Abstract
Heart failure is a life-threatening syndrome that is diagnosed in 3.6 million people worldwide each year. We propose a deep fusion learning model (DFL-IMP) that uses time series and category data from electronic health records to predict in-hospital mortality in patients with heart failure. We considered 41 time series features (platelets, white blood cells, urea nitrogen, etc.) and 17 category features (gender, insurance, marital status, etc.) as predictors, all of which were available within the time of the patient’s last hospitalization, and a total of 7696 patients participated in the observational study. Our model was evaluated against different time windows. The best performance was achieved with an AUC of 0.914 when the observation window was 5 days and the prediction window was 30 days. Outperformed other baseline models including LR (0.708), RF (0.717), SVM (0.675), LSTM (0.757), GRU (0.759), GRU-U (0.766) and MTSSP (0.770). This tool allows us to predict the expected pathway of heart failure patients and intervene early in the treatment process, which has significant implications for improving the life expectancy of heart failure patients.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Heart failure (HF) is a life-threatening syndrome diagnosed in 3.6 million people worldwide each year with 35% of patients dying within the first year and the rest within 5 years [1]. HF is increasingly common in China, with a standardized prevalence of HF of 1.38% in patients older than 35 years of age, about 50% than in the survey dated in 2000 [2]. A favorable prognosis of HF using deep learning techniques can delay disease progression and improve patients’ quality of life and life expectancy. Accurately predicting in-hospital mortality in HF can assist physicians in diagnosing, which plays an essential role in clinical decision-making [3].
Electronic health record (EHR) contains healthcare information about patients, including diagnoses, procedures, medications, laboratory measurements, and imaging data [4], typically used to develop clinical decision support systems [5]. For example, several studies have constructed EHR risk models for adverse event prediction in HF [6,7,8]. In these analyses, traditional machine learning algorithms such as logistic regression (LR), random forest (RF), support vector machine (SVM), decision tree (DT), etc. are applied to EHR data to identify early HF or predict patient outcomes. However, these models use only limited information based on traditional statistical methods, which has the potential limitation of information loss.
In recent years, deep learning has been used in bioinformatics and healthcare with great success [13, 14] and is used for risk prediction in different clinical situations [15,16,17]. However, most studies rarely consider longitudinal time series information regarding inpatient treatment trajectories. Recurrent neural network (RNN) has achieved better performance by capturing temporal patterns present in EHR longitudinal series data [18]. However, using only the time series features of the EHR as a single input model, without considering categorical features such as demographics or without additional processing of categorical features, results in insufficient features and thus biases the decision direction of the final model, limiting the accuracy of the model. Feature fusion techniques extract complementary and more complete information by fusing data from multiple modalities. The completeness of the data allows for better execution of machine learning models, thus improving the accuracy of decisions [19].
In this study, we propose a deep fusion learning model, DFL-IMP, for HF mortality prediction. The model refines time series features and category features in structured EHR data and fuses the refined features for analysis, fully exploiting different forms of data information under a single model. Specifically, in our DFL-IMP, we propose a novel GRU cell, stochastic-decay gate recurrent unit, called GRU-S, which introduced a stochastic decay factor to capture the information on time series features in EHR. The category features are input to the fully connected layer for feature dimensionality reduction. The reduced-dimension features are fused with the output of the last hidden layer of GRU-S. The overall features are then fed to the fully connected layer and further fed to the classifier to predict the in-hospital mortality of HF. In addition, to address the problem of many missing values commonly found in EHR serial data, in our model, we invoked a variational recurrent neural network (VRNN) to estimate the missing values of the serial data.
The remainder of this paper is organized as follows. We review the related work in Section 2. Section 3 introduces the data, formulates the problem, and presents our proposed approach in detail. The experimental setup and results using a real clinical dataset are presented in Section 4. We discuss in Section 5 and conclude our work and highlight future research directions in Section 6.
2 Related work
2.1 Machine learning in EHR applications
Early prognostic work of HF mainly relies on traditional machine learning modeling techniques, such as LR, RF, SVM, DT, etc. A large number of studies have shown that machine learning plays a crucial role in the prognostic study of HF based on EHR data. Konig et al. [9] used four machine learning algorithms (i.e., LR, RF, XGBoost, NNet) to predict in-patient mortality to explore whether managing routine data alone could improve future care for HF. The problem is that it ignores information about patients’ heart imaging, laboratory results, medication, and treatment-related data, making it impossible to add more objective measures of disease severity. Adler et al. [10] trained an enhanced DT algorithm to capture the correlation between patient characteristics and mortality. To capture the higher-order correlation between input variables, it excluded missing data, which may introduce additional bias. Angraal et al. [11] used five machine learning methods to predict the risk of death and hospitalization in HF with ejection fraction retention, but it only used the baseline characteristics of patients without follow-up data, and the model explored was not enough to be applied to a wider population. Davide et al. [12] applied 10 machine learning classifiers to predict the survival of patients, and the results showed that serum creatinine and ejection fraction were sufficient to predict the survival of patients, but the dataset used was only 299 pieces, and the data scale was small. In general, the above studies only use traditional statistical methods, such as LR model multivariate analysis, which may have the limitation of loss of information.
2.2 Deep learning in EHR applications
In recent years, an increasing number of scholars have applied deep learning techniques to medical research. One of the advantages of deep learning is that features and relationships can be learned automatically from given data without the need for feature engineering. Wang et al. [20] proposed a deep learning system based on feature rearrangement to predict heart failure mortality. This framework dealt with the problem of unbalanced datasets and achieved better feature representation. However, a limitation of their study is that only the aggregated features of events within a single observation window are used, ignoring the temporal relationship between events within the observation window.
