Abstract
Chronic obstructive pulmonary disease (COPD) is a common lung disease that can lead to restricted airflow and respiratory problems, causing a significant health, economic, and social burden. Detecting the COPD stage can provide a timely warning for prompt intervention in COPD patients. However, existing methods based on inspiratory (IN) and expiratory (EX) chest CT images are not sufficiently accurate and efficient in COPD stage detection. The lung region images are autonomously segmented from IN and EX chest CT images to extract the \(1,781\times 2\) lung radiomics and \(13,824\times 2\) 3D CNN features. Furthermore, a strategy for concatenating and selecting features was employed in COPD stage detection based on radiomics and 3D CNN features. Finally, we combine all the radiomics, 3D CNN features, and factor risks (age, gender, and smoking history) to detect the COPD stage based on the Auto-Metric Graph Neural Network (AMGNN). The AMGNN with radiomics and 3D CNN features achieves the best performance at 89.7\(\%\) of accuracy, 90.9\(\%\) of precision, 89.5\(\%\) of F1-score, and 95.8\(\%\) of AUC compared to six classic machine learning (ML) classifiers. Our proposed approach demonstrates high accuracy in detecting the stage of COPD using both IN and EX chest CT images. This method can potentially establish an efficient diagnostic tool for patients with COPD. Additionally, we have identified radiomics and 3D CNN as more appropriate biomarkers than Parametric Response Mapping (PRM). Moreover, our findings indicate that expiration yields better results than inspiration in detecting the stage of COPD.
Graphical Abstract
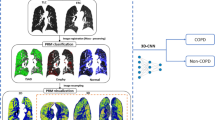
The workflow of this study. (a) The IN and EX lung region is segmented using a well trained U Net (R231). (b) Radiomics features are obtained through the PyRadiomics tool, and 3D CNN features are extracted using the frozen encoder in the pre trained Med3D model. (c) The IN+EX radiomics and IN+EX 3D CNN features are concatenated, and the Lasso algorithm is subsequently applied to screen for useful data. (d) The selected features and ri sk factors are sent to the AMGNN, resulting in COPD multi classification outcomes.

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Chronic obstructive pulmonary disease (COPD) is a progressive lung disease characterized by long-term respiratory symptoms and airflow limitation [1]. In 2030, it will become the third largest death factor worldwide, resulting in a heavy social and economic burden globally [2]. Therefore, improving the precision of COPD stage detection is imperative and pivotal for COPD management.
The classification of COPD stages utilizes the Global Initiative for Chronic Obstructive Lung Disease (GOLD) criteria, which range from stages 0 to IV [1]. For many years, the forced expiratory volume in 1 s/forced vital capacity (\(FEV_{1}\)/FVC) and \(FEV_{1}\%predicted\) have been the primary criterion for categorizing COPD in pulmonary function tests (PFTs) [3, 4]. Individuals with \(FEV_{1}\)/FVC ratios greater than 0.7 are classified as normal, whereas those with \(FEV_{1}\)/FVC ratios less than 0.7 are categorized as COPD patients. GOLD classification is based on \(FEV_{1}\)%predicted values, with GOLD 1 representing \(\ge \) 80%, GOLD 2 for 50% \(\le \) \(FEV_{1}\)%predicted < 80%, GOLD 3 for 30% \(\le \) \(FEV_{1}\)%predicted < 50%, and GOLD 4 for \(FEV_{1}\)%predicted < 30%. However, the accuracy of PFTs is constrained by the level of patient cooperation. Specifically, the measurement process of PFTs is complex, and it may be difficult for some patients to understand and comply with the requirements set by doctors [5]. In addition, PFTs cannot intuitively provide detailed anatomical information and morphological changes, such as subtypes of emphysema and bronchial wall thickening [6, 7].
Computed tomography (CT) has been deemed the most effective modality for characterizing and quantifying COPD [8]. The CT measurements of low-intensity areas exhibit a robust correlation with spirometry results [9, 10]. Compared to PFTs, the CT measurements of emphysema and small airway disease offer additional clinical insights [7, 11, 12]. In addition, the examination of COPD phenotypes that have been classified through the utilization of high-resolution computed tomography (HRCT) in a clinical context [13]. However, whether CT or HRCT is used, observing the pathological structures of small airways (<2 mm in diameter) remains challenging. In contrast to CT or HRCT images that only capture the IN phase, using both IN and EX CT images provides a comprehensive representation of pulmonary morphological changes across various respiratory phases. Validation studies have confirmed that both IN and EX chest CT images are reliable for identifying airway disease [14] and establishing their correlation with pulmonary ventilatory function [15], which is closely associated with the risk of COPD.
Despite the limitations of IN and EX chest CT images in visualizing the pathological structures of small airways, alternative methodologies have been developed that leverage these images as a foundation. In 2012, Galbán et al. proposed the PRM [16], a biomarker based on IN and EX chest CT images, demonstrating its potential to discriminate different COPD phenotypes. Specifically, the PRM can effectively distinguish functional small airways disease (\(PRM^{{f}SAD}\)), emphysema (\(PRM^{Emph}\)), and normal (\(PRM^{Normal}\)) regions. In 2020, Philip et al. confirmed that the PRM could detect the short-term progression of emphysema in severe cases of COPD [17]. In 2021, Ho et al. demonstrated that the PRM features might be utilized for COPD identification [18]. However, implementing the PRM approach is a challenging task that necessitates lung bronchus segmentation and registration of IN and EX chest CT images, as this significantly impacts the accuracy of COPD classification.
