Abstract
Selection of differentially expressed genes (DEGs) is a vital process to discover the causes of diseases. It has been shown that modelling of genomics data by considering relation among genes increases the predictive performance of methods compared to univariate analysis. However, there exist serious differences among most studies analyzing the same dataset for the reasons arising from the methods. Therefore, there is a strong need for easily accessible, user-friendly, and interactive tool to perform gene selection for RNA-seq data via machine learning algorithms simultaneously not to miss DEGs. We develop an open-source and freely available web-based tool for gene selection via machine learning algorithms that can deal with high performance computation. This tool includes six machine learning algorithms having different aspects. Moreover, the tool involves classical pre-processing steps; filtering, normalization, transformation, and univariate analysis. It also offers well-arranged graphical approaches; network plot, heatmap, venn diagram, and box-and-whisker plot. Gene ontology analysis is provided for both mRNA and miRNA DEGs. The implementation is carried out on Alzheimer RNA-seq data to demonstrate the use of this web-based tool. Eleven genes are suggested by at least two out of six methods. One of these genes, hsa-miR-148a-3p, might be considered as a new biomarker for Alzheimer’s disease diagnosis. Kidney Chromophobe dataset is also analyzed to demonstrate the validity of GeneSelectML web tool on a different dataset. GeneSelectML is distinguished in that it simultaneously uses different machine learning algorithms for gene selection and can perform pre-processing, graphical representation, and gene ontology analyses on the same tool. This tool is freely available at www.softmed.hacettepe.edu.tr/GeneSelectML.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Identification of differentially expressed genes (DEGs) by analyzing RNA-seq data is very important for the discovery of the mechanisms and pathways underlying the disease. Conventional statistical methods to find DEGs often apply univariate tests for each gene. Therefore, they do not take into account the correlations between genes and concordant or discordant effect between gene groups [1]. Also, these statistical methods generate a large number of false positives and false negatives due to the small biases included in the distribution estimates to predict DEGs from RNA-seq data [2]. In order to prevent these problems, machine learning algorithms can be used to find DEGs causing the disease. Wenric and Shemirani [1] used the permutation importance generated by the Random Forests algorithm to find DEGs in 12 datasets containing the samples of various cancers. Random Forests algorithm outperformed classical methods in most datasets. Wang et al. [3] compared three feature selection algorithms (Information Gain, Correlation Feature Selection, and ReliefF) using five classification algorithms (Logistic Regression, Classification via Regression, Random Forest, Logistic Model Trees, Random Subspace) to detect significant genes. Kakati et al. [2] proposed a deep neural network model called DEGnet in order to identify DEGs. Yu et al. [4] studied attaching the biological significance of regulatory information to differential expression analysis. For this purpose, they used Naive Bayes, Random Forest, and Support Vector Machine with radial basis kernel methods. Al-Obeidat et al. [5] proposed discrete filtering for RNA-seq gene expression data to carry out feature selection. They used Binary Artificial Bee Colony Algorithm and Support Vector Machine to select the fittest and relevant subset of features to classify tumor as malignant and benign samples.
As clear from these examples, there are many machine learning algorithms used for gene selection. These algorithms can propose quite different genes from each other. The proposed number of genes can also vary considerably from algorithm to algorithm. Therefore, in this paper, a web tool called GeneSelectML is developed by using shiny package [6]. GeneSelectML allows the users to discover DEGs using different machine learning algorithms simultaneously. This web tool is available at www.softmed.hacettepe.edu.tr/GeneSelectML and a snapshot of the main page is given in Fig. 1.
We have selected the machine learning algorithms among the ones that are shown to be successful in the literature. During the preliminary analysis, we have searched and tested many other algorithms as well. However, some of them are eliminated either due to unavailability of predict function or unstable results on a case study or reasons such as the discontinuation of the package in CRAN. The second criteria for the inclusion is the accessibility of the algorithm in R or Bioconductor. Since the web tool we developed is a shiny-based application, the algorithms should be available in R. There are other shiny-based web tools designed for various purposes for RNA-seq gene expression data such as pre-processing [7], discovering DEGs [8, 9] and conducting gene ontology analysis [10]. GeneSelectML is distinguished from other shiny-based web tools by the fact that it uses different machine learning algorithms simultaneously for gene selection and it can perform pre-processing, graphical representation and gene ontology analyses all on the same tool. There is also a software called CAMUR developed for RNA-seq datasets and uses machine learning algorithms for selecting significant genes [11]. However, this software needs the MySql database to run and is not appropriate for online use.