Recently, RNN models have been used effectively in many complex machine learning tasks, such as image captioning and language translation. The ability of RNNs to model high-dimensional non-linear long-term dependencies between elements has attracted the attention of researchers in healthcare, with a series of studies using RNN models to capture the temporal patterns present in EHR longitudinal series data for disease progression and risk prediction. Jun et al. [21] introduced a stochastic gradient variational Bayesian (SGVB) approach to RNN sequence models to capture the underlying sequence structure and generate missing values for multivariate time series data inference. Missing values expressed as variance were used as fidelity using uncertainty and a new uncertainty-gated stochastic sequence model was proposed for clinical time series prediction. Men et al. [22] used a long and short-term memory network with a time-aware and attention-based mechanism to classify multi-labeled diseases based on patients’ clinical attendance records, but with a single dataset and without using demographic data. Priyanga et al. [23] used a multilayer bidirectional LSTM algorithm for feature selection, the LCBWO algorithm for structural improvement and fast convergence, and the LSTM algorithm for predicting heart disease within 5 years, achieving high accuracy. Yoon et al. et al. [24] proposed the M-RNN, which uses a bidirectional RNN to estimate missing values. The M-RNN in which the estimated values are considered constant and cannot be updated sufficiently. It uses a bidirectional RNN-based consistency loss to prevent the propagation of errors. McGilvray et al. [25] integrated a deep learning model using time series and densely connected networks developed based on standard EHR data to assist clinicians to identify HF drug treatment non-response and predict death in a timely and accurate manner. Li et al. [38] proposed an MTSSP model for predicting survival, which interpolates missing values by combining mask and time interval information to obtain a global view using a bidirectional RNN architecture and a local view using a one-dimensional dilated convolution, combining a missing value complementation approach with a time series classification prediction approach. Shickel et al. [39] proposed a dynamic approach to label discrete and continuous patient data and proposed a transformer classifier that uses a joint embedding space to integrate different temporal patient measures. Six mortality and readmission outcomes were also predicted simultaneously. However, these studies focused on other clinical conditions and did not focus on the area of heart failure.
We further explored studies in the HF domain, where Chu et al. [26] used an adversarial learning scheme to distinguish generated feature vectors from true feature vectors, using the prediction of HF feature vectors as an adjunct to endpoint prediction, but its data used only part of the treatment trajectory, missing the value-rich outpatient information with follow-up data. Radhachandran et al. [27] developed a gradient-enhanced decision tree to predict 7-day mortality in AHF patients by applying continuous information from the first 8 h of the patient’s hospitalization.
It can be concluded from the above studies that previous studies in the field of heart failure are often limited by the dataset, where a single dataset or a small or unbalanced dataset may lead to a decrease in model accuracy. Most of the models using EHR time series have a single input, ignoring categorical features such as demographics and no additional features have been applied to them. Our research focuses on exploring the integration of time series data with categorical data for heart failure patients to predict their in-hospital mortality.
2.3 Feature fusion analysis of EHR
Feature fusion techniques improve decision accuracy by fusing data from multiple modalities to extract complementary and more comprehensive information for better execution of machine learning models [28]. Most studies [31,32,33] have shown that models using fusion perform better compared to the performance of a single model. Feature fusion plays a crucial role in medical decision making. Zhi et al. [29] proposed a multimodal fusion model based on a multilayer perceptron and a two-dimensional CNN to ingest EHR data and CT images for pulmonary embolism diagnosis. In addition, multidimensional scaling (MDS) algorithms were used to reduce the dimensionality of EHR data. Zheng et al. [30] used a novel longitudinal data fusion approach to model disease progression for chronic disease care. A temporal regularization term was designed to maintain the temporal inheritance of data at different time points, and data from both the source level and feature level were analyzed based on a sparse regularized regression approach. Li et al. [40] proposed an enhanced BEHRT model, Hi-BEHRT, for risk prediction. It allows integration of long EHR series from different modalities, addresses the shortcomings of common transformers in handling long series data and avoids the loss of important historical information in risk prediction, and has achieved superior results in four investigated (heart failure, diabetes, CKD and stroke) risk prediction tasks. Liu et al. [41] proposed a new multimodal PLM for jointly modeling unstructured and structured data in electronic medical records that learns cross-modal interactions while maintaining unimodal representation capabilities.
In summary, the above studies focus on data fusion in multiple modalities, ignoring the different forms of data types in a single modality. Instead, we argue that a granular analysis of different types of data in a single modality can help to mine more adequate information with fewer data. In addition, data from other modalities of medical clinics are not readily available.
3 Methods
3.1 Data preparation
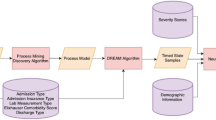
We used the Medical Information Mart for Intensive Care (MIMIC-IIIFootnote 1) dataset, an extensive medical center database with EHR data related to 53,423 adult patient admissions to intensive care units at the Beth Israel Deaconess Medical Center between 2001 and 2012. It includes vital signs, medications, laboratory measurements, observations, and notes recorded by nursing staff, fluid balance, procedure codes, diagnosis codes, imaging reports, length of stay, survival data, and more [36]. Criteria for the incident onset of HF were adopted from Gurwitz et al. [37], which relied on qualifying International Classification of Diseases, Ninth Revision (ICD-9) codes. Our target sample was HF patients with ICD-9 code 4280.
We selected 41 laboratory longitudinal measured time series features and 17 category features. Time series features are mainly laboratory measurements. These data share the same property that they change over time, and the distribution is nonuniform in time. For instance, a blood test is a discrete event that happens sometime during the admission. The category features are the patient’s gender, insurance, etc. Usually, it will not change during hospital admission. In all our observation windows, 41 time series features and 17 category features are included, and the number of features did not decrease due to the decrease in observation windows. Table 1 shows the information on several of the critical patient features we selected. Differences between the positive and negative classes were assessed for significance using a two-proportion t-test.
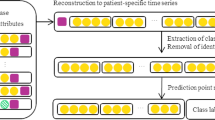
For the time series features in our data, we consider different observation and prediction windows to evaluate the performance of our model, each patient was considered for multiple observations, the observation window length lasted for days, choose one observation per day, corresponding to a time window. If the patient had multiple observations in a day, we used the average value to represent the observations for that day, and the unobserved values we set to Null. As shown in Fig. 1, the observation windows were selected as 5, 7, and 10 days, and the prediction windows were selected as 10, 20, and 30 days. The reason for choosing the window this way is that the data drives us to do so; an observation window that is too short would not take full advantage of the information in the time series data, and too long an observation window would exclude too many positive samples resulting in unbalanced data. As our goal was to predict patient death, for patients with multiple hospitalizations, we selected data from the patient’s last hospitalization. If the patient’s last hospitalization was less than the length of our observation window (5 days/7 days/10 days), this hospitalization data was excluded and the patient’s most recent hospitalization data was then selected. If all of the patient’s hospital admissions are less than our observation window, the patient is excluded. For category features, such as gender, and co-morbidities, that did not change during the hospital admission.