Using IN and EX chest CT images, Ho et al. have implemented ML classifiers to classify COPD and non-COPD cases. Based on this foundation, we have classified COPD from stages 0 to IV, utilizing radiomics and 3D CNN features, which are more suitable biomarkers than PRM in COPD stage detection. In 2012, radiomics emerged as an innovative approach for extracting more information from medical images by employing advanced feature analysis [19]. Radiomics features have also been employed in COPD for survival prediction [20, 21], COPD exacerbations [22], COPD early decision[23], and COPD stage detection [24, 25]. Moreover, 3D CNN features extracted by Med3D model [26] have been used in our previous study, which proved effective for COPD stage detection [24].
In the last decade, convolutional neural networks (CNNs) have played a significant role in the medical images field [27]. However, medical images are often of poor quality, difficult to acquire, and computationally expensive [28]. With the interpretability of CNNs being questioned, graph neural networks (GNNs) may provide a feasible direction for developing artificial intelligence with causal reasoning ability in the future [29, 30]. GNNs have evolved into the Graph Convolutional Network (GCN), inspired by CNNs. However, the fixed-graph structure of GCN, which utilizes the entire dataset, limits on its applicability and further development. To address these limitations while retaining the advantages of GNNs, in 2021, Song et al. proposed the Auto-Metric Graph Neural Network (AMGNN) based on a meta-learning strategy [31]. AMGNN has been successfully applied to Alzheimer’s disease classification, demonstrating strong performance. This further illustrates the superiority of AMGNN over GCN [31, 32]. Our experiments showed significant improvements in the AMGNN incorporating clinical data as risk factors (age, gender, and smoking history) compared to traditional ML classifiers. In summary, this study’s primary contributions are as follows:
-
We propose an effective CT imaging biomarkers selection method of AMGNN for COPD stage detection, achieving better performance at 89.7% accuracy, 90.9% precision, 89.5% F1-score, and 95.8% AUC. This method fully considers the imaging features of inspiratory and expiratory chest CT images. It uses the Lasso algorithm (a classic kind of feature selection technology) to determine the final node features of AMGNN for COPD detection.
-
The effectiveness of our proposed AMGNN model for COPD stage detection based on multidimensional imaging biomarkers has been verified, which is superior to single-dimensional radiomics and CNN features of inspiratory/expiratory chest CT images and even better than PRM imaging biomarkers. Based on chest CT images, this may provide a new view for COPD stage detection.
-
Compared with previous work on COPD identification (COPD and without COPD) based on chest CT images, our work (COPD stage detection) may become a more effective tool for COPD management of this fragile population.
2 Materials and methods
The materials used in the study are described in detail in Section 2.1, while the methods employed are explained thoroughly in Section 2.2.
2.1 Materials
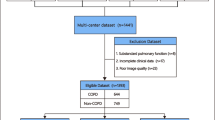
This study has received approval from the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (grant number: 201722). It has been registered on the website,Footnote 1 with the NCT number: NCT03240315. Before their participation, all subjects in the study provided written informed consent, which was signed and dated. The study involved the collection of IN and EX chest CT images and corresponding labels for 116 (29\(\times \)4) patients from the First Affiliated Hospital of Guangzhou Medical University from August 7, 2017, to February 15, 2022. Each four COPD stages comprised 29 patients, thus generating a balanced dataset. The abbreviation substitution of features is shown in Table 1.
The workflow of this study. a The IN and EX lung region is segmented using a well-trained U-Net (R231) [33]. b Radiomics features are obtained through the PyRadiomics tool [34], and 3D CNN features are extracted using the frozen encoder in the pre-trained Med3D model [26]. c The IN+EX radiomics and IN+EX 3D CNN features are concatenated, and the Lasso algorithm is subsequently applied to screen for useful data. d The selected features and risk factors are sent to the AMGNN, resulting in COPD multi-classification outcomes
2.2 Methods
The flowchart of the proposed method in this study is illustrated in Fig. 1. First, the IN and EX chest CT images were preprocessed to acquire lung region images by employing the U-Net (R231) [33]. Second, the IN and EX radiomics features are extracted using the PyRadiomics tool [34]. In addition, the IN and EX 3D CNN features are extracted from the lung region images employing the pre-trained Med3D model [26]. Third, the (IN+EX)_selected radiomics and (IN+EX)_selected 3D CNN features are concatenated. These processed features, along with the risk factors (age, gender, and smoking history), serve as input to the AMGNN for COPD stage detection.
2.2.1 Lung region segmentation
The primary step involves segmenting the lung region, as Fig. 1a illustrates. This IN and EX, lung region segmentation, is conducted via the employment of the U-Net (R231) [33], which was trained on a large and diverse dataset that covers a wide range of visual variability. The model can precisely extract the IN and EX lung regions through this process.
2.2.2 Feature extraction
Radiomics features were extracted from lung region images with the Hounsfield unit (HU) by PyRadiomics tool [34], as illustrated in Fig. 1b. Specifically, two steps are involved in extracting lung radiomics features from lung region images. To begin, the initial lung region images undergo a filtering process using wavelet and Laplacian of Gaussian (LoG) filters. This filtering generates derived lung region images. In the subsequent step, the original and derived lung region images are employed to compute the lung radiomics features according to predefined classes. These steps are automated using the PyRadiomics tool, an open-source Python package. For more comprehensive information regarding the process of the PyRadiomics tool, please refer to our previous studies [24, 35].
In addition, 3D CNN features were extracted using the pre-trained Med3D model [26], a 3D network designed to extract general medical 3D features. It accomplishes this task by constructing a comprehensive dataset called 3DSeg-8, which encompasses diverse modalities, target organs, and pathologies. The approach involves utilizing truncated transfer learning with the encoder backbone (ResNet-10) of the pre-trained Med3D model. The extraction of 3D CNN features is thoroughly elucidated in our prior publication [36].
