semple and Steel (2003).
For sufficiency and uniqueness, let \(\tilde{X}\) be the set of equivalence classes of X under identifying points \(x,y\in X\) with \(\rho (x,y)=0\). We define \(\tilde{\rho }\) on \(\tilde{X}\) through \(\tilde{\rho }([x],[y])=\rho (x,y)\) where [x], [y] are the equivalence classes of \(x,y\in X\). The triangle inequality for \(\rho \) implies that \(\tilde{\rho }\) is a well-defined metric on \(\tilde{X}\). Further, (4) is fulfilled for \(\tilde{\rho }\), too. Thus, there exists a weighted \(\tilde{X}\)-tree \(\tilde{\tau }=(V,E,\tilde{\mu },q)\) inducing \(\tilde{\rho }\) (Buneman 1971). Defining \(\mu =\tilde{\mu }\circ [\cdot ]\), the X-tree \(\tau =(V,E,\mu ,q)\) induces \(\rho \).
Let \(\tau '=(V',E',\mu ',q')\) be another weighted X-tree inducing \(\rho \). Since \(q'\) is strictly positive, any \(x,y\in X\) with \(\rho (x,y)=0\) must fulfil \(\mu '(x)=\mu '(y)\). Thus, there is a mapping \(\tilde{\mu }':\tilde{X}\rightarrow V'\) with \(\tilde{\mu }'=\mu '\circ [\cdot ]\). This gives us the weighted \(\tilde{X}\)-tree \(\tilde{\tau }'=(V',E',\tilde{\mu }',q')\) inducing the metric \(\tilde{\rho }\). By Theorem 7.1.8 of Semple and Steel (2003), \(\tilde{\tau }\) and \(\tilde{\tau }'\) are isomorphic. Thus, \(\tau \) and \(\tau '\) are isomorphic, too. \(\square \)
For tree-induced metrics \(\rho _\tau ,\rho _{\tau '}\), we can consider extensions \(\bar{d}\in E(\rho _\tau ,\rho _{\tau '})\) as being induced by a graph metric on \(X\cup X'\). Let us look at one example.
Example 1
We want to compare for \(X=\left\{ A,B,C,D\right\} \) the two unweighted X-trees

with corresponding distances \(\rho =\rho _\tau ,\rho '=\rho _{\tau '}\).
We want to derive possible extensions \(\bar{d}\) of \(\rho ,\rho '\). For this goal, denote \(\bar{d}(x,x')=\delta _x\), \(x=A,B,C,D\). Then, \(\delta _A,\delta _B,\delta _C,\delta _D\ge 0\) should be compatible with the distances on the weighted graph

see Theorem 6. One possible choice is \(\delta _A=0,\delta _B=1,\delta _C=1,\delta _D=0\), i.e.

is consistent. Obviously, we embedded now both \(\tau \) and \(\tau '\) isometrically into the metric space of the graph \(G_1\). We see \(D_\infty (\tau ,\tau ')\le 1\), \(D_2(\tau ,\tau ')\le \sqrt{2}\) and \(D_1(\tau ,\tau ')\le 2\). In fact, equality holds for \(D_1(\tau ,\tau ')\), see Example 2. Picturely, we look for graphs similar to \(G_1\) with “shortest” bridges between the left and the right sides. The meaning of “short” is given by the \(\ell ^i\)-norm.
In biological terms, the trees \(\tau \) and \(\tau '\) or their induced metrics \(\rho _\tau \) and \(\rho _{\tau '}\), respectively, entail certain (genetic) differences between the taxa \(A,B,C,\dots \) and \(A',B',C',\dots \). Those differences we try to match in a parsimonious (by minimisation), but consistent (\(\bar{d}\) is still a metric) way. For example, A and \(A'\) could stand for different individuals of taxon A and similarly for B, C, D, and we try to get a parsimonious yet consistent picture of possible mutations in the genealogy of those individuals.
It is an important general feature of the minimisation problem in (2) that we need not fix the whole extension \(\bar{d}\). It is enough to study the constraints on the variables \(\delta _x\), \(x=A,B,C,D\). This is elaborated in the next section.
4 Efficient Computation
As already mentioned after Lemma 1, we can compute \(\tilde{D}_1\) and \(\tilde{D}_\infty \) by solving a linear program and \(\tilde{D}_2\) by solving a quadratic program. Thus, we can compute the distance in a time polynomially bounded in \(n=\#X\) (Karmarkar 1984). In the naïve way, the linear (quadratic) program has the \(n^2\) variables \(\epsilon _{xy}=\bar{d}(x,y')\) and \(\mathrm {O}(n^3)\) constraints resulting essentially from the triangle inequalities in triangles of the form \(x,y,z'\) or similar. But, we can do the computation more efficiently. Observe that the objective function in (2) depends on the unknown values \((\delta _x)_{x\in X}\), \(\delta _x=\epsilon _{xx}=\bar{d}(x,x')\) only. The reformulation of the constraints forced by \(\bar{d}\) being a semimetric using that variables only is provided by the following theorem. We prove it in “Appendix”.
