Abstract
Lung cancer is one of the most critical diseases due to its significant death rate compared to all other types of cancer. The early diagnosis of lung cancer that improves the patient’s chance of surviving is mostly done in two phases: screening through CT scan imaging modality and, more importantly the medical expert’s reading of the scan, which is a time-consuming task and is vulnerable to errors. It is difficult to differentiate between malignant and benign nodules and biopsies are highly invasive, and patients with benign nodules may undergo unnecessary procedures. In this study, we propose a CNN-based computer-aided diagnosis system to automatically classify pulmonary nodules into benign or malignant. The proposed network architecture is based on AlexNet architecture that experiments with several types of layer ordering, hyperparameters, and functions for the various sides of the network. To build a well-trained model, several pre-processing steps are applied to the entire dataset, for instance segmentation, normalization, and zero centering. Finally, the proposed system obtained results with 98.7% accuracy, 98.6% sensitivity, and 98.9% specificity. The proposed model achieved superior performance compared to the AlexNet. The modifications in the original AlexNet is done to get a reasonable structure that has high nodule analysis sensitivity.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Lung cancer is one of the riskiest diseases that modern humanity can develop. It is listed as the second most common type of cancer in both genders combined. In females, breast cancer is more common, while in males, prostate cancer is more common. The mortality rate of colon, breast, and prostate cancers combined is by far less than the death rate of lung cancer, making lung cancer the major cause of cancer death among both males and females [1]. Diagnosing lung cancer in its early stages enhances the treatment time and the chance of the patient being cured, with a 10-year survival rate of about 90% [2, 3].
Early-stage diagnosis is mostly done through screening a patient’s lungs. A CT scan is one of the most-used modalities for that purpose and helps radiologists to identify lung cancer in the beginning phases. The use of CT imaging devices decreases the death rate of the high-risk group by up to 20%, because CTs have been shown to be a beneficial source of information for radiologists [4].
It’s easy to overlook a malignant nodule due to the lungs’ vascular structure. The radiologist must have specific characteristics and skills to make the procedure effective, such as experience, degree of skill, and concentration. One possible solution to reduce errors is to use more than one reader, where each reader examines the scan independently and the results are combined. However, this is a costly solution and puts more workload on the scan readers. A cheaper and much more reliable approach would be designing a computer system that can be used as a second opinion against a radiologist’s opinion in diagnosing lung nodules [5]. Such systems can be extremely useful to medical professionals because they can obtain more accurate results than human expertise and in a shorter time. For that, the use of a computerized detection and classification system has become increasingly necessary for medical laboratories [6].
However, developing a dependable automatic lung nodule analysis system requires an algorithm that can deal with the complex structure of the lungs and different behaviors of lung nodules, such as texture diversity, shape, and identical features of cancerous and non-cancerous nodules. Recently, deep learning algorithms have achieved great success in the medical imaging domain, as they supply a hands-free feature extraction and classification architecture that offers better results than a hand-based feature extraction system. Convolutional neural networks (CNNs) have shown state-of-the-art results in image classification tasks, and many investigators have tried to employ CNNs in designing lung nodule analysis tools [7]. We proposed a fully automatic CNN-based CADx system to classify lung nodules as benign or malignant in CT scan images. However, in medical imaging, especially when predicting cancer, it is very important for the system to achieve the lowest error rate for both false positives (FP) and false negatives (FN), since that would pose a threat to the patient’s life. Therefore, this study focuses on reducing both the FP and FN rates to make the system more realistic.
2 Related work
In recent years, several researchers have worked on building systems for automatic medical image analysis utilizing a diverse number of methods, starting with low-level pixel processing, passing by traditional machine learning algorithms that work on hand-craft features. The next step is applying techniques that automatically learn low- to high-level features from the data and done the diagnosing procedure on the learned features; these techniques are known as deep learning (DL) techniques. The most successful DL algorithm when it comes to image classification is CNN [8]. Since that, most published works use different CNN architectures and concepts to design systems for lung cancer analysis.
De Pinho Pinheiro et al. [6] investigated training a convolutional neural network by using swarm intelligence instead of traditional training techniques like gradient descent and back-propagation to check the effectiveness of this strategy and their experiments showed that the top swarm trained network operated 25% faster than the model that trained based on back-propagation. Furthermore, the work benefited from LIDC-IDRI dataset to train their designed model that could obtain up to 93.71% accuracy, 93.53% precision, 92.96% sensitivity, and 98.52% specificity.
Da Silva et al. [3] proposed a convolutional neural network based on particle swarm optimization methodology to decrease false positives in lung nodule detection that uses CT scan images in the LIDC-IDRI database. However, the experiments showed that their methodology could achieve 97.62% accuracy, 92.20% sensitivity, 98.64% specificity, and a ROC curve of 0.955.