A real life example dataset on Alzheimer’s disease is used in this study to illustrate the methods and the web tool. Alzheimer’s disease dataset is obtained from the Gene Expression Omnibus (GEO) Database [12]. It includes miRNAs obtained from 48 Alzheimer’s disease patients and 22 controls. miRNAs have important functions at the post-transcriptional level of gene expression in many pathological conditions including Alzheimer’s disease (AD) [13]. Previous studies showed that AD brains have significant miRNA alterations compared to healthy controls [14, 15]. Alzheimer’s disease, major cause of dementia, is a progressive neurodegenerative disorder which is forecasted to affect 1 in 85 people globally in 2050 [16]. Moreover, there is no curative treatment for AD and the pathological mechanism is not fully known. Therefore, there are needs for new biomarkers and treatment strategies. With the recent advent of new “-omics” based technologies, large amount of data is being generated. In order to analyze and interpret quickly for diagnostic and therapeutic use, there is a need for user-friendly and fast tools. Following the uploading of data to the web tool, pre-processing steps including filtering, normalization, transformation, and univariate analysis are carried out. Different machine learning methods are applied to the pre-processed dataset simultaneously and the DEGs are discovered. Moreover, network plot, heatmap, venn diagram, and box-and-whisker plot can be obtained, and gene ontology analysis can be conducted.
The sections of this paper are organized as follows: Section 2 introduces the proposed methodology, including the pre-processing procedures, machine learning techniques, and development of web tool. Section 3 provides the implementation of GeneSelectML web-based tool on Alzheimer’s disease data, its findings, a case study based on this dataset, and the validation of the tool on a different dataset. Finally, the paper is concluded with a summary of the main findings explored during our study.
2 Methods
2.1 Pre-processing
RNA-seq data must go through some pre-processing steps. These steps can be generalized as filtering, normalization, transformation, and univariate analysis.
2.1.1 Filtering
It is recommended to filter low expressed genes before analysis. Filtering can be done in different ways. The following filtering methods are available in the web tool:
-
i)
Genes with all readings lower than a specified threshold can be eliminated.
-
ii)
Genes with “near-zero variances” can be excluded.
These filtering methods are available in genefilter [17] and caret [18] packages, respectively.
2.1.2 Normalization
Normalization is applied to RNA-seq data to minimize bias that may arise from technical processes. The number of readings required in RNA-seq data is determined by the minimum amount of RNA species of interest. Sequencing depth can be increased for the purposes such as identifying genes with low expression levels, identifying very small fold changes between different situations and detecting new transcripts. However, different sequencing depth values may lead to underestimation or overestimation of gene expression levels [19]. Another source of variation is gene length. Longer genes may have higher readings, i.e., expression levels, than genes with shorter sequences due to differences in their size [20].
Normalization aims to make the samples comparable by reducing the effect of such bias factors. Many methods have been developed for the normalization of RNA-seq data. The methods are generally based on scaling the data according to a calculated normalization factor.
Median ratio normalization
Consider a gene expression matrix with samples at rows \((i=1,..,n)\) and genes as columns \((g=1,\ldots ,p)\). This matrix contains raw gene read counts \(X_{ig}\). For each gene, a reference sample is created by taking the geometric mean in all samples. Then, the ratio of the sample of interest to the reference sample is calculated for each gene. Finally, by taking the median of the rates, the normalization factor is calculated for the relevant sample. Normalized values are obtained by dividing the read counts of the gene by the normalization factor for each sample.
The normalization factor \((d_{i})\) for each sample can be calculated as follows [21]:
This method can be applied to the data using the DESeq2 package [22] in R Bioconductor.
Trimmed mean of M values normalization (TMM)
Genes with very low or high expression levels are removed from the dataset based on M values. M values (\(M_{ig}\)) are trimmed by 30% as default [23]. Weight values (\(w_{ig}\)) are calculated for the remaining genes. Then, normalization factor is calculated based on these weights. The transformed normalization factor is calculated as follows:
where \(p'\) indicates the number of genes after trimming.
TMM with singleton pairing normalization (TMMwsp)
This method is a type of TMM which performs better for the data containing the zeros with high proportion. In TMM method, a sample is chosen as a reference sample. The fold changes and absolute expression levels are obtained relative to the reference sample. The genes which take the value of zero in both corresponding and reference samples are discarded. Unlike the TMM method, TMMwsp method makes a correction by using the total read number of these genes.
Upper quartile normalization
Transcripts with zero value are removed from the dataset and normalized over the 75th percentile values of the remaining values. Therefore, this method is, unfortunately, affected by the genes with high expression levels.
2.1.3 Transformation
Normalizing data may not be sufficient to apply feature selection methods since the expression levels can be distributed in a wide range in RNA-seq data. The logarithmic transformation is also be used in such a situation. With logarithmic transformation, data with a less skewed distribution and fewer excessive values are obtained than untransformed data. The logarithmic transformation may be undefined as the count values for a gene can be zero under some conditions. To avoid this situation, transformation is performed after a prior count of 1 is added.