Theorem 2
(quadrangle inequalities) Let \(\rho ,\rho '\in M(X)\) and \((\delta _x)_{x\in X}\in \mathbb {R}_{\ge 0}^X\) be given. Then, there exists a \(\bar{d}\in E(\rho ,\rho ')\) with
if and only if for all \(x\ne y\in X\) the following inequalities are fulfilled:
Consequently, \(\tilde{D}_i(\rho ,\rho ')\) solves the program
Observe that \(x=y\) in the second line yields \(\delta _x\ge 0\). Thus, \(\tilde{D}_i(\rho ,\rho ')\) can be obtained as solution of a linear (quadratic) program in the n variables \(\delta _{x}=\bar{d}(x,x')\) with \(\mathrm {O}(n^2)\) constraints.
Example 2
Let us continue Example 1 and compute \(D_i(\tau ,\tau ')\) exactly. Since \(\rho (A,D)=\rho '(A,D)\) and \(\rho (B,C)=\rho '(B,C)\), the nontrivial constraints from the upper part of (5) read as
Consequently,
The graph \(G_1\) from Example 1 realises this lower bound.
For \(i=\infty \), (7) immediately shows
or \(D_\infty (\tau ,\tau ')\ge \frac{1}{2}\). This lower bound is realised by \(\delta _A=\delta _B=\delta _C=\delta _D=\frac{1}{2}\):

For \(i=2\), (7) gives \(\delta _A^2+\delta _C^2\ge \frac{(\delta _A+\delta _C)^2}{2}=\frac{1}{2}\). The same calculation for B, D yields
or \(D_2(\tau ,\tau ')\ge 1\). \(G_2\) realises this lower bound, too.
The upper bounds on the absolute differences were not used in the example. Interestingly, they are not important in general:
Theorem 3
For \(\rho ,\rho '\in M(X)\) \(\tilde{D}_i(\rho ,\rho ')\) is the solution of the convex program
In Isbell (1964), see also Dress (1984), the set of these constraints for \(\rho '=0\) was studied thoroughly. Our proof is obtained by adapting some arguments from Isbell (1964).
Proof
By sublevel compactness for \(\left||\cdot \right||_i\) there exists a minimal point \(\delta \in \mathbb {R}_{\ge 0}^X\) of (8). We show by contradiction that for this \(\delta \) the second part of (5) is fulfilled as well. This implies coincidence of the solutions of (8) and (6).
So let us fix \(x,y\in X\) with
We define \(\delta ^*\in \mathbb {R}_{\ge 0}^X\) by
Clearly, \(0\le \delta ^*_z\le \delta _z\) for all \(z\in X\) with strict second inequality for \(z=x\). Thus, \(\left||\delta ^*\right||_i<\left||\delta \right||_i\).
First we see
Fix now an arbitrary \(u\in X\), \(u\ne x,y\). The triangle inequality shows
Thus, \(\delta ^*\) fulfils all constraints from (8). \(\left||\delta ^*\right||_i<\left||\delta \right||_i\) contradicts that \(\delta \) is optimal for (8). \(\square \)
5 Comparison to Other Metrics
First we compare our metrics to the pathwise difference metrics, defined by
on M(X). On T(X), we set \(D^{pd}_i(\tau ,\tau ')= \tilde{D}^{pd}_i(\rho _\tau ,\rho _{\tau '})\). \(D^{pd}_1\) and \(D^{pd}_2\) were defined in Williams and Clifford (1971) and Steel and Penny (1993), respectively. \(\tilde{D}^{pd}_\infty \) is just the distortion of the identity map in the theory of metric spaces (Burago et al. 2001; Lang et al. 2013). To your knowledge, it was used in Huggins et al. (2012) under the name k-interval cospeciation the first time.
Abbreviating \(n=\#X\) we have standard estimates between our metrics for different i, resulting from similar estimates for the norms \(\left||\cdot \right||_i\), first.
Lemma 4
For \(\rho ,\rho '\in M(X)\), it holds
Theorem 4
For \(\rho ,\rho '\in M(X)\), it holds
(12) reminds of the fact that the Gromov–Hausdorff distance of two compact metric spaces is one half of the infimum over the distortions of correspondences between the two spaces (Burago et al. 2001, Theorem 7.3.25). Further, (12) shows that we need not solve a linear program for computing \(\tilde{D}_\infty \).
Proof
We choose a minimal point \(\delta \) of (8), i.e. \(\left||\delta \right||_i=\tilde{D}_i(\varrho ,\varrho ')\). Like in Example 2 we get for all \(x\ne y\in X\)
Let \(i=\infty \). Taking the maximum of (15) over all \(\left\{ x,y\right\} \in \left( {\begin{array}{c}X\\ 2\end{array}}\right) \) yields
Now define \(\bar{\delta }\in \mathbb {R}_{\ge 0}^X\) by setting
The constraints from (8) are clearly fulfilled for \(\bar{\delta }\). Evaluating \(\left||\bar{\delta }\right||_\infty \) gives
and (12) is proved.