Overall, we extracted a total of 13,112 EHR data from the MIMIC-III database for patients with HF. After excluding non-compliant patients and after data pre-processing, 7696/6296/4472 samples remained, with sample sizes varying by observation window, as shown in Table 2, with sample sizes varying by observation window. Patients were included in the positive class only if the death date occurred within the selected prediction windows for mortality prediction. Patients with mortality dates later than the observation window were included in the negative class, as they were still alive within the prediction window. Surviving patients with no documented mortality were also included in the negative class.
Since the data values of the time series are continuous, to prevent the influence of outliers on the study results, we remove the outliers with a Winsorize process. As each variable has other evaluation indicators, there are different units of magnitude, and they are in different magnitudes. Z-score normalization is performed for all datasets so that each feature is in the same order of magnitude for a comprehensive comparison. For the category features, we use one-hot coding to transform the category variables.
3.2 Problem formulation
We define a patient sample with \(D\) time series features and \(Q\) category features (time-invariant features). Given a time series feature \(X\) , which is observed at \(T\) time points, we denote it as \(X=\left[{x}_{1},\cdots ,{x}_{d},\cdots ,{x}_{D}\right]=\left[\begin{array}{ccc}{x}_{1}^{1}& \cdots & {x}_{D}^{1}\\ \vdots & & \vdots \\ {x}_{1}^{T}& \cdots & {x}_{D}^{T}\end{array}\right]\), where \({x}_{d}^{t}\) is the observed value of the feature \(d\) at the moment \(t\). Given a category feature \(C\), which is denoted as \(C=\left[{c}_{1},\cdots ,{c}_{q},\cdots ,{c}_{Q}\right]\), \({c}_{q}\) denotes the observed value of the feature \(q\).
We introduce the masking vector \(M\) that marks the observed and missing values in the time series, denoted as \(M=\left({m}_{1},\dots ,{m}_{d},\dots ,{m}_{D}\right)\), if \({x}_{d}^{t}\) is observed, then \({m}_{d}^{t}=1\); otherwise, \({m}_{d}^{t}=0\). Based on the mask vector, we define a new time series containing missing values \(\widetilde{X}=\left({\widetilde{x}}_{1},\dots ,{\widetilde{x}}_{d},\dots ,{\widetilde{x}}_{D}\right)\), as follows:
where \(\ast\) indicates the missing values, and the initial \(\ast\) is set to 0. In addition, we define the time interval \(\Delta =\left({\delta }_{1},...,{\delta }_{d},...,{\delta }_{D}\right)\) as the difference between the time stamp of the last observed value and the current timestamp with the following equation:
where \({s}_{d}^{t}\) represents the time stamp \(t\) observed by the feature \(d\), assuming that the timestamp of the first observation is 0 (i.e.\({s}_{1}=0\)).
For \(N\) samples, given the dataset \(V={\left\{\left({\widetilde{X}}^{(n)},{M}^{(n)},{\Delta }^{(n)}\right)\circ {C}^{(n)}\right\}}_{n=1}^{N}\), \(\circ\) denotes the matrix juxtaposition. \(Y\) is the predictive label for whether the patient died in the hospital (1 for death and 0 for survival), denoted as:\({Y}_{n}=\left[{y}_{1},\cdots ,{y}_{n}\right]\).
3.3 Model description
The proposed DFL-IMP consists of three main components: (1) VRNN missing value imputation, (2) GRU-S, and (3) mortality prediction of feature fusion. The idea of the bidirectional recurrent neural network is applied in our model, specifically, the inputs of DFL-IMP are both the serial forward direction \(X=\left\{{x}_{1},{x}_{2},\dots ,{x}_{D}\right\}\) and backward direction \({X}^{^{\prime}}=\left\{{x}_{1}^{^{\prime}},{x}_{2}^{^{\prime}},\dots ,{x}_{D}^{^{\prime}}\right\}\). The final result is the average of the forward DFL-IMP and backward DFL-IMP calculations. The overall architecture of the DFL-IMP is shown in Fig. 2.
3.3.1 VRNN missing value imputation
We adopt VRNN [34] to fill in the missing values in the time series. Figure 3 shows the flowchart of the model, which consists of three main steps: (1) prior, (2) inference, and (3) generation. VRNN cyclically updates the hidden layer state as follows:
The derivation of the formula is detailed in Appendix A.
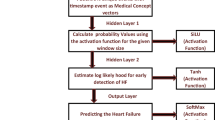
3.3.2 GRU-S
Our proposed model GRU-S is based on the GRU-U [21] made improvement; the model architecture is shown in Fig. 4. We propose two different decay factors, the stochastic decay factor and the time decay factor, which mainly address the problem of input variables disappearing over time due to the long-term absence of EHR time series data.