2.2.3 Feature processing
In Fig. 1c, the feature processing involves concatenation and selection. Specifically, the Lasso method is employed to select IN and EX radiomics features from a matrix size of 1781\(\times \)2, and IN and EX 3D CNN features from a matrix size of 13824\(\times \)2. Our previous research [24] has demonstrated that applying Lasso improves the accuracy of COPD classification. To achieve COPD stage detection, the risk factors (k=3) and node features (d=44) are concatenated, resulting in the proposed lung combination vector of size (3+44). The mathematical form of the Lasso is given by Eq. 1.
where matrix \(\hat{\beta }^{lasso}\) denotes the (IN+EX)_selected radiomics/3D CNN feature. \(x_{ij}\)(the independent variable) denotes the (IN+EX) radiomics/3D CNN features. \(y_i\)(the dependent variable) denotes 116 patients with COPD stage: GOLD 0, I, II, III-IV. \(\lambda \) denotes the penalty parameter (\(\lambda \) \(\ge \)0). \(\beta _j\) denotes the regression coefficient. n denotes the number of subjects (116). p denotes the size of the (IN+EX) radiomics (\(1781\times 2\))/3D CNN features (\(13824\times 2\)), i \(\in \) \(\left[ 1, n \right] \), and j \(\in \) \(\left[ 0, p \right] \). We employ LassoCV with 10-fold cross-validation (CV) iterations 100,000 times, employing coordinate descent to fit the model and select the optimal penalty coefficient \(\lambda \). Initially, we split the dataset into training and testing sets. Then, we applied the feature selection function LassoCV exclusively to the training set, selecting the related features. Subsequently, we extracted corresponding features from the testing set. Consequently, the specific count of selected features is detailed in Fig. 3.
2.2.4 COPD stage detection
In Fig. 1d, we utilize the AMGNN based on meta-learning for COPD stage detection. The AMGNN layer and the pipeline of meta-learning are illustrated in Fig. 2.
As shown in Fig. 2a, the AMGNN mainly consists of two steps: calculating the auto-metric adjacency matrix with probability constraint and updating the node. (i) The AMGNN inputs a matrix V, comprising risk factors (age, gender, and smoking history) and (IN+EX)_selected (radiomics+3D CNN) features. Initially, the absolute difference between each patient at the same risk factor position is computed to connect distinct nodes, forming three adjacency matrices. These matrices are then weighted and summed to yield the edge constraint matrix E. The computation method for matrix E is given by Eq. 2.
where \(e_{i,j} \in (0, 1)\) represents the edge weight between nodes \(v_i\) and \(v_j\). \(e_{i,j}\) is the element of the i-th row and j-th column of the edge constraint matrix \(E \in R^{N\times N}\). \(e_{i,j}^{k}\) equals \(\eta _i^k - \eta _j^k\). K denotes the total number of risk factors, and k signifies the k-th risk factor feature.
Next, the \(\text {N}\times 44\) matrix is replicated N times to create an \(\text {N}\times \text {N}\times 44\) matrix, and the absolute difference is computed between this matrix and its transpose. Subsequently, this matrix is sent to the CNNs to learn the similarity metric between different nodes. This process results in deriving the edge weight matrix W. Finally, the final auto-metric adjacency matrix A with probability constraint is computed by multiplying the matrices E and W. (ii) After obtaining the adjacency operator family, the model uses each operator in the family to update the nodes. The updated results are then accumulated. Matrix A is multiplied with \(V^{(l)}\), and the resulting product is passed through a fully connected layer to obtain W. The formula for updating nodes is given by Eq. 3.
where \(V^{(l)}\) represents the nodes in the layer l. The trainable parameters \(\theta _B^{(l)}\epsilon R^{f\times f'}\) are represented as a fully connected layer. Leaky−ReLU is a nonlinear activation function.
This layer is repeated twice in the network. The output of the second fully connected layer is then input into the softmax function to get the final result of the AMGNN. The number of AMGNN layers is 2. In the first layer, the input channels are 2\(\times \)(numbers of features + numbers of risk factors + numbers of classification), and the output channels are 48. In the second layer, the input channels are 2\(\times \)(numbers of features + numbers of risk factors + numbers of classification + 48), and the output channels are 4. We optimized the network parameters using the Adam optimizer, and the learning rate was set to 0.001.
Figure 2b shows the training strategy based on meta-learning, which outlines the model training process. Samples are randomly extracted from the training dataset, and various small graphs are formed using these samples. In the training dataset, each graph consists of 20 nodes, with 5 nodes representing each COPD stage and one node being unknown. These smaller graphs are fed into the model for analysis. Using the probability-constrained AMGNN layer, information is effectively exchanged and updated among the nodes. As a result, the class of the unknown node is determined with impressive accuracy. For a more comprehensive understanding of the AMGNN layer based on meta-learning, please refer to the detailed description provided in the referenced study [31].
3 Experiments and results
Figure 3 presents the experimental design adopted in our study, which involved conducting four experiments to assess the effectiveness of different input features for COPD stage detection. These features include radiomics features, 3D CNN features, radiomics+3D CNN features, and PRM features. Each experiment evaluated two types of models: the AMGNN and several ML classifiers, namely Random Forest (RF) [37], Support Vector Machine (SVM) [38], Multi-Layer Perceptron (MLP) [39], Logistic Regression (LR) [40], Gradient Boosting (GB) [41], and Linear Discriminant Analysis (LDA) [42]. The performance of the models was assessed using various metrics, including accuracy, precision, F1-score, and Area Under the Curve (AUC).