Let the normalized gene be denoted by \(X_{ig}^{'}\). In this case, the transformed genes can be represented as follows:
2.1.4 Filtering with univariate analysis
Univariate analysis can be used to reduce the size of the dataset and identify genes that differ significantly between groups. Our web tool has two alternatives to carry out the univariate analysis for each gene by comparing two groups with Student’s t-test using the colttests function or by calculating AUC with the rowpAUCs function in the genefilter package [17]. If the Student’s t-test is selected, the genes are ordered from the smallest p-value to the highest according to the test result. If the AUC method is chosen instead of the Student’s t-test, the genes are ordered from the highest AUC value to the least. In both methods, the specified number of genes at the top of the ranking are selected by the user. Together with the p-values obtained as a result of Student’s t-test, adjusted p-values are also calculated according to the Benjamini-Hochberg (FDR) [24] or Benjamini-Yekutieli [25] correction methods. The default is set to Benjamini-Hochberg method.
2.2 Machine learning algorithms
Six different machine learning methods have been used in our web tool. These methods will be explained in the next four sub-titles.
2.2.1 Biosigner algorithm
Rinaudo et al. [26] proposed a four-step algorithm for selecting the important genes and provided the algorithm in biosigner R package. These steps involve constructing a model by using bootstrap sub-samples, ranking the genes by their importance, eliminating the non-significant ones, and deciding on the final model. The models are based on the Partial Least Squares-Discriminant Analysis, Random Forest, and Support Vector Machines. Our web tool provides the list of genes selected by any of these three models.
2.2.2 GMDH-type neural network algorithm
GMDH-type neural network algorithm is a heuristic self organizing system to learn complex relation between exploratory variables and dependent variable. In its architecture, some neurons performing better compared to the rest of the neurons in each layer, called living cells, continue their ways until the decrease in performance across layers. At last neuron, one neuron is selected to obtain predicted output. The features contributing model performance are selected at the end. The algorithm is available in GMDH2 package [27].
2.2.3 Determan’s optimal gene selection algorithm
Our web tool uses the Support Vector Machines, Random Forest, and Elastic Net Generalized Linear Models within the Determan’s algorithm [28] for gene selection. This algorithm uses bootstrap for measuring feature selection stability and uses cross-validation or leave-one-out procedures to avoid the overfitting problem. The average of cross-validation results is used to calculate performance measures (such as accuracy, sensitivity, etc.) and these measures are then used to obtain the list of best genes. The algorithms are available in omicsMarkeR package [28].
2.2.4 Data mining algorithm for RNA-Seq data
Chiesa et al. [29] developed a data mining algorithm for RNA-seq data and implemented it in DaMiRseq package. It is possible to normalize, select genes, and classify via this algorithm. We use the gene selection procedures, specifically DaMiR.Fsort and DaMiR.FBest functions, of this algorithm in our web tool.
Genes are first ranked based on RReliefF [30] or standardized RReliefF scores in DaMiR.Fsort function. RReliefF is a filtering algorithm that can also take the correlation between genes into account. These ranked genes are then used to pick the best subset via DaMiR.FBest function. The user can either provide the number of selected genes, or algorithm can automatically pick the best subset by using a threshold on the scaled importance scores. Our web tool uses the later approach.
2.3 Evaluation of model performances
The performances of models are obtained through a confusion matrix between the predicted and actual class labels. In our case, Table 1 presents 2-by-2 classification table where the predicted and actual class labels are provided in the rows and columns, respectively. Various performance measures can be obtained using a confusion matrix. We assess the model performance with accuracy, kappa, Matthews correlation coefficient (MCC), sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), balanced accuracy, Youden index, detection rate, detection prevalence, and F1 measure. Calculation of these measures is presented in Table 2. It is recommended to use fivefold or tenfold cross-validation to avoid overfitting in machine learning algorithms [31, 32]. In this study, we use fivefold cross-validation to prevent overfitting and improve model performances. We obtain the performance measures based on test set for each fold. Then, we report the mean of the performance measures obtained in five folds.
2.4 Web tool development
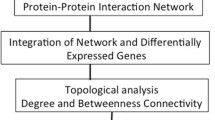
The tool is developed using R software. This tool is designed into seven parts; data upload, pre-processing, methods, selected genes, pathway analysis, visualization, and gene ontology analysis. In Data upload part, researchers can upload raw count gene expressions in .txt format. The raw data must be a n\(\times\)(1+p) dimensional data matrix, where n refers to the total number of samples, p refers to the total number of genes. The first column must be the output variable. The data must include a header indicating gene names. In Pre-processing part, caret [18] and genefilter [17] packages are used for filtering. DESeq2 [22] and edgeR [33] packages are utilized for normalization. The number of genes is reduced using univariate analyses, Student’s t-test or calculating AUC with genefilter package [17]. In Methods part, biosigner [26], GMDH2 [27], omicsMarkeR [28], and DaMiRseq [29] packages are used for the selection of DEGs. In this process, five-fold cross-validation is carried out to validate the models. The process is paralleled with doParallel package [34] to overcome high-volume computational load. The tool can recognize whether the data type is miRNA or mRNA with miRNAmeConverter package [35]. ReactomePA package [36] is used to obtain the pathway analysis of the genes suggested by the models. multiMiR package [37] is used to identify target genes for miRNA datasets prior to pathway analysis. ComplexHeatmap [38], igraph [39], venn [40], and graphics [41] packages are utilized for the visualization of the genes suggested by models in Visualize part. For gene ontology analysis, topGO [42], mirnatab [43], and miRNAtap.db [44] packages are used. Annotation of genes is provided by using org.Hs.eg.db package [45]. All analysis steps of GeneSelectML web tool are presented in Fig. 2.