For \(i=1\), we sum (13) over all \(\left\{ x,y\right\} \in \left( {\begin{array}{c}X\\ 2\end{array}}\right) \). This yields
the lower bound in (10). From Lemma 4, we derive the upper bound:
For \(i=2\), the lower bound in (11) is proved similarly to \(i=1\) using (14). The upper bound follows again from the previous lemma:
\(\square \)
We want to demonstrate now that the new metrics are biologically meaningful. Especially we show that an NNI (nearest neighbour interchange) move is a relatively small step in tree space \(T^2_1(X)\) when measured by these metrics. An NNI move (Allen and Steel 2001) is given by

or

where  denote different subtrees. The minimal number of NNI moves to reach \(\tau '\in T_1^2(X)\) from \(\tau \in T_1^2(X)\) is their NNI distance \(D^{NNI}(\tau ,\tau ')\) (Robinson 1971).
denote different subtrees. The minimal number of NNI moves to reach \(\tau '\in T_1^2(X)\) from \(\tau \in T_1^2(X)\) is their NNI distance \(D^{NNI}(\tau ,\tau ')\) (Robinson 1971).
Theorem 5
Consider \(\tau ,\tau '\in T^2_1(X)\) with \(D^{NNI}(\tau ,\tau ')=1\). Then,
Consequently, for all \(\tau ,\tau '\in T^2_1(X)\)
Note that these formulae give estimates of the gradient of the metrics \(D_i\) in the sense of Lin et al. (2012).
Proof
Specifically we consider

Let \(A'\) be the set of labels mapped into the subtrees  and let \(B'\) be the set of labels mapped into the subtrees
and let \(B'\) be the set of labels mapped into the subtrees  . Then,
. Then,
Theorem 4 yields directly \(D_\infty (\tau ,\tau ')=\frac{1}{2}\).
For \(\delta \in \mathbb {R}_{\ge 0}^X\) fulfilling the constraints in (8), define \(\delta ^*\in \mathbb {R}_{\ge 0}^X\) by
Neither permutations of labels in \(A'\) nor in \(B'\) change the absolute difference of the metrics. Thus, \(\delta ^*\) fulfils the constraints, too. By convexity, \(\left||\delta ^*\right||_i\le \left||\delta \right||_i\) for \(i=1,2\).
For \(i=1\) optimisation among all vectors of the form, \(\delta ^*\) means to solve the linear program
Its solution is \(1-\tilde{\delta }_{B}=\tilde{\delta }_{A}=\left\{ \begin{array}{l}1 \#A' <\#B'\\ 0 \#A' \ge \#B' \end{array} \right. \) with objective value
For \(i=2\), we have to solve
Now the minimum is realised by \(\tilde{\delta }_{A}=\frac{\#B'}{n}\) and \(\tilde{\delta }_{B}=\frac{\#A'}{n}\) with value
The definition of the NNI distance and Lemma 4 imply the second hypothesis. \(\square \)
Note that different NNI moves have different \(D_i\)-length for \(i=1,2\) in general.
Now we show that the \(D_i\)-distance of \(\tau ,\tau '\in T^2_1(X)\) which are one NNI move apart is small compared to the diameter of the space \(T^2_1(X)\). For the latter, we have upper bounds which hold on \(T_1(X)\) as well.
Lemma 5
For all \(\tau ,\tau '\in T_1(X)\), it holds
Proof
All shortest paths in \(\tau \) and \(\tau '\) have at least one and at most \(n-1\) edges. Thus, \(D^{pd}_\infty (\tau ,\tau ')\le {n-2}\). By Theorem 4, \(D_\infty (\tau ,\tau ')\le \frac{n-2}{2}\). Lemma 4 implies the other two bounds. \(\square \)
We want to show now that the estimates in Lemma 5 have the correct order in the number of taxa n. The examples are caterpillars, i.e. binary trees for which all interior vertices form a chain.
Lemma 6
Let us be given \(n=4m+1\) for some \(m\in \mathbb {N}\), \(m\ge 1\), \(X=\left\{ 1,2,\dots ,4m,4m+1\right\} \). Suppose \(\tau \) is an unrooted (binary) caterpillar tree with cherries \(\left\{ 1,2\right\} \) and \(\left\{ 4m,4m+1\right\} \):

and \(\tau '\) is obtained from \(\tau \) by reversing the order of the even labels, i.e. 2j is interchanged with \(2(2m+1-j)\) for \(j=1,\dots ,2m\):

Then,
Proof
It is easy to see that for \(1\le x<y\le n=4m+1\)
By construction,
First, the formula for \(D_\infty (\tau ,\tau ')\) follows immediately from Theorem 4.
Continuing, (8) contains the constraints \(\delta _{2j-1}+\delta _{2j}\ge \left|4(m-j)+2\right|\) and \(\delta _{4m+2-2j}+\delta _{4m+3-2j}\ge \left|4(m-j)+2\right|\), \(1\le j\le m\). Summing up these constraints gives the lower bound for \(D_1(\tau ,\tau ')\).
Applying the inequality \(a^2+b^2\ge \frac{(a+b)^2}{2}\) to the same constraints and again summing up these inequalities yield the lower bound for \( D_2(\tau ,\tau ')\). \(\square \)
6 Local Properties
From Theorem 3, we obtain for all \(\rho ,\rho ',\rho ''\in M(X)\)
In general, the sum of two tree-induced semimetrics is not a tree-induced semimetric. But, if \(\rho ',\rho ''\) result from simple manipulations of the edge lengths in the tree \(\tau \) corresponding to \(\rho =\rho _\tau \), some computations are possible. They inform us about the local behaviour of the metrics \(D_1,D_2\).