3.1 Detection based on PRM features
In Experiment 1, the input consists of PRM features, which are essential biomarkers extracted from IN and EX chest CT images. The PRM features were automatically derived using Imbio’s Lung Density Analysis software.Footnote 2 These features reflect changes in lung function and involve segmenting the lung region into functional units. PRM features have proven utility in characterizing COPD pulmonary function and its severity. Therefore, we included PRM features in the analysis to investigate their potential contribution to COPD stage detection. The results of PRM_original features and PRM_selected features obtained through Lasso are presented in Fig. 8a. The findings indicate the following:
Firstly, the AMGNN outperformed other ML classifiers, demonstrating improvements of 6.9\(\%\), 13.0\(\%\), 13.0\(\%\), 16.4\(\%\), 15.6\(\%\), and 12.1\(\%\) in accuracy when using PRM_selected features. Similarly, there were enhancements of 3.2\(\%\), 11.3\(\%\), 11.3\(\%\), 19.4\(\%\), 13.1\(\%\), and 11.7\(\%\) in precision and 5.3\(\%\), 12.7\(\%\), 13.5\(\%\), 18.5\(\%\), 14.5\(\%\), and 11.9\(\%\) in F1-score. Additionally, AUC showed 2.1\(\%\), 1.9\(\%\), 8.4\(\%\), 4.0\(\%\), 5.8\(\%\), and 2.2\(\%\) improvements.
Secondly, the PRM_selected features exhibited better performance compared to the PRM_original features when applied with the AMGNN, showcasing improvements of 1.8\(\%\) in accuracy, 1.7\(\%\) in precision, 1.9\(\%\) in F1-score, and 1.3\(\%\) in AUC.
In Table 2, we have carried out supplementary experiments for fairness. The number of original PRM features is 54. To maintain parity, we have extracted the top 54 features based on Lasso regression coefficients from the (IN+EX) selected (Radiomics+3D CNN) features. And, the number of selected PRM features is 9. We have extracted the top 9 features based on Lasso regression coefficients from the (IN+EX) selected (Radiomics+3D CNN) features. The results show that when the number of PRM features and (radiomics+3D CNN) features is the same, the accuracy of (radiomics+3D CNN) features is higher. When the number of selected PRM features and selected (radiomics+3D CNN) features is the same, the accuracy of radiomics+3D CNN is higher. The results indicate that the effectiveness of PRM is relatively poor because PRM relies on specific threshold values for defining regions within the lung images. These thresholds may need to be carefully selected, and variations in thresholding can impact the results.
3.2 Detection based on radiomics features
In Experiment 2, as depicted in Fig. 8b, the input for the model consisted of radiomics features extracted from the IN and EX chest CT images. The evaluation metrics for COPD stage detection based on radiomics features using the AMGNN and ML classifiers are presented in Fig. 8b. The results indicate the following:
Firstly, the AMGNN demonstrated superior performance compared to other ML classifiers, exhibiting improvements of 10.3\(\%\), 17.2\(\%\), 20.7\(\%\), 17.2\(\%\), 24.1\(\%\), and 17.2\(\%\) in accuracy when using (IN+EX)_selected radiomics features. Similarly, there were improvements of 8.7\(\%\), 10.2\(\%\), 15.9\(\%\), 8.5\(\%\), 21.2\(\%\), and 7.2\(\%\) in precision and 10.2\(\%\), 13.7\(\%\), 19.9\(\%\), 22.5\(\%\), 24.2\(\%\), and 21.5\(\%\) in F1-score. Additionally, AUC showed improvements of 3.7\(\%\), 7.2\(\%\), 12.3\(\%\), 7.6\(\%\), 9.5\(\%\), and 8.4\(\%\).
Secondly, the (IN+EX)_selected radiomics features outperformed the other six types of features when applied with the AMGNN, showcasing improvements of 20.7\(\%\), 17.2\(\%\), 13.8\(\%\), 10.3\(\%\), 6.9\(\%\), and 3.4\(\%\) in accuracy. Moreover, there were enhancements of 35.4\(\%\), 12.9\(\%\), 11.4\(\%\), 6.3\(\%\), 3.5\(\%\), and 3.9\(\%\) in precision and 29.1\(\%\), 17.6\(\%\), 14.9\(\%\), 9.7\(\%\), 8.5\(\%\), and 3.8\(\%\) in F1-score. Finally, the AUC displayed improvements of 13.0\(\%\), 9.9\(\%\), 10.7\(\%\), 5.3\(\%\), 2.4\(\%\), and 3.5\(\%\).
3.3 Detection based on 3D CNN features
In Experiment 3, depicted in Fig. 8c, we assessed the effectiveness of using 3D CNN features as input. We used the ResNet-10 as the encoder backbone to generate 512 3D CNN feature maps with a size of \(3\times 3\times 3\). Therefore, each participant has 13,824 (512\(\times \)3\(\times \)3\(\times \)3\(=\)13,824) 3D CNN features obtained by flattening the 512 3D CNN feature maps. The evaluation metrics for COPD stage detection based on 3D CNN features using the AMGNN and ML classifiers are presented in Fig. 8c. The results indicate the following:
Firstly, the AMGNN outperformed the other ML classifiers when using (IN+EX)_selected 3D CNN features, showcasing improvements of 3.5\(\%\), 6.9\(\%\), 10.4\(\%\), 13.8\(\%\), 27.6\(\%\), and 20.7\(\%\) in accuracy. Similarly, enhancements were 7.2\(\%\), 9.5\(\%\), 12.8\(\%\), 18.0\(\%\), 27.8\(\%\), and 23.1\(\%\) in precision and 2.3\(\%\), 5.6\(\%\), 9.8\(\%\), 14.0\(\%\), 25.1\(\%\), and 18.8\(\%\) in F1-score. Additionally, AUC displayed 0.5\(\%\), 1.6\(\%\), 5.2\(\%\), 4.2\(\%\), 11.7\(\%\), and 5.6\(\%\) improvements.