Firstly, we investigated the effectiveness of the stochastic decay factor propagation in the GRU hidden state \({h}_{t}\). Since the imputation value are stochastic, we capture stochastically in VRNN with a stochastic estimate of \({\sigma }_{x,t}\), as:
We introduced a training decay rate in the model using a negative exponential rectifier to make the stochastic decay factor \({d}_{t}\in (\mathrm{0,1}]\) monotonically decreasing, as follows:
where \({s}_{t}\) is a stochastic factor and \({W}_{d}\) is a diagonal matrix to ensure that the decay factors of each variable are independent of each other. The stochastic decay factor \({d}_{t}\) is fed into our network by updating the state of \({{\varvec{h}}}_{t-1}\):
Secondly, we consider a time decay factor \({\gamma }_{t}\) to decay the input features. The calculation is similar to the stochastic decay factor as follows:
\({W}_{\gamma }\), \({b}_{\gamma }\) is the trainable model parameter, and \({\delta }_{t}\) is the time interval. Based on the time decay factor \({\gamma }_{t}\), \({\widetilde{x}}_{t}\) and the estimated mean values \({\mu }_{x,t}\) from VRNN weighted calculation to obtain the time decay estimate \({c}_{t}\) as follows:
where \({m}_{t}\) is the mask vector, and \({\mathcal{H}}^{decay}\) is a fully connected layer. We finally express the imputation estimates in the following equation:
where \({\mathcal{H}}^{cor}\) is a feature regression layer to calculate the relationship between features, \({\mathcal{H}}^{pool}\) is a neural network layer. From this, we can obtain the hidden layer state \({h}_{t}\) as follows:
In GRU-S, the information is controlled by resetting gate \(r\) and updating gate \(u\) with the following equations:
where \(\sigma\) is a nonlinear activation function. It is worth noting that \({x}_{t}\) and \({h}_{t-1}\) in the original GRU formula have been replaced with \({\mathcal{H}}^{x}\left({\widehat{x}}_{t}\right)\circ {\mathcal{H}}^{z}\left({z}_{t}\right)\) and \({h}_{t-1}^{^{\prime}}\). In addition, we input the masking vector \({m}_{t}\) additionally to the model.
3.3.3 Mortality prediction of feature fusion
Feature fusion is the process of combining data and knowledge from different sources to maximize useful information content. It improves the reliability or discriminant capability and offers the opportunity to minimize the data retained [35]. There are three main types of integration strategies, namely early fusion, joint fusion, and late fusion [19]. In this work, joint fusion was adopted. For time series features, we take the output of the features from the last hidden layer of GRU-S, and for category, we use a fully connected layer (FC) to extract features. Specifically, a new feature vector is composed which consists of time series features extracted using GRU-S, and category features extracted by FC, as shown in Fig. 2. We directly concatenate the features extracted from the two networks.
Firstly, we extract category features as follows:
where \({C}_{one-hot}\) is the category features after one-hot encoding, which is because our category features are mainly composed of patient demographic features (gender, insurance, etc.) and patient co-morbidities, and these feature values are discrete and disordered, and using one-hot encoding to process the data will make the calculation of the distance between features more reasonable. \(\mathcal{H}\) is a fully connected layer that extracts and reduces the dimensionality of the features and alleviates the sparsity and high dimensionality of the one-hot coding input vector. \(\mathrm{ tanh}\) is a nonlinear activation function.
Secondly, since the last GRU-S hidden state contains temporal information encoded across all time steps, we use the last GRU-S hidden state \({h}_{t}\) to fuse with the category features as follows:
where \(\oplus\) is a matrix concatenation, the fused features \({V}_{t}\) contain richer data information, and \({V}_{t}\) is fed into a fully connected layer, followed by a sigmoid activation function, which is used to perform our binary classification task. This is shown below:
where \({W}_{o}\) is a classifier parameter.
3.4 Training and testing
We use a joint learning strategy throughout defining the loss function of the model as a composite function with four components: (1) VRNN loss \({\mathcal{L}}_{VRNN}\); (2) consistency loss \({\mathcal{L}}_{cons}\); (3) masked imputation value loss \({\mathcal{L}}_{imp}\); and (4) classification loss \({\mathcal{L}}_{bce}\).
-
(1)
We refer to the loss calculation function of VAE to define the VRNN loss \({\mathcal{L}}_{VRNN}\), the main objective is to integrate the reconstruction error and Kullback–Leibler (KL) divergence of \(N\) samples over the time series, as follows:
$${\mathcal{L}}_{VRNN}=\sum_{n=1}^{N}\sum_{t=1}^{T}(-KL(q({z}_{t}|{\widetilde{x}}_{1:t}^{(n)},{z}_{1:t-1})\parallel p({z}_{t}|{\widetilde{x}}_{1:t}^{(n)},{z}_{1:t-1}))+\mathrm{log }p({\widetilde{x}}_{t}^{(n)}{|}_{1:t},{\widetilde{x}}_{1:t-1}^{(n)}))$$(18) -
(2)
Consistency loss is defined as the difference between the estimate of VRNN in the forward DFL-IMP \({\widehat{x}}_{t}\) and the estimate of VRNN in the backward DFL-IMP \({\widehat{x}}_{t}^{^{\prime}}\) by time variation, calculated from the mean absolute error (MAE).
$${\mathcal{L}}_{cons}=\frac{1}{N}\sum_{n=1}^{N}\left|{\widehat{X}}^{^{\prime}}{}^{(n)}-{\widehat{X}}^{(n)}\right|$$(19) -
(3)
Masked imputation value loss \({\mathcal{L}}_{imp}\), we calculate the masked MAE between the original sample \({X }\) as the ground truth and the input sample \(\widehat{X}\), \({M}_{imp}\) is the initial masking matrix.
$${\mathcal{L}}_{imp}=\frac{1}{N}\sum_{n=1}^{N}\left|{X}^{(n)}\odot {M}_{imp}^{(n)}-{\widehat{X}}^{(n)}\odot {M}_{imp}^{(n)}\right|$$(20) -
(4)
Classification loss \({\mathcal{L}}_{bce}\), we calculate from the binary cross entropy loss.
$${\mathcal{L}}_{bce}=\frac{1}{N}\sum_{n=1}^{N}(-[{y}_{n}\mathrm{log}\sigma ({p}_{n})+(1-{y}_{n})\mathrm{log}(1-\sigma ({p}_{n}))]$$(21)
\({y}_{n}\) is the label, \({p}_{n}\) is the predicted probability and \(\sigma\) is the sigmoid activation function.
Therefore, all losses are defined by integrating the forward and reverse losses and the composite loss is defined as \(\mathcal{L}={\omega }_{1}\left({\mathcal{L}}_{VRNN}+{\mathcal{L}}_{VRNN}^{^{\prime}}\right)+{\omega }_{2}{\mathcal{L}}_{cons}+{\omega }_{3}\left({\mathcal{L}}_{imp}+\right.\left.{\mathcal{L}}_{imp}^{^{\prime}}\right)+{\omega }_{4}\left({\mathcal{L}}_{bce}+{\mathcal{L}}_{bce}^{^{\prime}}\right)\), where \({\omega }_{1}\), \({\omega }_{2}\), \({\omega }_{3}\), and \({\omega }_{4}\) are the hyperparameters that control the loss ratio. We optimize all parameters of the model in an end-to-end manner through this composite loss.