First we compute the influence of changing one edge length. Usually edges of an X-tree are described by the split they induce on X. Deleting an edge decomposes the tree into two connected components. Then, the induced split is the corresponding bipartition of X. Bipartitions are denoted A|B with \(A,B\subset X\), \(A\cup B= X\), \(A\cap B=\emptyset \).
Example 3
Consider for \(l>0\) the unresolved weighted X-trees

Thus, A|B is a split of X and l is the length of the edge inducing this split. We are interested in \(D_i(\tau _{A,B}^l,\tau _{A,B}^{l'})\). By the preceding considerations, these are the distances between two weighted X-trees displaying the same splits with the same length except the split A|B, where the lengths are l and \(l'\).
We see that the constraints from (8) turn into
Using the same arguments as in the proof of Theorem 5, we may assume that
for some \(a,b\in \mathbb {R}_{\ge 0}\) with \(a+b\ge \left|l-l'\right|\).
For \(i=1\), we find
The minimal value of the latter function of a in \(\left[ 0,\left|l-l'\right|\right] \) is
It is attained at \(\left|l-l'\right|-b=a=\left\{ {\begin{array}{l} 0 \#A\ge \#B \\ \left|l-l'\right|\#A\le \#B \end{array}}\right. \).
Similarly, we have to minimise for \(i=2\)
Now the minimum is attained at \(a=\frac{\#B}{\#A+\#B}\left|l-l'\right|\), \(b=\frac{\#B}{\#A+\#B}\left|l-l'\right|\) as
Summarisingly, we observe that different splits of a tree may contribute with different strengths to the distance. This differs from the behaviour of the geodesic distance.
Note that the above computations imply estimates for the Robinson–Foulds metric similar to Theorem 5.
Now we change two edge lengths simultaneously. Let \(\tau _0\in T(X)\) denote the tree with one vertex and without edges. Thus, the label function \(\mu \) maps to a single point and \(\rho _{\tau _0}=0\).
Example 4
Let \(l,l'>0\) and pairwise disjoint \(A,B,C\subset X\), \(A\cup B\cup C=X\), be given. We want to compute \(D_i(\tau _0,\tau _{A,B,C}^{l,l'})\) where

This tree captures the “difference” of two trees with the same shape which differ in the lengths of two edges.
Again, symmetry allows us to consider only \(\delta \in \mathbb {R}_{\ge 0}^X\) with
for some \(a,b,c\in \mathbb {R}_{\ge 0}\) which fulfil now
This yields a linear program or a quadratic program in \(\mathbb {R}_{\ge 0}^3\).
For computing \( D_1(\tau _0,\tau _{A,B,C}^{l,l'})\), we want
under the constraints (16). We know that this minimum is achieved in a corner of the feasible set. But, we see easily that not all inequalities in (16) could be equalities unless \(b=0\). Thus, at least one of a, b, c must be zero and we obtain the minimal value as
A distinction of cases whether \(\#A\gtreqless \#B+\#C\) and \(\#C\gtreqless \#A+\#B\) gives us in any case one of the values as minimum. Thus, in any case, \( D_1(\tau _0,\tau _{A,B,C}^{l,l'})\) is a linear combination of l and \(l'\), i.e. some weighted \(\ell ^1\)-distance.
The computation of \( D_2(\tau _0,\tau _{A,B,C}^{l,l'})\) would mean solving the quadratic program
under the constraints (16). For this problem, we only know that the solution is the projection of the null vector onto the affine hyperspace determined by some face of the feasible set. This projection is linear in l and \(l'\). This means that \( \left( D_2(\tau _0,\tau _{A,B,C}^{l,l'})\right) ^2\) is the minimum of five quadratic functions in \(l,l'\). Since the algebra is rather tedious, we stop here now with the indication that this minimum is just a single quadratic function similar to the linear case before. A numerical test for several cardinalities and random lengths \(l,l'\) showed that the parallelogram equality is fulfilled in all considered situations (data not shown, see https://math-inf.uni-greifswald.de/fileadmin/uni-greifswald/fakultaet/mnf/mathinf/liebscher/phylodistpaper4.R). Thus, the local geometry under \(D_2\) seems to be euclidean. Note that the previous example showed that \(D_2\) is not a version of the geodesic distance from Billera et al. (2001) for unrooted trees.
7 Implementation and Numerical Examples
We did a small simulation study on a Intel\(\circledR \) i7-7700 3,60GHz PC running Ubuntu 16.04 to get some empirical insight extending our mathematical results.
The different metrics were implemented in R (R Core Team 2017) and form now the gromovlab package (Liebscher 2015). For the geodesic distance, the implementation by the package distory (Chakerian and Holmes 2017) was used. To root trees, the first taxon was marked as outgroup. The weighted matching distance was implemented using the package lpSolve (Berkelaar et al. 2015). Random (weighted and unweighted) trees were generated by the function rtree of the R package ape (Paradis et al. 2004). This function generates uniformly distributed binary trees with uniformly in [0, 1] distributed edge lengths in the weighted case. To avoid a potential bias, labels were randomly permuted afterwards. Random caterpillars were generated by random labelling of the caterpillar tree generated by the stree function of the package ape. The corresponding R-script can be downloaded from https://math-inf.uni-greifswald.de/fileadmin/uni-greifswald/fakultaet/mnf/mathinf/liebscher/phylodistpaper4.R.