Secondly, the (IN+EX)_selected 3D CNN features performed better than the other six types of features when applied with the AMGNN, demonstrating improvements of 24.2\(\%\), 17.3\(\%\), 17.3\(\%\), 13.8\(\%\), 6.9\(\%\), and 3.5\(\%\) in accuracy. Furthermore, improvements of 37.0\(\%\), 22.1\(\%\), 12.5\(\%\), 14.9\(\%\), 8.5\(\%\), and 6.4\(\%\) in precision, and 28.7\(\%\), 17.8\(\%\), 18.7\(\%\) 12.8\(\%\), 4.9\(\%\), and 1.7\(\%\) in F1-score were noted. Finally, the AUC showed improvements of 12.7\(\%\), 12.1\(\%\), 12.5\(\%\), 9.6\(\%\), 2.9\(\%\), and 4.9\(\%\).
In Table 3, We conducted experiments to explore the impact of using a larger model, specifically Resnet-18, Resnet-34, and Resnet-50, on our classification performance. Despite the larger model’s increased complexity, we observed that the best results were achieved using Resnet-10 in our specific context.
The Box-Plot of p-value as derived from the Pearson correlation coefficient based on the Radiomics + 3D CNN features. a EX_selected (Radiomics + 3D CNN) features, b IN_selected (Radiomics + 3D CNN) features, c (IN_selected + EX_selected) (Radiomics + 3D CNN) features, and d (IN + EX)_selected (Radiomics + 3D CNN) features
3.4 Detection based on radiomics and 3D CNN features
In Experiment 4, depicted in Fig. 8d, we aimed to verify whether the concatenation of radiomics and 3D CNN features could further improve the model’s performance.
During Lasso feature selection, we conducted 100,000 iterations of 10-fold cross-validation to calculate the regression coefficients for each feature. We retained features with non-zero regression coefficients and removed those with zero regression coefficients. Figure 4 presents the regression coefficients based on the Radiomics + 3D CNN features. Figure 5 presents the Box-Plot of p-value as derived from the Pearson correlation coefficient based on the Radiomics + 3D CNN features.
The findings of the COPD stage detection, utilizing IN and EX radiomics+3D CNN features with the AMGNN and ML classifiers, are depicted in Fig. 8d for Experiment 4. The results indicate the following:
Firstly, the AMGNN showcased superior performance compared to other ML classifiers when using (IN+EX)_selected (Radiomics+3D CNN) features, demonstrating improvements of 6.9\(\%\), 17.3\(\%\), 17.3\(\%\), 24.2\(\%\), 13.8\(\%\), and 24.2\(\%\) in accuracy. Similarly, there were enhancements of 7.0\(\%\), 14.7\(\%\), 14.0\(\%\), 14.7\(\%\), 14.1\(\%\), and 19.7\(\%\) in precision and 6.7\(\%\), 16.7\(\%\), 17.5\(\%\), 25.0\(\%\), 13.9\(\%\), and 23.4\(\%\) in F1-score. Additionally, AUC displayed improvements of 5.1\(\%\), 2.9\(\%\), 9.3\(\%\), 9.9\(\%\), 6.7\(\%\), and 10.2\(\%\).
Secondly, the (IN+EX)_selected (radiomics+3D CNN) features outperformed the other six types of features when applied with the AMGNN, demonstrating improvements of 24.2\(\%\), 17.3\(\%\), 17.3\(\%\), 13.8\(\%\), 10.4\(\%\), and 4.0\(\%\) in accuracy. Furthermore, there were enhancements of 25.1\(\%\), 35.6\(\%\), 18.5\(\%\), 15.3\(\%\), 8.9\(\%\), and 4.4\(\%\) in precision and 27.2\(\%\), 26.8\(\%\), 18.9\(\%\), 14.8\(\%\), 10.6\(\%\), and 3.9\(\%\) in F1-score. Additionally, AUC showed improvements of 14.1\(\%\), 8.9\(\%\), 9.0\(\%\), 6.6\(\%\), 5.4\(\%\), and 2.5\(\%\).
The results indicate that combining radiomics and 3D CNN features has significantly improved the performance. Figure 6 depicts the ROC of different classifiers based on the (IN+EX)_selected (radiomics+3D CNN) features are depicted. The final result is shown in Fig. 7, depicting the confusion matrix for the AMGNN and ML classifiers based on the (IN+EX)_selected (radiomics+3D CNN) features.
According to the findings presented in Table 4, it was discovered that the performance of the fusion of radiomics and 3D CNN was superior. The four feature types’ comparative analysis is conducted using the AMGNN. Additionally, in Fig. 8, we delve into a comparative analysis of the four feature types.
4 Discussion
According to the findings of this study, the AMGNN based on meta-learning has exhibited remarkable performance in COPD stage detection based on (IN+EX)_selected (radiomics+3D CNN) features, compared to RF, SVM, MLP, LR, GB, and LDA.
4.1 Merits of the AMGNN based on meta-learning
The AMGNN based on meta-learning exhibits a remarkable capacity for overcoming three significant challenges in COPD stage detection: (i) identifying the connections among different patients; (ii) minimizing the time-consume for diagnosis; (iii) possessing sustainable learning capabilities from newly acquired patient data.