There are two example datasets available in the web tool, Alzheimer’s disease data (miRNA) and Kidney chromophobe data (mRNA), to help users learn the usage of the tool. Also, there is a toy data available in .txt format just to learn how to upload the data. There exist two panels of the interface: sidebar and main panels. Researchers can specify the arguments of the methods in the sidebar panel. The parameters not decided by user are set to the defaults of the original algorithms. The results of the specified models are provided in the main panel. After the process is completed in pre-processing and methods parts, summary of the process is provided in summary under methods tab. Selected genes are listed based on genes and methods in two sub-tabs of selected genes tab. In this tab, there are two options to continue to pathway analysis, graphical approaches, and gene ontology analysis. Users can choose the genes suggested by at least one method or at least two methods. After this choice, the results of the pathway analysis can be downloaded via the download link from this tab. There exist various graphical approaches in visualize tab including a number of options for editing plots. A gene ontology analysis is conducted in GO tab. All results including tables and plots can be downloaded in different file formats. A detailed manual of the tool is available in the web page of the tool.
3 Results
3.1 Implementation of the tool
In this section, we analyze Alzheimer RNA-seq data to demonstrate the use of this web-based tool. The dataset is uploaded via Data upload tab (Fig. 3). After uploading the data, the dataset is pre-processed in Pre-processing tab with four steps: filtering with conventional ways, normalization, transformation, and filtering with univariate analysis. In Methods tab, we construct six models presented in Section 2.2. Selected genes are provided in Selected genes tab. Users can specify the genes selected by at least one or two method(s) to continue Visualize and GO tabs. There exist four graphical approaches; network plot, heatmap, venn diagram, and box-and-whisker plot in Visualize tab. Finally, we perform gene ontology analysis of DEGs in GO tab.
3.2 Dataset
We analyze Alzheimer RNA-seq dataset [13] for this illustration. This dataset includes a cohort of 70 samples — 48 Alzheimer’s disease patients and 22 controls — and 503 features (i.e., miRNAs). The data can be found at GEO with accession number GSE46579 [46]. We load the dataset to the tool using Data upload tab (Fig. 3) before starting analysis.
3.3 Pre-processing
Dimension reduction is the essential step to improve the performance of methods for diagnosing DEGs. In this part, we use near-zero variances filtering. Then, the data are normalized using median ratio normalization. After normalizing the data, logarithmic transformation is applied. The number of genes is reduced to 200 using univariate analysis (i.e., Student’s t-test).
3.4 Gene selection and classification performance
We construct six machine learning algorithms after pre-processing stage is completed. We report the selected genes in two ways. One is providing the genes based on methods. The other one is reporting the list of genes (Table 4). In Table 4, there exist a list of genes, the frequency and percent of methods suggesting the corresponding gene, the regulation status and the names of methods suggesting the corresponding gene. Twenty-four genes, of which 11 genes are selected by at least two methods, are suggested by at least one method.
The classification performances of the methods are presented in Table 3. The results show that SVM performs better than the other methods with respect to the most of performance measures. It is important to point out that DaMirseq performs best for the classification of Alzheimer patients when the sensitivity is assessed. The algorithm classifies 100\(\%\) of the persons having Alzheimer’s disease. For SVM, sensitivity is obtained as 0.950. The method classifies 95\(\%\) of the persons having Alzheimer’s disease. GLMNET outperforms other algorithms in terms of MCC, is one of the best in terms of detection rate and is competitive with others in most of the measures.
In Selected genes tab, users can specify the genes selected by at least one or two method(s). We select the genes suggested by at least two methods for further analysis.
3.5 Visualization
This web-based tool offers well-arranged graphical approaches; network plot (Fig. 4a), heatmap (Fig. 4b), venn diagram (Fig. 4c), and box-and-whisker plot (Fig. 4d). The network plot shows whether the correlation exists between selected genes in a way that the correlation is positive or negative. The tool offers the users to color positive and negative correlations. In our case, we color blue for positive correlation and color red for negative correlation if the magnitude of correlation is larger than 0.6. Researchers can draw the heatmap of selected genes with class labels. The tool also provides venn diagram which shows the number of genes selected by methods and their intersections. Users can draw box-and-whisker plot to compare the groups with respect to each of the selected genes.