Some testing showed in the \(\ell ^1\)-case best performance in terms of computing time for the dual simplex algorithm. The computing time for obtaining the distance between random trees of size \(n=100\) was around 0.1 s. This compares to the computing times of the geodesic distance and the weighted matching distance, see Fig. 2. Of course, computations of the Robinson–Foulds distance and the pathwise difference metrics are faster.
Boxplot of computing times for different metrics (logarithmic scale) for \(10^3\) random weighted X-trees with \(n=\#X=100\). From left: \(D_1\), \(D_2\), \(D^{pd}_\infty \), \(D^{pd}_1\), \(D^{pd}_2\), the geodesic, the Robinson–Foulds and the weighted matching distance. All times were rounded to milliseconds
We also compared the values of \(D_i\), \(i=1,2,\infty \) with the pathwise difference metrics [see (9)], the geodesic distance and the Robinson–Foulds metric, for random weighted binary X-trees with \(n=10\) leaves. The resulting scatterplots are presented in Fig. 3. One can observe correlations only among the different Gromov-type metrics and among the different pathwise difference metrics. There is not much correlation to the geodesic distance. This shows that our metrics differ essentially from the pathwise difference and the geodesic metrics. Further, it is not determined by any of those metrics in the sense of Coons and Rusinko (2016).
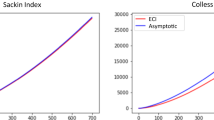
Similar pictures are found for random unweighted trees, see Fig. 4. Now there is a strong correlation to the weighted matching distance. Interestingly, \(D_1 \) turns out to be integer-valued now, see Fig. 5. That is surprising since the matrix corresponding to the linear program (8) is not totally unimodular in the sense of Hoffman and Kruskal (2010), it contains the \(3\times 3\) submatrix \( \begin{pmatrix} 1&{}1&{}0\\ 1&{}0&{}1\\ 0&{}1&{}1 \end{pmatrix}\) with determinant \(-2\). Note that for general integer-valued metrics, \(\tilde{D}_1\) assumes also half-integer values (https://math-inf.uni-greifswald.de/fileadmin/uni-greifswald/fakultaet/mnf/mathinf/liebscher/phylodistpaper4.R). The lower bound from Lemma 6 computes to \(\frac{(n-1)^2}{4}\approx 21\). Obviously, it is not sharp. Random caterpillars provide a similar distribution with only even values and more extreme to the right (data not shown) (https://math-inf.uni-greifswald.de/fileadmin/uni-greifswald/fakultaet/mnf/mathinf/liebscher/phylodistpaper4.R).
Frequency tables of the \(D_1\) metric for \(10^3\) random unweighted trees with \(n=10\). The formal lower bound on the diameter of T(X) from Lemma 6 is added as dotted line
8 Discussion
We constructed three different well motivated metrics on the space M(X) of semimetrics on the taxon set X. This leads to at least two new efficiently computable metrics for comparing unrooted, but possibly weighted, phylogenetic X-trees. There is an obvious interpretation of the metrics by parsimonious consistent matching of the differences entailed by the two trees, see Example 2. Since we showed that NNI moves are small in these metrics compared to the whole space of binary trees, these metrics surely capture some biological similarity. We think this rather abstract approach to tree metrics is valuable and could generalise well. One direction could be the extension to rooted trees. We should then just measure the distance of the induced metrics on \(X\cup \left\{ root\right\} \). Another generalisation could focus phylogenetic networks.
In general, we follow Steel and Penny (1993) in arguing that there is no universal metric for phylogenetic trees which suits perfectly for all purposes. We think that every application has its own choice, and we added a further choice to this portfolio. Yet, we should discuss further properties of phylogenetic metrics to guide the users. Monotonicity as considered in Allen and Steel (2001), Lemma 2.2 is a start in this direction. Here we want to discuss some results of the present paper and possible extensions only.
It looks interesting to extend the metric to tree shapes, with allowing the labels to be permuted. But computation of the general Gromov–Hausdorff distance is NP-hard (Pardalos and Wolkowicz 1994), and the same result holds for tree-induced metrics (Agarwal et al. 2015).
One important topic which raised up already in Bogdanowicz and Giaro (2012), Lin et al. (2012), Gavryushkin and Drummond (2016), and Kendall and Colijn (2016) is the question how to weight the edges of the trees. We computed the influence of different splits on our metric in Example 3. If those weights do not fit the intention of the user, one could change the tree-induced metrics by rescaling the edges of the trees in an objective way. Using weighted \(\left||\cdot \right||_i\) norms can account for uneven taxon sampling or rooting the tree. The principle of the computations would remain the same. Note that we met already such weights in Examples 3 and 4. Further, also a Kantorovich–Wasserstein approach similar to Mémoli (2007) might be feasible if the weights of the taxa differ between the trees. Thus, our approach is natural, but can be well adjusted to the needs of applications.