3.5 Experimental setup
In our mortality prediction task, missing value imputed and outcome prediction are performed simultaneously during training. We trained our model using the RAdam optimizer with an initial learning rate of 0.001, epoch set to 80, learning rate decay set to every 10 epochs, decay 0.01 times, and batch size set to 64. The composite loss parameter \({\omega }_{1}\), \({\omega }_{2}\), \({\omega }_{3}\), and \({\omega }_{4}\) is set to \(1{e}^{-5}\), 1, \(1{e}^{-2}\) and 1. We set masking scenarios is 5% of the observations that were additionally masked for dataset. We selected the final optimal model based on the performance of the validation set.
4 Results
Our baseline model consists of three main components: machine learning models (LR, RF, SVM), deep learning models (LSTM, GRU), and published approaches from the literature (GRU-U [21], MTSSP [38]). LR is a linear classifier with a linear combination of features as independent variables, RF is an algorithm that integrates multiple decision trees, and SVM is a binary classification model defined on the feature space with the largest interval. The above model is difficult to process sequence data, the model is prone to underfitting problems, and the experimental results are not very ideal. The time series model LSTM learns long-term dependencies through three gate mechanisms, while the GRU greatly improves the training speed by discarding forgetting gates, but these two models are less effective when there is a large amount of missing data.
Our proposed model, DFL-IMP, captures missing time series through the attenuation mechanism of GRU-S and introduces the VRNN model to interpolate missing values, which solves the problem of missing time series values in medical data of heart failure well. In addition, we fused category features and extracted features using FC to make full use of different forms of data information in patients with HF.
For all models, we used a five-fold cross-validation strategy, with 60% of the data trained, 20% used for validation, and the performance of the trained models evaluated on the remaining 20% of the data (the final test set), which was not used during training. The experiment was repeated five times and the final performance was the mean and standard deviation of these five repetitions. AUC (area under the ROC curve), accuracy, precision, F1 score, and specificity were used as evaluation metrics.
The algorithms were implemented in Python 3.7. LSTM and GRU were trained using Pytorch 1.10.0 for model training and implemented using the Torch.nn.Module library. LR, RF, and SVM were implemented using Python Scikit-Learn 0.24.2.
4.1 Performance by DFL-IMP
We evaluated the validity of the proposed model DFL-IMP, based on the observation window shown in Fig. 1.
First, we investigated the effect on the AUC values obtained by the HF mortality prediction model by changing the observation and prediction windows. We set one of the windows by default and conducted experiments by changing the value of the other window, as shown in Fig. 5. The results show that the performance of our model decreases as the length of the observation window increases (Fig. 5(a)) and increases as the prediction window increases (Fig. 5(b)).
With an observation window of 5 days and a prediction window of 30 days, our model achieves an AUC of 0.914. Machine learning models have AUCs around 0.7, specifically: LR (0.708), RF (0.717), and SVM (0.675). Our model improved by 20.6% over LR and 19.7% over RF. The AUC values for the deep learning models were approximately 0.75 for LSTM (0.757) and GRU (0.759), GRU-U (0.766), and MTSSP (0.770). Our model improved by 15.7% over LSTM, 14.8% over GRU-U, and 14.4% over MTSSP. Overall, our model results are around 0.9 for all evaluation windows, significantly better than other benchmark models. Secondly, we investigated the model’s performance in terms of accuracy, precision, F1 score, and specificity under specific prediction windows, as shown in Table 3. When the observation window is 5 days and the prediction window is 30 days, the accuracy (0.928), precision (0.867), and F1 score (0.734) of our model all rank first, although the specificity (0.982) of our model is not as good as SVM (0.999), but in general, our model metrics outperform other models.
In addition, we explored the ROC curves for all the models evaluated over a 7-day observation window and a 30-day prediction window, as shown in Fig. 6, and the results show that our models achieved excellent results.
From the results, our model achieves such excellent results mainly due to the refinement analysis and fusion processing of time series data with category data, as well as the inclusion of a stochastic attenuation factor in GRU-S to capture the missing data. Specifically, firstly, the missing time series data are captured by the stochastic attenuation in GRU-S to reduce the non-fidelity of the information passed downstream. We also introduce VRNN to interpolate the sequence data to reduce the information loss of the missing time series, capturing the trend information well. Secondly, the patient demographic information is captured by extracting category features through FC. Our proposed model provides significant improvements in all metrics, fully explores patients’ survival health information, and more accurately predicts patient survival outcomes. This has important implications for researchers and provides a reference for future research work.
4.2 Performance by GRU-S
To analyze the effectiveness of our proposed GRU-S in handling time series data, we analyzed the performance of HF mortality prediction based on time series features. We evaluated GRU-S according to the window shown in Fig. 1, and the results are shown in Table 4. The results show that the deep learning model outperforms the traditional machine learning model in handling time series data, while our proposed GRU-S outperforms MTSSP and GRU-U in most cases. When the observation window is 5 days and the prediction window is 30 days, the AUC value of GRU-S reaches 0.775, while the AUC value of MTSSP is only 0.770, GRU-U (0.766), GRU (0.759), and LSTM (0.757). The GRU-S model worked best and outperformed the other benchmark models in most evaluation windows.
It is shown that the update and reset gates of the GRU-S control the inflow of more useful information and the discarding of unnecessary information, respectively. For time series with missing values in the EHR, the impact of low-fidelity input data on the prediction task is reduced by adding a stochastic decay factor to the data input and hidden layers, effectively propagating randomness within the GRU unit gates at each timestamp, thus allowing the estimated data to be combined with this randomness in a non-linear manner.