Graphical approaches in GeneSelectML web tool. (a) indicates the correlation between selected genes with a magnitude greater than 0.60. Blue color states positive correlation while red color states negative correlation. (b) represents standardized values based on rows. Genes are given in the rows, samples are given in the columns. (c) demonstrates the number of genes selected by algorithms and their intersections. (d) shows the distribution of the expression values by groups for the gene of interest. If Student’s t-test is selected as univariate analysis, p-values are added to the bottom of the plot
3.6 Findings on Alzheimer RNA-seq data
In the Alzheimer study, we analyze 503 genes of 48 AD and 22 healthy controls. The number of genes is reduced to 200 after pre-processing. Out of these 200 genes, 11 of them are found to be differentially expressed by two or more algorithms in our GeneSelectML web tool, and an additional of 13 genes are detected as DEGs by one algorithm (Table 4). Out of 11 DEGs, only three of them are upregulated. The results highlight the strength of using a tool which incorporates many methods. For instance, using only the DaMirseq would detect 11 genes as significant instead of 24 DEGs. Similarly, using only OmicsMarkeR-GLMNET would miss 15 genes found by other methods. Our web tool is able to list a combination of genes detected by many algorithms in a reasonable time. The computational time for the process, including data upload, filtering, normalization, transformation, univariate analysis, and applying six different machine learning algorithms, is approximately 300 seconds.
By gene ontology analysis, we analyze the biological process of 11 miRNAs which are proposed via at least two algorithms. We find that decreased miRNAs affect positive regulation of phosphorylation, cell cycle, setting macromolecules, regulation of locomotion, and increased miRNAs affect positive regulation of nucleobase, chromosome organization processes.
The miRNA proposed by four different machine learning algorithms is has-miR-628-3p. It has been shown that hsa-miR-628-3p is related to many cancers and it promotes apoptosis in lung cancer cell cultures [47]. Similar to current study results, a previous study, which analyzed more than 1200 miRNAs in AD temporal cortex, showed that expression levels of hsa-miR-628-3p, has-miR-1234, hsa-miR-144, and hsa-miR-148b were decreased in AD samples [48].
In the reference study, from which the dataset of the current study is obtained, they selected 12 differentially expressed miRNA [46]. Four of these 12 miRNAs, specifically, miR-151a-3p, brain-miR-112, let-7f-5p, and hsa-miR-1285-5p, are found as differentially expressed miRNAs in the long list of our current study. Moreover, Satoh et al. [49] also analyzed the same miRNA dataset using omiRas web tool. They identified 27 differentially expressed miRNAs [49]. Seven of 11 miRNAs proposed by at least two algorithms in our study were also identified as differentially expressed miRNAs in the Satoh’s study. These common genes include has-let-7a-5p, has-let-7g-5p, has-miR-144-5p, has-miR-151a-3p, hsa-let-7f-5p, has-miR-148a-3p, and has-miR-148b-5p. In fact, all of the differentially expressed miRNAs, except one, in our short list, were also found significant by at least one of the references [46, 48,49,50,51]. The only exception is that our tool also detects hsa-miR-148a-3p.
3.7 A case study based on Alzheimer’s disease dataset
A case study is conducted to reveal the capacity of the web tool for suggesting DEGs. For this purpose, a dataset is simulated based on Alzheimer’s disease dataset from the negative binomial distribution using the ssizeRNA package [52] in R. The number of genes is taken as 503 and the number of observations is 70. Two hundred genes remain after near-zero variance filtering and Student’s t-test results. A response variable including treatments and controls is generated to obtain a binary outcome. The rate of the treatments is taken as 0.686 (48/70). As the distribution parameters, a mean vector and a dispersion vector are specified based on the Alzheimer’s disease dataset. That is, the mean vector is obtained as the arithmetic mean for each gene, taking into account the control group in the Alzheimer’s disease data. The dispersion parameter is taken 0.1 for each gene. Ten of the genes are simulated statistically significant between the groups. Our tool proposes 17 genes in long list, of which 10 of them are placed in short list. All genes in short list are the genes that are simulated to be statistically significant between two groups. That means all of the significant 10 genes are suggested by at least two methods. Thus, the tool suggests 100% of the DEGs with at least two methods and also 7 additional genes with a single method.