We compared the new metrics with the NNI metric, the pathwise difference metrics, the Robinson–Foulds metric (see Example 3). Not many of the estimates are tight. So it would be valuable to get tight lower and upper bounds in these cases and for the quartet, SPR-, TBR-, maximum parsimony, weighted matching and geodesic metrics as well. It is important to know more about the 1-neighbourhoods on \(T_1^2(X)\), e.g. whether there are islands in the sense of Bogdanowicz and Giaro (2012). Our numerical study in Sect. 7 is still sparse.
We expect the diameter between two unweighted X-trees to be realised by caterpillar trees. The simulation result in Fig. 5 points into this direction. We would like to know why \(D_1\) takes integer values only on \(T_1^2(X)\).
The geometry induced by \(D_2\) needs to be explored. Is it locally fully euclidean? How do the geodesics look like?
Outside phylogenetics, there should be applications to other kinds of finite labelled metric spaces. At the moment, we are only aware of the papers of F.Memoli, e.g. Mémoli (2007), which deal with \(\ell ^p\)-type Gromov–Hausdorff metrics.
In spite of these many open questions, we are sure that this work is just the start of studying this interesting kind of metrics.
References
Agarwal PK, Fox K, Nath A, Sidiropoulos A, Wang Y (2015) Computing the Gromov–Hausdorff distance for metric trees. In: Elbassioni K, Makino K (eds) Algorithms and computation. Lecture Notes in Computer Science, vol 9472, pp 529–540. Springer, Berlin. arXiv:1509.05751
Allen BL, Steel M (2001) Subtree transfer operations and their induced metrics on evolutionary trees. Ann Comb 5:1–15
Benner P, Bačak M, Bourguignon P-Y (2014) Point estimates in phylogenetic reconstructions. Bioinformatics 30:i534–i540
Berkelaar M et al (2015) lpSolve: Interface to “Lp_solve” v. 5.5 to solve linear/integer programs. R package version 5.6.13. https://CRAN.R-project.org/package=lpSolve
Bernstein DI (2017) L-infinity optimization to Bergman fans of matroids with an application to phylogenetics. arXiv:1702.05141
Bernstein DI, Long C (2017) L-infinity optimization to linear spaces and phylogenetic trees. arXiv:1702.05127
Billera LJ, Holmes SP, Vogtmann K (2001) Geometry of the space of phylogenetic trees. Adv Appl Math 27(4):733–767
Bogdanowicz D, Giaro K (2012) Matching split distance for unrooted binary phylogenetic trees. IEEE/ACM Trans Comput Biol Bioinform 9(1):150–160
Bonet ML, St. John K (2010) On the complexity of uSPR distance. IEEE/ACM Trans Comput Biol Bioinform 7(3):572–576
Bourque M (1978) Arbres de Steiner et reseaux dont certains sommets sont a localisation variable. PhD thesis, Montreal
Brodal GS, Fagerberg R, Pedersen CNS (2001) Computing the quartet distance between evolutionary trees on time \({\rm O}(n\log ^2n)\). In: Proceedings of the 12th international symposium on algorithms and computation (ISAAC). Lecture Notes in Computer Science, vol 2223, pp 731–737. Springer
Buneman P (1971) The recovery of trees from measures of dissimilarity. In: Kendall DG, Tautu P (eds) Mathematics in the archeological and historical sciences. Edinburgh University Press, Edinburgh, pp 387–395
Buneman P (1974) A note on the metric properties of trees. J Comb Theory 17(1):48–50
Burago D, Burago Y, Ivanov S (2001) A course in metric geometry. Graduate studies in mathematics, vol 33. American Mathematical Society, Providence
Chakerian J, Holmes S (2017) Distory: distance between phylogenetic histories. R package version 1.4.3. http://CRAN.R-project.org/package=distory
Coons JI, Rusinko J (2016) A note on the path interval distance. J Theor Biol 398:145–149
Cristina J (2008) Gromov–Hausdorff convergence of metric spaces, Helsinki. http://www.helsinki.fi/~cristina/pdfs/gromovHausdorff.pdf. Accessed 2 Feb 2015
DasGupta B, He X, Jiang T, Li M, Tromp J, Zhang L (1997) On distances between phylogenetic trees. In: Proceedings of the eighth ACM/SIAM symposium discrete algorithms (SODA ’97), pp 427–436
Day WHE (1985) Optimal algorithms for comparing trees with labeled leaves. J Classif 2(1):7–28
Dress A (1984) Trees, tight extensions of metric spaces, and the cohomological dimension of certain groups: a note on combinatorial properties of metric spaces. Adv Math 53(3):321–402
Dress A, Holland B, Huber KT, Koolen J, Moulton V, Weyer-Menkoff J (2005) \(\Delta \)-additive and \(\Delta \)-ultra-additive maps, Gromov’s trees and the Farris transform. Discrete Appl Math 146:51–73
Edwards DA (1975) The structure of superspace. In: Stavrakas NM, Allen KR (eds) Studies in topology. Academic Press, New York, pp 121–133
Estabrook GF, McMorris FR, Meacham CA (1985) Comparison of undirected phylogenetic trees based on subtrees of four evolutionary units. Syst Zool 34(2):193–200
Fischer M, Kelk S (2016) On the maximum parsimony distance between phylogenetic trees. Ann Comb 20(1):87–113
Gavryushkin A, Drummond A (2016) The space of ultrametric phylogenetic trees. J Theor Biol 403:197–208