The AMGNN possesses an auto-metric capability that exceeds that of GNNs. It enables the model to automatically calculate the edge constraint matrix based on the risk factors. This leads to a superior understanding of complex relationships between patients with diverse COPD stages. Additionally, the AMGNN demonstrates higher training efficiency compared to CNNs, as it can accomplish 500 epochs of training within approximately 10 min when running on a single core of the Intel (R) Xeon (R) Gold 5218 2.30GHz CPU and a GPU of the NVIDIA A100. This remarkable performance significantly enhances the efficiency of COPD diagnosis.
Furthermore, we employed a training strategy based on meta-learning in our model. This strategy ensures that the model can effectively learn from the small graph and transfer the learned knowledge to the following graph. Meta-learning has the ability to learn how to learn, as it utilizes past knowledge and experience to guide the learning of new tasks, which is in contrast to most current deep learning (DL), which typically requires training from scratch. In addition, based on its ability to learn how to learn, the meta-learning to our model can adapt to novel COPD datasets. Therefore, the AMGNN may surpass other models when trained on other additional central datasets.
4.2 Strategies of data concatenation and selection
A series of experiments were conducted to evaluate the efficacy of radiomics, 3D CNN, and radiomics+3D CNN features. Regardless of the feature types, the same conclusion is manifested.
Initially, when utilizing IN and EX features separately for the AMGNN and ML classifiers, the accuracy was observed to be below 70\(\%\). Afterward, we individually attempted feature selection by the Lasso for IN and EX, resulting in improved accuracy, as shown in Fig. 9a, b. However, the accuracy still did not exceed 80\(\%\). Subsequently, both IN+EX and IN_selected + EX_selected features were attempted separately. The IN+EX features (high dimension) results revealed that the accuracy did not improve compared to utilizing either IN or EX features. Despite containing more elements, the IN+EX features are highly sparse, and helpful information is limited, making it challenging for learning models to extract relevant information compared to IN/EX features.
In contrast, the IN_selected + EX_selected features (low dimension) showed improved accuracy compared to using IN_selected/EX_selected features. Hence, we must extract useful information from the IN+EX features. The final results showed that (IN+EX)_selected achieved the best performance using the AMGNN, surpassing the IN features in accuracy, precision, F1-score, and AUC metrics, respectively. Therefore, data concatenation and selection strategies as valuable tools based on IN and EX chest CT images can enhance the efficacy of the AMGNN and ML classifiers in COPD stage detection.
4.3 Expiration surpasses inspiration in COPD stage detection
Our findings indicate that the utilization of EX chest CT images, as opposed to IN chest CT images, results in the superior performance of the AMGNN and ML classifiers, as depicted in Fig. 9c. These observations carry significant implications for enhancing the clinical management of COPD patients. Healthcare professionals may prioritize using EX chest CT images for diagnosing and monitoring COPD patients, potentially leading to more effective treatment strategies and improved patient outcomes. Based on our analysis of the model results, we can conclusively state that EX demonstrates greater effectiveness than IN in detecting COPD stages. Nevertheless, from a clinical standpoint, it is evident that further research in this field is imperative to clarify these findings.
4.4 More suitable biomarker in COPD stage detection compared to PRM
To predict the COPD stage, this study evaluated four different types of features: radiomics, 3D CNN, radiomics+3D CNN, and PRM. According to the findings presented in Table 4, it is evident that the performance of PRM features did not achieve the highest level of optimization. The widespread use and applicability of PRM in diagnosing and staging lung diseases are well-established. This method offers the distinct advantage of providing local information about lung diseases and correlations with clinical and biological markers. Our study found that the PRM features did not exceed the expected benefits. In contrast, our research demonstrated that radiomics+3D CNN features proved more effective for COPD stage detection, improving 18.1\(\%\) accuracy, 13.8\(\%\) precision, 19.2\(\%\) F1-score, and 9.9\(\%\) AUC compared to PRM_selected features. Moreover, PRM requires lung bronchus segmentation and registration of IN and EX chest CT images, which are challenging tasks and can directly impact the precision of the \(PRM^{Normal}\), \(PRM^{{f}SAD}\) and \(PRM^{Emph}\) regions. However, our proposed approach requires only the segmentation of the lung region, which is a straightforward task. Therefore, all four types of features used in our study surpass PRM features, with radiomics+3D CNN features exhibiting the most impressive performance. These findings suggest that radiomics+3D CNN features possess the potential to serve as a novel biomarker for the diagnosis of COPD stage.
4.5 Limitations and future direction
It is essential to acknowledge three limitations of this study. Firstly, the datasets used were limited in scale and obtained from a single center. However, the meta-learning strategy allows continuous learning with new datasets, minimizing the need to train from scratch. Secondly, the utilization of the AMGNN requires diligent attention to the joint input of both training and testing data. Lastly, our study primarily focused on the engineering and algorithm aspects. Future research should delve into the clinical significance of our approach within these domains.
5 Conclusion
In this research paper, we introduced a novel approach for detecting COPD stages utilizing (IN+EX)_selected (radiomics+3D CNN) features with the auto-metric graph neural network (AMGNN). When compared to machine learning (ML) classifiers, the AMGNN integrated with risk factors (age, gender, and smoking history) achieved the highest performance, with an accuracy of 89.7\(\%\), precision of 90.9\(\%\), F1-score of 89.5\(\%\), and an AUC of 95.8\(\%\). Additionally, the three types of biomarkers-radiomics, 3D CNN, and radiomics+3D CNN-outperformed the predictive capability of PRM features in detecting COPD stages. Moreover, we discovered that the expiratory phase outperforms the inspiratory phase in COPD stage detection. Consequently, our proposed method overcame the limitations of traditional pulmonary function tests (PFTs) and held the potential to become an effective tool for managing COPD.