3.8 Implementation on KICH dataset
Kidney Chromophobe (KICH) dataset is used to demonstrate the validity of GeneSelectML web tool on a different dataset. This dataset is obtained via TCGAbiolinks R/Bioconductor package [53] and includes mRNAs from 66 tumor samples and 25 matched-normal samples. The number of genes is 19,947. Near-zero variance filtering, median ratio normalization and logarithmic transformation are applied to the data, respectively. Student’s t-test is performed as univariate analysis. The number of genes decreased to 200 after pre-processing. Of the remaining 200 genes, 10 genes are found to be differentially expressed by two or more algorithms in our GeneSelectML web tool and all of them are downregulated. Additionally, 14 genes are selected as DEGs by one method. Zhang et al. [54] analyzed the same dataset in their study and displayed the top 100 DEGs. Eight of 10 genes proposed by at least two algorithms in our study were also identified as DEGs in Zhang’s study. These common genes are RALYL, IRX1, UGT2A3, UGT3A1, UPK1B, DACH2, SLC9A3, and UNCX. One of the remaining two genes, UMOD gene, was found significant in the references [55,56,57]. MYH8 gene is the only exception our tool proposed in the short list.
4 Conclusion
Diagnosing DEGs is the crucial step to explore the reasons of diseases. Rather than univariate analysis, modelling the data considering the relationship among genes improves the prediction performance. However, there exist critical distinctions among studies analyzing the same dataset for the causes arising from a variety of methods.
In this study, the objective is to minimize these risks arising from the methods. GeneSelectML is a web-based platform which brings various gene selection algorithms together for RNA-seq data. All steps can be conducted using separate R packages, but the process might be distractive and time consuming for the inexperienced researchers in R programming language.
GeneSelectML is a user-friendly, comprehensive, and freely available tool for gene selection through machine learning algorithms that can deal with high performance computation. Currently, GeneSelectML tool involves six machine learning algorithms for gene selection. These are Biosigner, GMDH, OmicsMarkeR-GLMNET, OmicsMarkeR-SVM, OmicsMarkeR-RF, and DaMirseq algorithms. The tool also offers the users easy-to-use pre-processing steps; filtering, normalization, transformation, and univariate analysis. Moreover, there exists a user-friendly interface for graphical approaches; network plot, heatmap, venn diagram, and box-and-whisker plot. Also, gene ontology analysis is provided for the selected genes.
In this study, we construct aforementioned machine learning algorithms on GSE46579 dataset to explore the features for Alzheimer’s disease as well as to show the implementation of the tool. Eleven features are found to be differentially expressed by at least two methods. One of these features, hsa-miR-148a-3p, might be considered as a new biomarker for Alzheimer’s disease diagnosis. Of course, this finding needs clinical assessment and verification. Also, KICH dataset is used to demonstrate the validity of GeneSelectML web tool on a different dataset.
GeneSelectML will be periodically updated as the R packages are updated and the novel approaches are developed. This tool is freely available at www.softmed.hacettepe.edu.tr/GeneSelectML.
References
Wenric S, Shemirani R (2018) Using supervised learning methods for gene selection in rna-seq case-control studies. Front Genet 9:297
Kakati T, Bhattacharyya DK, Kalita JK (2019) Degnet: Identifying differentially expressed genes using deep neural network from rna-seq datasets. In: International conference on pattern recognition and machine intelligence. Springer, pp 130–138
Wang L, Xi Y, Sung S, Qiao H (2018) Rna-seq assistant: machine learning based methods to identify more transcriptional regulated genes. BMC Genomics 19(1):1–13
Yu Z, Wang Z, Yu X, Zhang Z (2020) Rna-seq-based breast cancer subtypes classification using machine learning approaches. Computational Intelligence and Neuroscience
Al-Obeidat F, Rocha Á, Akram M, Razzaq S, Maqbool F (2021) (cdrgi)-cancerdetection through relevant genes identification. Neural Computing and Applications, pp 1–8
Chang W, Cheng J, Allaire J, Xie Y, McPherson J et al (2017, version 1.0.1) shiny: web application framework for R, R Package. https://cran.r-project.org/web/packages/shiny/index.html. Accessed 15 July 2021
Guo W, Tzioutziou NA, Stephen G, Milne I, Calixto CP, Waugh R, Brown JW, Zhang R (2021) 3d rna-seq: a powerful and flexible tool for rapid and accurate differential expression and alternative splicing analysis of rna-seq data for biologists. RNA Biol 18(11):1574–1587