4.3 Performance by feature fusion strategy
First, we explore whether data integration can improve the accuracy of the prediction models. For the machine learning models LR, RF and SVM, we concat mean of time series features and category features, fed into the model to predict patient mortality. For LSTM, GRU, GRU-U and MTSSP, we applied our fusion ideas to these baseline models by fusing the RNN final hidden layer state with the category features. The experimental results under all evaluation windows are shown in Table 5. Our model works best with an observation window of 5 days and a prediction window of 30 days, with an AUC value of 0.914, compared to LR (0.898), RF (0.900), SVM (0.881), LSTM (0.909), GRU (0.910), GRU-U (0.913), and MTSSP (0.792). Notably, with an observation window of 5 days and a prediction window of 20 days, GRU-U achieves an AUC of 0.902 after fusing features, which is higher than our model (0.900); with an observation window of 10 days and a prediction window of 20 days, GRU-U achieves an AUC of 0.891 after fusing features, which is higher than our model (0.890). However, in most cases, our model performed better than the other benchmark models.
Figure 7 compares the model performance before and after data integration, with the same window settings as in Fig. 5 by default. With an observation window of 5 days and a prediction window of 30 days, the AUC of LR can reach 0.898 after data integration, which is 19% higher than before integration, the AUC of LSTM can reach 0.909, which is 15.2% higher than before integration, and the AUC of GRU-U can reach 0.913, which is 14.7% higher than before integration, and MTSSP can achieve an AUC of 0.792, 2.2% higher than before integration. Our model achieves an AUC of 0.914, 13.9% higher than before integration. The experimental results show that data integration of time series features with category features is effective in improving the accuracy of the models compared to prediction models using a single time series feature or a single aggregated feature.
In addition, we explored the effectiveness of fine-grained analysis of the data on the sequence models, as shown in Fig. 8. “Model + Fusion” denotes our fusion strategy, which explicitly treats time series data separately from category data using different models. “Model + Integration” means that the category data are concatenated to the time series data based on the length of the time series data and input to a separate model. The results show that our model achieves an AUC of 0.914 under the applied fusion strategy, while under data integration the AUC is 0.911. GRU-U achieves an AUC of 0.913 under the applied fusion strategy, while under data integration, the AUC is 0.896. LSTM achieves an AUC of 0.909 under the applied fusion strategy, while under data integration the AUC is 0.881. The results show that the model fusion strategy is particularly effective in improving outcomes. We believe that refined analysis of healthcare data can help improve prediction accuracy.
The study shows that feature fusion in heart failure mortality prediction research questions, introducing multiple forms of patient electronic health data helps to fully exploit patient information and obtain a closer approximation to the patient’s true physical condition. In addition, with the joint fusion strategy we used, the model better achieved interactions between data in different formats, improving the accuracy of the model for heart failure prediction tasks.
4.4 Ablation studies
In this study, we performed two sets of ablation experiments under two different observation windows to validate the results obtained by our method.
Firstly, we conducted an ablation study of the proposed GRU-S and fusion strategy, and the results are presented in Table 6. Our proposed DFL-IMP model exhibited degraded performance by ablating the GRU-S and fusion strategies. With an observation window of 5 days, the model performance was 0.775 when GRU-S was applied without the fusion strategy, and conversely, the model performance reached 0.910. It can be concluded that the gain in classification performance by applying the GRU-S model was not as significant as the fusion strategy. The model performance is best when both are applied and worst when neither is applied. It can be seen that the DFL-IMP model is not a single module that improves the algorithm performance, but the best result is produced by the combination.
Secondly, we used a convolutional neural network (CNN) instead of the FC used in the category features and the results of the study are shown in Table 7. With an observation window of 5 days, the AUC value reached 0.914 when using FC, whereas when using CNN, the AUC value was only 0.887. This indicates that FC performs better than CNN. We speculate that this is because FC mainly maps vectors, while CNN is mainly used for feature extraction and is good at extracting Euclidean structured data. For the category features after preprocessing (one-hot) in the experiment, convolution cannot extract feature information effectively, while FC can learn effective mapping transformations.
5 Discussion
In this study, it was first hypothesized that a refined analysis of time series and category data in the electronic health record would help to improve the accuracy of in-hospital mortality prediction models for patients with HF. The prediction performance results for all models are shown in Fig. 5 and Table 3. The proposed model, DFL-IMP, is considerably more effective after the introduction of category data and significantly outperforms other baseline models (LR, RF, SVM, LSTM, GRU, GRU-U, MTSSP). This leads to the conclusion that mining the hidden information contained in each of the different forms of data and introducing auxiliary information (i.e., demographic information) is crucial to the HF mortality prediction model.
Secondly, we believe that for a large number of missing values in medical time series data, the model adopts additional processing mechanisms to help extract information from the data and improve prediction accuracy. As can be seen from Table 4, our proposed GRU-S outperforms other sequence models (LSTM, GRU, GRU-U, MTSSP), 0.9% increase compared to GRU-U, achieving good results. This suggests that propagating uncertainty within the GRU unit gate reduces the loss of information in the time series due to missing values, allowing the estimated data to be combined with randomness in a non-linear manner. This helps to reduce the impact of low-fidelity input data on mortality prediction results. That is, in the presence of missing values, key information in the sequence data can still be captured.
For the first time, we use a feature fusion strategy to construct a deep learning model to predict in-hospital mortality in patients with heart failure. In our evaluation results, we confirmed that our model results represent the most advanced predictive performance that can be achieved by applying a deep learning model that uses feature fusion strategies to discover complex relationships in EHR data. For small differences in window length, the model effect does not differ much. For the same prediction window, the prediction effect decreases with the increase of the observation window. Under the same observation window, the prediction effect of the model increases with the increase of the prediction window, as shown in Fig. 5(b), and Fig. 7(b). This is due to our dataset; the longer the observation window (the shorter the prediction window), the more positive samples we exclude, the more unbalanced the data and the decreasing model effect. This suggests that our model prefers a more balanced dataset and works relatively better on a more balanced dataset. We expect that model performance will benefit from using a more balanced sample of cases.
Limitations of this study focus on three aspects. Firstly, the length of our time series feature observation window is short and insufficient, and the performance of the model is limited by the equilibrium state of the dataset. Secondly, our fusion is a simple fusion without further exploration of other fusion strategies. Third, we mined different forms of data in a single mode without combining multiple modes, such as chest radiographs, medical prescriptions, and medications, to obtain adequate prognostic information on heart failure.