References
Singh D, Agusti A, Anzueto A, Barnes PJ, Bourbeau J, Celli BR, Criner GJ, Frith P, Halpin DM, Han M, et al (2019) Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease: the gold science committee report 2019. Eur Respir J 53(5)
Naghavi M, Abajobir AA, Abbafati C, Abbas KM, Abd-Allah F, Abera SF, Aboyans V, Adetokunboh O, Afshin A, Agrawal A, et al (2017) Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the global burden of disease study 2016. The Lancet 390(10100):1151–1210
Fabbri L, Pauwels R, Hurd S (2007) Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: gold executive summary updated 2003. COPD: J Chron Obstruct Pulmon Dis 1(1):105–141. https://doi.org/10.1081/copd-120030163
Fortis S, Comellas A, Make BJ, Hersh CP, Bodduluri S, Georgopoulos D, Kim V, Criner GJ, Dransfield MT, Bhatt SP (2019) Combined forced expiratory volume in 1 second and forced vital capacity bronchodilator response, exacerbations, and mortality in chronic obstructive pulmonary disease. Ann Am Thorac Soc 16(7):826–835
Flesch JD, Dine CJ (2012) Lung volumes: measurement, clinical use, and coding. Chest 142(2):506–510
Fan L, Xia Y, Guan Y, Zhang T-f, Liu S-y (2014) Characteristic features of pulmonary function test, CT volume analysis and MR perfusion imaging in COPD patients with different HRCT phenotypes. Clin Respir J 8(1):45–54
Lynch DA, Austin JH, Hogg JC, Grenier PA, Kauczor H-U, Bankier AA, Barr RG, Colby TV, Galvin JR, Gevenois PA et al (2015) Ct-definable subtypes of chronic obstructive pulmonary disease: a statement of the Fleischner Society. Radiology 277(1):192–205
Lynch DA (2014) Progress in imaging COPD, 2004-2014. Chronic Obstr Pulm Dis J COPD Found 1(1):73
Schroeder JD, McKenzie AS, Zach JA, Wilson CG, Curran-Everett D, Stinson DS, Newell Jr JD, Lynch DA (2013) Relationships between airflow obstruction and quantitative CT measurements of emphysema, air trapping, and airways in subjects with and without chronic obstructive pulmonary disease. AJR Am J Roentgenol 201(3):460
Bhatt SP, Washko GR, Hoffman EA, Newell Jr JD, Bodduluri S, Diaz AA, Galban CJ, Silverman EK, San José Estépar R, Lynch DA (2019) Imaging advances in chronic obstructive pulmonary disease. insights from the genetic epidemiology of chronic obstructive pulmonary disease (copdgene) study. Am J Respir Crit Care Med 199(3):286–301
Subramanian DR, Gupta S, Burggraf D, Vom Silberberg SJ, Heimbeck I, Heiss-Neumann MS, Haeussinger K, Newby C, Hargadon B, Raj V, et al (2016) Emphysema-and airway-dominant COPD phenotypes defined by standardised quantitative computed tomography. Eur Respir J 48(1):92–103
McDonough JE, Yuan R, Suzuki M, Seyednejad N, Elliott WM, Sanchez PG, Wright AC, Gefter WB, Litzky L, Coxson HO, et al (2011) Small-airway obstruction and emphysema in chronic obstructive pulmonary disease. N Engl J Med 365(17):1567–1575
Fujimoto K, Kitaguchi Y, Kubo K (2006) Honda T. Clinical analysis of chronic obstructive pulmonary disease phenotypes classified using high-resolution computed tomography. Respirology 11(6):731–740
Cho JL, Villacreses R, Nagpal P, Guo J, Pezzulo AA, Thurman AL, Hamzeh NY, Blount RJ, Fortis S, Hoffman EA, et al (2022) Quantitative chest CT assessment of small airways disease in post-acute sars-cov-2 infection. Radiology 304(1):185–192
Wu F, Chen L, Huang J, Fan W, Yang J, Zhang X, Jin Y, Yang F (2021) Zheng C. Total lung and lobar quantitative assessment based on paired inspiratory-expiratory chest CT in healthy adults: correlation with pulmonary ventilatory function. Diagnostics 11(10):1791
Galbán CJ, Han MK, Boes JL, Chughtai KA, Meyer CR, Johnson TD, Galbán S, Rehemtulla A, Kazerooni EA, Martinez FJ et al (2012) Computed tomography-based biomarker provides unique signature for diagnosis of COPD phenotypes and disease progression. Nat Med 18(11):1711–1715
Konietzke P, Wielpütz MO, Wagner WL, Wuennemann F, Kauczor H-U, Heussel CP, Eichinger M, Eberhardt R, Gompelmann D (2020) Weinheimer O. Quantitative CT detects progression in COPD patients with severe emphysema in a 3-month interval. Eur Radiol 30:2502–2512
Ho TT, Kim T, Kim WJ, Lee CH, Chae KJ, Bak SH, Kwon SO, Jin GY, Park E-K (2021) Choi S. A 3D-CNN model with CT-based parametric response mapping for classifying COPD subjects. Sci Rep 11(1):1–12
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, Van Stiphout R.G, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48(4):441–446
Yun J, Cho YH, Lee SM, Hwang J, Lee JS, Oh Y-M, Lee S-D, Loh L-C, Ong C-K, Seo JB, et al (2021) Deep radiomics-based survival prediction in patients with chronic obstructive pulmonary disease. Sci Rep 11(1):15144
Cho YH, Seo JB, Lee SM, Kim N, Yun J, Hwang JE, Lee JS, Oh Y-M, Do Lee S, Loh L-C, et al (2021) Radiomics approach for survival prediction in chronic obstructive pulmonary disease. Eur Radiol 31:7316–7324