Gromov M (1981) Groups of polynomial growth and expanding maps. Publ Math IHÉS 53:53–73
Guénoche A, Leclerc B, Makarenkov V (2004) On the extension of a partial metric to a tree metric. Discrete Math 276:229–248
Hoffman AJ, Kruskal J (2010) Introduction to integral boundary points of convex polyhedra. In: Jünger M et al (eds) 50 years of integer programming, 1958–2008. Springer, Berlin, pp 49–50
Huggins P, Owen M, Yoshida R (2012) First steps toward the geometry of cophylogeny. In: Hibi T (ed) Harmony of Gröbner bases and the modern industrial society. World Scientific, Singapore, pp 99–116
Isbell JR (1964) Six theorems about injective metric spaces. Commun Math Helv 39(1):65–76
Karmarkar N (1984) A new polynomial-time algorithm for linear programming. Combinatorica 4(4):373–395
Kelk S, Fischer M (2017) On the complexity of computing MP distance between binary phylogenetic trees. Ann Comb 21(4):573–604
Kendall M, Colijn C (2016) Mapping phylogenetic trees to reveal distinct patterns of evolution. Mol Biol Evol 33(10):2735–2743
Lang U, Pavón M, Züst R (2013) Metric stability of trees and tight spans. Arch Math 101(1):91–100
Liebscher V (2015) gromovlab: Gromov–Hausdorff type distances for labeled metric spaces. R package version 0.7-6. http://CRAN.R-project.org/package=gromovlab
Lin Y, Rajan V, Moret BME (2012) A metric for phylogenetic trees based on matching. IEEE/ACM Trans Comput Biol Bioinform 9(4):1014–1022
Lin B, Sturmfels B, Tang X, Yoshida R (2017) Convexity in tree spaces. SIAM J Discrete Math 31(3):2015–2038
Mémoli F (2007) On the use of Gromov–Hausdorff distances for shape comparison. In: Symposium on point based graphics, Prague, Sept 2007
Moulton V, Wu T (2015) A parsimony-based metric for phylogenetic trees. Adv Appl Math 66:22–45
Nye TMW (2011) Principal components analysis in the space of phylogenetic trees. Ann Stat 39(5):2716–2739
Owen M, Provan J (2011) A fast algorithm for computing geodesic distances in tree space. IEEE/ACM Trans Comput Biol Bioinform 8(1):2–13
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20(2):289–290
Pardalos PM, Wolkowicz H (eds) (1994) Quadratic assignment and related problems. DIMACS series in discrete mathematics and theoretical computer science, vol 16. AMS, Providence, RI. Papers from the workshop held at Rutgers University, New Brunswick, New Jersey, May 20–21, 1993
Pattengale ND, Gottlieb EJ, Moret BM (2007) Efficiently computing the Robinson–Foulds metric. J Comput Biol 14(6):724–735
Penny D, Hendy MD (1985) The use of tree comparison metrics. Syst Biol 34(1):75–82
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, version 3.4.3, Vienna, Austria. http://www.R-project.org/
Robinson DF (1971) Comparison of labeled trees with valency three. J Comb Theory 11:105–119
Robinson DF, Foulds LR (1979) Comparison of weighted labelled trees. In: Combinatorial mathematics VI. Lecture Notes in Mathematics, vol 748, pp 119–126. Springer, Berlin
Robinson DF, Foulds LR (1981) Comparison of phylogenetic trees. Math Biosci 53:131–147
Semple C, Steel MA (2003) Phylogenetics. Oxford University Press, Oxford
Sokal RR, Rohlf FJ (1962) The comparison of dendrograms by objective methods. Taxon 11:33–40
Steel MA, Penny D (1993) Distributions of tree comparison metrics—some new results. Syst Biol 42(2):126–141
Tuzhilin AA (2016) Who invented the Gromov–Hausdorff distance? arXiv:1612.00728
Villar S, Bandeira AS, Blumberg AJ, Ward R (2016) A polynomial-time relaxation of the Gromov–Hausdorff distance. arXiv:1610.05214
Whidden C, Beiko RG, Zeh N (2016) Fixed-parameter and approximation algorithms for maximum agreement forests of multifurcating trees. Algorithmica 74(3):1019–1054
Williams WT, Clifford HT (1971) On the comparison of two classifications of the same set of elements. Taxon 20:519–522
Zaretskii KA (1965) Constructing a tree on the basis of a set of distances between the hanging vertices (in Russian). Uspekhi Mat Nauk 20(6):90–92
Acknowledgements
First of all, I have to thank Mareike Fischer for introducing me to the world of phylogenetic distances. She helped also a lot for getting a clear notation. Second, I’m very grateful to Jürgen Eichhorn who unconsciously draw my attention to metrics between metric spaces. Third, I’d like to thank Michelle Kendall for her inspiring talk at the Portobello conference 2015 and additional discussion later. Fourth, I thank Mike Steel for many interesting discussions, useful hints, his kind hospitality during my stay in Christchurch 2010, and for the organisation of the amazing 2015 workshop in Kaikoura with an inspiring and open atmosphere. Further, Miroslav Bačak, Andrew Francis, Alexander Gavryushkin, Stefan Grünewald, Marc Hellmuth and Giulio dalla Riva gave useful hints and inspiration in many discussions. The questions and hints of five anonymous referees regarding previous versions of this manuscript helped to improve it substantially.