6 Conclusion and future work
This paper aims to apply deep learning methods to in-hospital mortality prediction in HF using classification methods. We propose a new model DFL-IMP to refine the analysis of time series features and category features in EHR data and perform feature fusion modeling. In this, GRU-S is proposed to capture time series features, FC to capture category features and VRNN to estimate missing values of time series features. The experimental results show that our proposed model DFL-IMP dramatically improves the accuracy of in-hospital mortality decisions in HF, which helps physicians to make timely interventions in high-risk patients, delay the progression of HF disease, and improve the quality of life and life expectancy of patients.
In future work, we will combine data from clinical hospitals, select samples with fewer missing values, and choose observation windows of appropriate length. We will also introduce a priori knowledge of clinical medicine and use statistical analysis to find the impact of specific features on the results to enhance the interpretability of deep learning. Further, we will explore whether other advanced fusion strategies can improve model performance, as well as consider multimodal data fusion studies, e.g., physician’s orders versus chest films, to produce more clinically meaningful models.
Notes
Available at https://mimic.physionet.org/
Abbreviations
- EHR:
-
Electronic health record
- HF:
-
Heart failure
- LR:
-
Logistic regression
- RF:
-
Random forest
- SVM:
-
Support vector machine
- LSTM:
-
Long short-term memory
- GRU:
-
Gate recurrent unit
References
Chen J, Aronowitz P (2022) Congestive heart failure[J]. Medical Clinics 106(3):447–458. https://doi.org/10.1016/j.mcna.2021.12.002
Wang H, Chai K, Du M et al (2021) Prevalence and incidence of heart failure among urban patients in China: a national population-based analysis[J]. Circ Heart Fail 14(10):e008406. https://doi.org/10.1161/CIRCHEARTFAILURE.121.008406
Błaziak M, Urban S, Wietrzyk W et al (2022) An artificial intelligence approach to guiding the management of heart failure patients using predictive models: a systematic review[J]. Biomedicines 10(9):2188. https://doi.org/10.3390/biomedicines10092188
Kao DP (2022) Electronic health records and heart failure[J]. Heart Fail Clin 18(2):201–211. https://doi.org/10.1016/j.hfc.2021.12.004
Reimer AP, Dai W, Smith B et al (2021) Subcategorizing EHR diagnosis codes to improve clinical application of machine learning models[J]. Int J Med Inform 156:104588. https://doi.org/10.1016/j.ijmedinf.2021.104588
Lv H, Yang X, Wang B et al (2021) Machine learning-driven models to predict prognostic outcomes in patients hospitalized with heart failure using electronic health records: retrospective study[J]. J Med Internet Res 23(4):e24996. https://doi.org/10.2196/24996
Javeed A, Khan S U, Ali L et al (2022) Machine Learning-Based Automated Diagnostic Systems Developed for Heart Failure Prediction Using Different Types of Data Modalities: A Systematic Review and Future Directions[J]. Computational and Mathematical Methods in Medicine 2022:9288452. https://doi.org/10.1155/2022/9288452
Benke KK (2019) Data Analytics and Machine Learning for Disease Identification in Electronic Health Records[J]. JAMA Ophthalmol 137(5):497–498. https://doi.org/10.1001/jamaophthalmol.2018.7055
König S, Pellissier V, Hohenstein S et al (2021) Machine learning algorithms for claims data-based prediction of in -hospital mortality in patients with heart failure[J]. ESC Heart Fail 8(4):3026–3036. https://doi.org/10.1002/ehf2.13398
Adler ED, Voors AA, Klein L et al (2020) Improving risk prediction in heart failure using machine learning[J]. Eur J Heart Fail 22(1):139–147. https://doi.org/10.1002/ejhf.1628
Angraal S, Mortazavi BJ, Gupta A et al (2020) Machine learning prediction of mortality and hospitalization in heart failure with preserved ejection fraction[J]. JACC Heart Fail 8(1):12–21. https://doi.org/10.1016/j.jchf.2019.06.013
Davide C, Giuseppe J (2020) Machine learning can predict survival of patients with heart failure from serum creatinine and ejection fraction alone[J]. BMC Medical Informatics and Decision Making 20(1):16. https://doi.org/10.1186/s12911-020-1023-5
Xie F, Yuan H, Ning Y et al (2022) Deep learning for temporal data representation in electronic health records: A systematic review of challenges and methodologies[J]. J Biomed Inform 126:103980. https://doi.org/10.1016/j.jbi.2021.103980
Penso M, Solbiati S, Moccia S et al (2022) Decision Support Systems in HF based on Deep Learning Technologies[J]. Current Heart Failure Reports 19(2):38–51. https://doi.org/10.1007/s11897-022-00540-7
Haq IU, Chhatwal K, Sanaka K et al (2022) Artificial intelligence in cardiovascular medicine: current insights and future prospects[J]. Vasc Health Risk Manag 18:517. https://doi.org/10.2147/VHRM.S279337
Doppalapudi S, Qiu R G, Badr Y (2021) Lung cancer survival period prediction and understanding: Deep learning approaches[J]. International Journal of Medical Informatics 148:104371
Gupta VK, Gupta A, Kumar D et al (2021) Prediction of COVID-19 confirmed, death, and cured cases in India using random forest model[J]. Big Data Min Anal 4(2):116–123. https://doi.org/10.26599/BDMA.2020.902001
Tong R, Lei L, Zhou Y et al (2019) Representation learning for clinical time series prediction tasks in electronic health records[J]. BMC Med Inform Decis Mak 19(Suppl 8):259. https://doi.org/10.1186/s12911-019-0985-7
Amal S, Safarnejad L, Omiye JA et al (2022) Use of Multi-Modal Data and Machine Learning to Improve Cardiovascular Disease Care[J]. Frontiers in Cardiovascular Medicine 9:840262. https://doi.org/10.3389/fcvm.2022.840262