Liang C, Xu J, Wang F, Chen H, Tang J, Chen D, Li Q, Jian W, Tang G, Zheng J, et al (2021) Development of a radiomics model for predicting COPD exacerbations based on complementary visual information. In: TP41. TP041 diagnosis and risk assessment in COPD, pp 2296–2296. American Thoracic Society
Yang Y, Li W, Guo Y, Liu Y, Li Q, Yang K, Wang S, Zeng N, Duan W, Chen Z, et al (2022) Early COPD risk decision for adults aged from 40 to 79 years based on lung radiomics features. Frontiers in Medicine 9
Yang Y, Li W, Guo Y, Zeng N, Wang S, Chen Z, Liu Y, Chen H, Duan W, Li X, et al (2022) Lung radiomics features for characterizing and classifying COPD stage based on feature combination strategy and multi-layer perceptron classifier. Math Biosci Eng 19(8):7826–7855
Li Z, Liu L, Zhang Z, Yang X, Li X, Gao Y (2022) Huang K. A novel CT-based radiomics features analysis for identification and severity staging of COPD. Acad Radiol 29(5):663–673
Chen S, Ma K, Zheng Y (1904) Transfer learning for 3D medical image analysis. arXiv preprint arXiv
Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK (2018) Medical image analysis using convolutional neural networks: a review. J Med Syst 42:1–13
Tajbakhsh N, Jeyaseelan L, Li Q, Chiang JN, Wu Z (2020) Ding X. Embracing imperfect datasets: a review of deep learning solutions for medical image segmentation. Med Image Anal 63:101693
Zhou J, Cui G, Hu S, Zhang Z, Yang C, Liu Z, Wang L, Li C (2020) Sun M. Graph neural networks: a review of methods and applications. AI open 1:57–81
Wu Z, Pan S, Chen F, Long G, Zhang C, Philip SY (2020) A comprehensive survey on graph neural networks. IEEE Trans Neural Netw Learn Syst 32(1):4–24
Song X, Mao M (2021) Qian X. Auto-metric graph neural network based on a meta-learning strategy for the diagnosis of Alzheimer’s disease. IEEE J Biomed Health Inform 25(8):3141–3152
McCombe N, Bamrah J, Sanchez-Bornot JM, Finn DP, McClean PL, Wong-Lin K, (ADNI) ADNI (2022) Alzheimer’s disease classification using cluster-based labelling for graph neural network on heterogeneous data. Healthc Technol Lett 9(6):102–109
Hofmanninger J, Prayer F, Pan J, Röhrich S, Prosch H (2020) Langs G. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur Radiol Exp 4(1):1–13
Van Griethuysen JJ, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RG, Fillion-Robin J-C, Pieper S, Aerts HJ. Computational radiomics system to decode the radiographic phenotype. Cancer Res 77(21):104–107 (2017)
Yang Y, Li W, Kang Y, Guo Y, Yang K, Li Q, Liu Y, Yang C, Chen R, Chen H et al (2022) A novel lung radiomics feature for characterizing resting heart rate and COPD stage evolution based on radiomics feature combination strategy. Math Biosci Eng 19:4145–4165
Yang Y, Chen Z, Li W, Zeng N, Guo Y, Wang S, Duan W, Liu Y, Chen H, Li X, et al (2022) Multi-modal data combination strategy based on chest HRCT images and PFT parameters for intelligent dyspnea identification in COPD. Frontiers in Medicine 9
Qi Y (2012) Random forest for bioinformatics. In: Ensemble machine learning: methods and applications, pp 307–323. Springer
Jakkula V (2006) Tutorial on support vector machine (SVM). School of EECS, Washington State University 37(2.5):3
Wan S, Liang Y, Zhang Y, Guizani M (2018) Deep multi-layer perceptron classifier for behavior analysis to estimate Parkinson’s disease severity using smartphones. IEEE Access 6:36825–36833
LaValley MP (2008) Logistic regression. Circulation 117(18):2395–2399
Ayyadevara VK (2018) Gradient boosting machine. Pro machine learning algorithms: a hands-on approach to implementing algorithms in python and R, pp 117–134
Balakrishnama S (1998) Ganapathiraju A. Linear discriminant analysis-a brief tutorial. Inst Signal Inf Process 18(1998):1–8
Acknowledgements
Thanks to the Department of Radiology, the First Affiliated Hospital of Guangzhou Medical University, for providing the dataset.
Funding
This work was supported by the National Key Research and Development Program of China [grant numbers 2022YFF0710800, 2022YFF0710802]; the National Natural Science Foundation of China [grant number 62071311]; the Stable Support Plan for Colleges and Universities in Shenzhen of China [grant number SZWD2021010]; and the Special Program for Key Fields of Colleges and Universities in Guangdong Province (Biomedicine and Health) of China [grant number 2021ZDZX2008].
Author information
Authors and Affiliations
Contributions
XD: methodolog, software, data analysis. WL: methodology, conceptualization. YY: methodology. SW: data analysis. NZ: data analysis. JX: medical guidance. HH: language polishing. ZC: data analysis. XM: investigation. YG: investigation. RC: conceptualization. YK: conceptualization.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deng, X., Li, W., Yang, Y. et al. COPD stage detection: leveraging the auto-metric graph neural network with inspiratory and expiratory chest CT images. Med Biol Eng Comput 62, 1733–1749 (2024). https://doi.org/10.1007/s11517-024-03016-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-024-03016-z