Author information
Authors and Affiliations
Corresponding author
A On Semimetric Extensions
A On Semimetric Extensions
Several times we met the problem whether a partial dissimilarity on X, i.e. a map \(q:E\rightarrow \mathbb {R}_{\ge 0}\), \(E\subseteq \left( {\begin{array}{c}X\\ 2\end{array}}\right) \), has an extension to a semimetric on X. This seems to be a well-known problem, one folklore solution I found in Guénoche et al. (2004). For our needs, the following reformulation proved more useful.
We call a cycle \(p=x_0x_1\dots x_m\), \(x_0=x_m\), in a graph (X, E) induced, if it is simple (\(x_i\), \(i=0,\dots ,m-1\), are different) and chordless (\(\left\{ x_i,x_j\right\} \notin E\), \(0\le i,j\le m-1\), \(2\le \left|i-j\right|\le m-2 \)).
Theorem 6
If the graph \(G=(X,E)\) is connected, then \(q:E\rightarrow \mathbb {R}_{\ge 0}\) extends to a semimetric on X if and only if for all induced cycles p of G and all edges e in p
Proof
By Guénoche et al. (2004), Proposition 2.1, q has a semimetric extension if and only if for all \(\left\{ x,y\right\} \in E\) \(q(\left\{ x,y\right\} )=d^q_G(x,y)\). \(d^q_G\) was introduced in (3).
Let there be an extension of q to a semimetric. Fix an induced cycle \(p=x_0x_1\dots x_{m-1} x_m\), \(x_m=x_0\), and the edge \(e=\left\{ x_0,x_1\right\} \) in p. We obtain
Now assume (17) is fulfilled, but there is no extension to a semimetric. Thus, we find \(\left\{ x,y\right\} \in E\) such that \(q(\left\{ x,y\right\} )>d^q_G(x,y)\). This means there is a path \(\tilde{p}=x_0x_1\dots x_{m-1}\), \(x_0=x\), \(x_{m-1}=y\), such that
We may assume w.l.o.g. that m is minimal. Thus, \(x_i\), \(i=0,\dots ,m-1\) are different. Setting \(x_m=x_0\), \(e=\left\{ x,y\right\} =\left\{ x_0,x_{m-1}\right\} \), the (simple) cycle \(p=x_0x_1\dots x_m\) violates (17). Suppose now that p has a chord, say \(\left\{ x_i,x_j\right\} \). Since m is minimal, we know
and
Substituting the first inequality into the right hand side of the second one yields
This contradiction shows that p is an induced cycle and completes the proof. \(\square \)
We can use this result for the
Proof of Theorem 2
We apply Theorem 6 to \(X\cup X'\), \(E=\left( {\begin{array}{c}X\\ 2\end{array}}\right) \cup \left( {\begin{array}{c}X'\\ 2\end{array}}\right) \cup \left\{ \left\{ x,x'\right\} :x\in X\right\} \) and \(q:E\rightarrow \mathbb {R}_{\ge 0}\) given by
Induced cycles in \((X\cup X',E)\) are either triangles in X, triangles in \(X'\) or quadrangles \(x,y,y',x',x\). For the two former, (17) is equivalent to the triangle inequalities for \(\rho ,\rho '\). For the latter, (17) is the same as (5). \(\square \)
The following result was used in the proof of Theorem 1.
Lemma 7
Suppose X, Y, Z are disjoint sets and there are given \(d_1\in M(X\cup Y)\) and \(d_2\in M(Y\cup Z)\) such that \(d_1|_{\left( {\begin{array}{c}Y\\ 2\end{array}}\right) }=d_2|_{\left( {\begin{array}{c}Y\\ 2\end{array}}\right) }\). Then, there exists a \(d\in M(X\cup Y\cup Z)\) such that \(d|_{\left( {\begin{array}{c}X\cup Y\\ 2\end{array}}\right) }=d_1\) and \(d|_{\left( {\begin{array}{c}Y\cup Z\\ 2\end{array}}\right) }=d_2\).
Proof
Now we apply the theorem to the graph \(\left( X\cup Y\cup Z,\left( {\begin{array}{c}X\cup Y\\ 2\end{array}}\right) \cup \left( {\begin{array}{c}Y\cup Z\\ 2\end{array}}\right) \right) \) with
Since both \(X\cup Y\) and \(Y\cup Z\) are complete in this graph, the only induced cycles are triangles. The triangle inequalities for \(d_1,d_2\) show (17). \(\square \)
Rights and permissions
About this article
Cite this article
Liebscher, V. New Gromov-Inspired Metrics on Phylogenetic Tree Space. Bull Math Biol 80, 493–518 (2018). https://doi.org/10.1007/s11538-017-0385-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-017-0385-z