Su W, Sun J, Shimizu K, Kadota K (2019) Tcc-gui: a shiny-based application for differential expression analysis of rna-seq count data. BMC Res Notes 12(1):1–6
Weber C, Hirst MB, Ernest B, Baskir H, Tristan CA, Chu PH, Singeç I (2022) Sequin: rapid and reproducible analysis of rna-seq data in r/shiny. bioRxiv
Ge SX, Jung D, Yao R (2020) Shinygo: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36(8):2628–2629
Cestarelli V, Fiscon G, Felici G, Bertolazzi P, Weitschek E (2016) Camur: Knowledge extraction from rna-seq cancer data through equivalent classification rules. Bioinformatics 32(5):697–704
Clough E, Barrett T (2016) The gene expression omnibus database. In: Statistical genomics. Springer, pp 93–110
Nagaraj S, Zoltowska KM, Laskowska-Kaszub K, Wojda U (2019) Microrna diagnostic panel for Alzheimer’s disease and epigenetic trade-off between neurodegeneration and cancer. Ageing Res Rev 49:125–143
Kumar P, Dezso Z, MacKenzie C, Oestreicher J, Agoulnik S, Byrne M, Bernier F, Yanagimachi M, Aoshima K, Oda Y (2013) Circulating mirna biomarkers for Alzheimer’s disease. PLoS One 8(7):e69807
Riancho J, Vázquez-Higuera JL, Pozueta A, Lage C, Kazimierczak M, Bravo M, Calero M, González A, Rodríguez E, Lleó A, Sánchez-Juan P (2017) Microrna profile in patients with Alzheimer’s disease: Analysis of mir-9-5p and mir-598 in raw and exosome enriched cerebrospinal fluid samples. Journal of Alzheimer’s Disease 57(2):483–491
Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM (2007) Forecasting the global burden of Alzheimer’s disease. Alzheimer’s & Dementia 3(3):186–191
Gentleman R, Carey V, Huber W, Hahne F (2019, version 1.68.0) genefilter: genefilter: methods for filtering genes from high-throughput experiments, R Package. https://www.bioconductor.org/packages/release/bioc/html/genef ilter.html. Accessed 15 July 2021
Kuhn M (2020, version 6.0-86) caret: Classification and Regression Training, R Package. https://cran.r-project.org/web/packages/caret/index.html. Accessed 15 July 2021
Piao Y, Ryu KH (2017) Detection of differentially expressed genes using feature selection approach from rna-seq. In: 2017 IEEE international conference on big data and smart computing (BigComp). IEEE, pp 304–308
Abbas-Aghababazadeh F, Li Q, Fridley BL (2018) Comparison of normalization approaches for gene expression studies completed with high-throughput sequencing. PLoS One 13(10):e0206312
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Nature Precedings, pp 1–1
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Zyprych-Walczak J, Szabelska A, Handschuh L, Górczak K, Klamecka K, Figlerowicz M, Siatkowski I (2015) The impact of normalization methods on rna-seq data analysis. BioMed Research International 2015
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol) 57(1):289–300
Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Annals of Statistics pp 1165–1188
Rinaudo P, Boudah S, Junot C, Thevenot E (2016) Biosigner: a new method for the discovery of significant molecular signatures from omics data. Front Mol Biosci 3:26
Dag O, Karabulut E, Alpar R (2019) GMDH2: Binary classification via gmdh-type neural network algorithms - R package and web-based tool. International Journal of Computational Intelligence Systems 12(2):649–660
Determan C (2015) Optimal algorithm for metabolomics classification and feature selection varies by dataset. International Journal of Biology 7(1):100–115
Chiesa M, Colombo G, Piacentini L (2018) Damirseq-an r/bioconductor package for data mining of rna-seq data: normalization, feature selection and classification. Bioinformatics 34(8):1416–1418
Robnik-Sikonja M, Kononenko I (1997) An adaptation of relief for attribute estimation in regression. In: Machine Learning: Proceedings of the Fourteenth International Conference (ICML’97). Morgan Kaufmann Publishers Inc., pp 296–304
Toussi CA, Haddadnia J, Matta CF (2021) Drug design by machine-trained elastic networks: predicting ser/thr-protein kinase inhibitors’ activities. Mol Divers 25(2):899–909
Fushiki T (2011) Estimation of prediction error by using k-fold cross-validation. Stat Comput 21(2):137–146
McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor rna-seq experiments with respect to biological variation. Nucleic Acids Res 40(10):4288–4297. https://doi.org/10.1093/nar/gks042
Corporation M, Weston S (2020, version 1.0.16) doParallel: Foreach Parallel Adaptor for the ’parallel’ Package, R Package. https://cran.r-project.org/web/packages/doParallel/index.html. Accessed 15 July 2021
Haunsberger SJ, Connolly NMC, Prehn JHM (2016) miRNAmeConverter: an r/bioconductor package for translating mature mirna names to different mirbase versions. Bioinformatics. https://doi.org/10.1093/bioinformatics/btw660