Wang Z, Zhu Y, Li D et al (2020) Feature rearrangement based deep learning system for predicting heart failure mortality[J]. Comput Methods Prog Biomed 191:105383. https://doi.org/10.1016/j.cmpb.2020.105383
Jun E, Mulyadi A W, Choi J, et al (2020). Uncertainty-Gated Stochastic Sequential Model for EHR Mortality Prediction[J]. IEEE Transactions on Neural Networks and Learning Systems 32(9):4052–4062. https://doi.org/10.1109/TNNLS.2020.3016670
Men L, Ilk N, Tang X et al (2021) Multi-disease prediction using LSTM recurrent neural networks[J]. Expert Syst Appl 177:114905. https://doi.org/10.1016/j.eswa.2021.114905
Priyanga P, Pattankar VV, Sridevi S (2021) A hybrid recurrent neural network-logistic chaos-based whale optimization framework for heart disease prediction with electronic health records[J]. Comput Intell 37(1):315–343. https://doi.org/10.1111/coin.12405
Yoon J, Zame W R, van der Schaar M (2018) Estimating Missing Data in Temporal Data Streams Using Multi-directional Recurrent Neural Networks[J]. IEEE Transactions on Biomedical Engineering 66(5):1477–1490. https://doi.org/10.1109/TBME.2018.2874712
McGilvray MMO, Heaton J, Guo A et al (2022) Electronic health record-based deep learning prediction of death or severe decompensation in heart failure patients[J]. Heart Fail 10(9):637–647. https://doi.org/10.1016/j.jchf.2022.05.010
Chu J, Dong W, Huang Z (2020) Endpoint prediction of heart failure using electronic health records[J]. J Biomed Inform 109:103518. https://doi.org/10.1016/j.jbi.2020.103518
Radhachandran A, Garikipati A, Zelin N S, et al (2021) Prediction of short-term mortality in acute heart failure patients using minimal electronic health record data[J]. BioData Mining 14(1):23. https://doi.org/10.1186/s13040-021-00255-w
Zheng G, Han G, Soomro NQ (2019) An inception module CNN classifiers fusion method on pulmonary nodule diagnosis by signs[J]. Tsinghua Sci Technol 25(3):368–383. https://doi.org/10.26599/TST.2019.9010010
Zhi Z, Elbadawi M, Daneshmend A, et al (2022) Multimodal Diagnosis for Pulmonary Embolism from EHR Data and CT Images[C]. 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). IEEE, pp 2053–2057. https://doi.org/10.1109/EMBC48229.2022.9871041
Zheng Yi, Xiangpei Hu (2020) Healthcare predictive analytics for disease progression: a longitudinal data fusion approach. J Intell Inf Syst 55(2):351–369. https://doi.org/10.1007/s10844-020-00606-9
Niu K, Lu Y, Peng X et al (2022) Fusion of sequential visits and medical ontology for mortality prediction[J]. J Biomed Inform 127:104012. https://doi.org/10.1016/j.jbi.2022.104012
Lin M, Wang S, Ding Y, et al (2021) An empirical study of using radiology reports and images to improve ICU-mortality prediction[C]. 2021 IEEE 9th International Conference on Healthcare Informatics (ICHI). IEEE, Victoria, BC, Canada, pp 497–498. https://doi.org/10.1109/ICHI52183.2021.00088
Hess LM, Han Y, Zhu YE et al (2021) Characteristics and outcomes of patients with RET-fusion positive non-small lung cancer in real-world practice in the United States[J]. BMC Cancer 21(1):1–12
Chung J , Kastner K , Dinh L et al (2015) A recurrent latent variable model for sequential data[J]. Advances in neural information processing systems 28
Ross A, Jain A (2003) Information fusion in biometrics[J]. Pattern Recogn Lett 24(13):2115–2125. https://doi.org/10.1016/S0167-8655(03)00079-5
Alistair LS, Johnson EW, Pollard TJ (2016) Data descriptor: MIMIC-III a freely accessible critical care database[J]. Thromb Haemost 76(2):258–262. https://doi.org/10.1038/sdata.2016.35
Gurwitz JH, Magid DJ, Smith DH et al (2013) Contemporary prevalence and correlates of incident heart failure with preserved ejection fraction[J]. Am J Med 126(5):393–400. https://doi.org/10.1016/j.amjmed.2012.10.022
Li B, Shi Y, Cheng L, et al (2022) MTSSP: Missing value imputation in multivariate time series for survival prediction[C]. 2022 International Joint Conference on Neural Networks (IJCNN). Padua, Italy, pp 1–8. https://doi.org/10.1109/IJCNN55064.2022.9892806.
Shickel B, Silva B, Ozrazgat-Baslanti T et al (2022) Multi-dimensional patient acuity estimation with longitudinal EHR tokenization and flexible transformer networks[J]. Front Digital Health 4:1029191. https://doi.org/10.3389/fdgth.2022.1029191
Li Y, Mamouei M, Salimi-Khorshidi G et al (2022) Hi-BEHRT: Hierarchical Transformer-Based Model for Accurate Prediction of Clinical Events Using Multimodal Longitudinal Electronic Health Records[J]. IEEE J Biomed Health Inform27(2):1106–1117. https://doi.org/10.1109/JBHI.2022.3224727
Liu S, Wang X, Hou Y et al (2022) Multimodal Data Matters: Language Model Pre-Training Over Structured and Unstructured Electronic Health Records[J]. IEEE Journal of Biomedical and Health Informatics 27(1):504–514. https://doi.org/10.1109/JBHI.2022.3217810
Acknowledgements
The paper supported by National Major Scientific Research Instrument Development Project (62027819): High-speed Real-time Analyzer for Laser Chip’s Optical Catastrophic Damage Process; The General Object of National Natural Science Foundation(62076177): Research on Risk Assessment Model for Heart Failure Incorporating Multi-modal Big Data; Shanxi Province key technology and generic technology R\&D project (2020XXX007): Energy Internet Integrated Intelligent Data Management and Decision Support Platform; Key research and development program of Shanxi Province NO.202102020101006.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, M., Hao, X., Zhao, J. et al. Predicting heart failure in-hospital mortality by integrating longitudinal and category data in electronic health records. Med Biol Eng Comput 61, 1857–1873 (2023). https://doi.org/10.1007/s11517-023-02816-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02816-z