Yu G, He QY (2016) Reactomepa: an r/bioconductor package for reactome pathway analysis and visualization. Mol BioSyst 12(2):477–479
Ru Y, Kechris KJ, Tabakoff B, Hoffman P, Radcliffe RA, Bowler R, Mahaffey S, Rossi S, Calin GA, Bemis L et al (2014) The multimir r package and database: integration of microrna-target interactions along with their disease and drug associations. Nucleic Acids Res 42(17):e133–e133
Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32(18):2847–2849
Csardi G, Nepusz T et al (2006) The igraph software package for complex network research. InterJournal, Complex Systems 1695(5):1–9
Dusa A (2021, version 1.10) venn: Draw Venn Diagrams, R Package. https://cran.r-project.org/web/packages/venn/index.html. Accessed 15 July 2021
R Core Team (2020) R: A language and environment for statistical computing. https://www.R-project.org/
Alexa A, Rahnenfuhrer J (2020, version 2.40.0) topGO: Enrichment analysis for gene ontology, R Package. https://bioconductor.org/packages/release/bioc/html/topGO.htm l. Accessed 15 July 2021
Pajak M, Simpson TI (2020, version 1.22.0) miRNAtap: miRNAtap: microRNA Targets - Aggregated Predictions, R Package. https://bioconductor.org/packages/release/bioc/html/miRNAtap. html. Accessed 15 July 2021
Pajak M, Simpson TI (2016, version 0.99.10) miRNAtap.db: Data for miRNAtap, R Package. https://bioconductor.org/packages/release/data/annotation/htm l/miRNAtap.db.html. Accessed 15 July 2021
Carlson M (2020, version 3.11.4) org.Hs.eg.db: Genome wide annotation for Human, R Package. https://bioconductor.org/packages/release/data/annotation/htm l/org.Hs.eg.db.html. Accessed 15 July 2021
Leidinger P, Backes C, Deutscher S, Schmitt K, Mueller SC, Frese K, Haas J, Ruprecht K, Paul F, Stähler C, Lang CJG, Meder B, Bartfai T, Meese E, Keller A (2013) A blood based 12-mirna signature of Alzheimer disease patients. Genome Biol 14(7):R78
Pan J, Jiang F, Zhou J, Wu D, Sheng Z, Li M (2018) Hsp90: A novel target gene of mirna-628-3p in a549 cells. Biomed Res Int
Pichler S, Gu W, Hartl D, Gasparoni G, Leidinger P, Keller A, Meese E, Mayhaus M, Hampel H, Riemenschneider M (2017) The mirnome of alzheimer’s disease: consistent downregulation of the mir-132/212 cluster. Neurobiol Aging 50:167.e1-167.e10
Satoh JI, Kino Y, Niida S (2015) Microrna-seq data analysis pipeline to identify blood biomarkers for Alzheimer’s disease from public data. Biomarker Insights 10:21–31
Keller A, Backes C, Haas J, Leidinger P, Maetzler W, Deuschle C, Berg D, Ruschil C, Galata V, Ruprecht K, Stähler C, Würstle M, Sickert D, Gogol M, Meder B, Meese E (2016) Validating Alzheimer’s disease micro rnas using next-generation sequencing. Alzheimer’s & Dementia 12(5):565–576
Li QS, Cai D (2021) Integrated mirna-seq and mrna-seq study to identify mirnas associated with Alzheimer’s disease using post-mortem brain tissue samples. Front Neurosci 15:260
Ran B, Peng L (2019, version 1.3.2) ssizeRNA: Sample size calculation for RNA-Seq experimental design, R Package. https://cran.r-project.org/web/packages/ssizeRNA/index.html. Accessed 15 July 2021
Mounir M, Lucchetta M, Silva TC, Olsen C, Bontempi G, Chen X, Noushmehr H, Colaprico A, Papaleo E (2019) New functionalities in the tcgabiolinks package for the study and integration of cancer data from gdc and gtex. PLoS Comput Biol 15(3):e1006701
Zhang W, Xu Y, Zhang J, Wu J (2020) Identification and analysis of novel biomarkers involved in chromophobe renal cell carcinoma by integrated bioinformatics analyses. BioMed research international
Liu H, Tang C, Yang Y (2021a) Identification of nephrogenic therapeutic biomarkers of wilms tumor using machine learning. Journal of Oncology 2021
Liu Y, Huang Q, Sun H, Chang Y (2021b) A causality-inspired feature selection method for cancer imbalanced high-dimensional data. bioRxiv
Grewal JK, Tessier-Cloutier B, Jones M, Gakkhar S, Ma Y, Moore R, Mungall AJ, Zhao Y, Taylor MD, Gelmon K et al (2019) Application of a neural network whole transcriptome-based pan-cancer method for diagnosis of primary and metastatic cancers. JAMA Netw Open 2(4):e192597–e192597
Acknowledgements
The authors are very grateful to four anonymous reviewers for their constructive comments and suggestions which helped to improve the quality of this paper.
Funding
This study is supported by Hacettepe University Scientific Research Projects Coordination Unit with project number THD-2020-18545.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dag, O., Kasikci, M., Ilk, O. et al. GeneSelectML: a comprehensive way of gene selection for RNA-Seq data via machine learning algorithms. Med Biol Eng Comput 61, 229–241 (2023). https://doi.org/10.1007/s11517-022-02695-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-022-02695-w