Naqi et al. [9] suggested a method for detecting and classifying pulmonary nodules in CT scans that consists of four main stages. The initial one is applying optimal gray-level thresholding to extract lung regions that are calculated by using fractional-order Darwinian particle swarm optimization, then proposing a state-of-the-art nodule detection model that performed based upon geometric fit in parametric shape including the geometrical characteristics of the nodules, the following step was designing a hybrid texture characteristic descriptor for representing candidate nodules that combine the 2D and 3D nodule information, and the last step was developing a deep learning classification model that builds upon stacked autoencoder and softmax which is applied for false-positive reduction. Moreover, the study utilizes a large public dataset which is LIDC/IDRI, and finally, their approach could decrease the rate of false positives into 2.8 per scan and a sensitivity of 95.6%.
G.S. et al. [10] work on a deep convolutional neural network model for classifying pulmonary nodule on CT scan images. They apply the focal loss to the training procedure in order to boost the accuracy of the classifier. The model trained on LUNA16 grand challenge dataset, and the experiments show the ability of focal loss that could achieve 97.2% accuracy, 96.0% sensitivity, and 97.3% specificity.
The suggested study by Xie et al. [11] is an attempt to design a model that can be trained on both labeled and unlabeled data to classify lung nodules into malignant-benign named a semi-supervised adversarial classification (SSAC). The model consists of three parts which are an adversarial autoencoder-based unsupervised reconstruction network R, supervised classifier C, and transition layers that learn to allow the adaption of the image representation capability learned by R to C. However, they aimed to use three SSACs to describe the overall look of the nodules, heterogeneity in shape, and texture, all that through expanding their model to the multi-view knowledge-based collaborative learning. The model was evaluated on the LIDC-IDRI dataset and could obtain 92.53% accuracy and 95.81% AUC.
3 Data preparation
The original model that was specially designed for this work was trained using a combination of two separate datasets, namely Data Science Bowl 2017 (DSB) [12] and the Lungx challenge dataset. The DSB dataset consists of 1595 low-dose CT scans provided by the National Cancer Institute (NCI). Each scan contains a set of 2D slices of the chest cavity that vary based on the patient and the modality. The images were in the DICOM file format and had a different slice thickness (Bowl 2017 Kaggle,” n.d.). The Lungx, or SPIE-AAPM-NCI Lung CT, challenge was conducted in 2015 and covered lung nodule classification into benign and malignant tasks. The challenge provided the participants with 70 thoracic CT scans, with a 1.0-mm slice thickness.
The scans contained 83 nodules (13 scans each consisting of 2 nodules and the remaining scans consisting of only one nodule) with 42 benign and 41 malignant [13].
To prepare the DSB dataset, 250 scans with 124 benign and 126 malignant nodules were chosen. Next, the annotations for these scans were separated and stored in a new CSV file, and then the preparation procedure continued using two operations.
The initial operation reads the annotations. Each scan was read according to its ID. Afterward, all scans went through two pre-processing steps, which were converting the pixel values to the Hounsfield unit (HU) and resampling, by applying a linear transformation of the normal units found in CT data (a typical dataset ranges from 0 to 4000), which multiplying each pixel value with the slope value and then adding result to intercept value. The Rescale Slope and Rescale Intercept are stored in DICOM header and getting a fixed value of 1 and − 1024, respectively. Then, segmentation and normalizing are applied to image data. Normalization is a procedure of reforming the scope of pixel values intensity. Thus, the pixel values are approximately between − 1000 and 1000, every pixel value over 400 are different intensity bones, so the images are regularized among − 1000 to 400 as it is a frequent used threshold for this type of images; afterward, the images normalized to values between 0 and 1.
Again the data has been normalized by applying zero centering which is subtracting each image pixel values from a mean (average) value. To find the mean value, it requires to average the pixel value of all images in the entire dataset that is a tricky work, but interestingly there is a commonly used value for a mean around 0.25, so all pixel values of every image are subtracted from this value. Figure 1 shows segmented slice, normalization, and zero centering.
Subsequently, since the lung CT scan was taken from the neck to the abdomen and most likely the lung nodule did not appear in a few of the first and last slices, to avoid these slices, the first and last 40 slices were excluded. Out of the remaining slices, 80 slices were randomly selected from each scan.
The segmentation algorithm applied to this study is a “marker-based watershed” method, and the major advantage of this technique is the internal and external markers that preserve the candidate nodules that fall at the corners; the method has been applied in two separate fractions.
The first fraction is generating an internal marker that identifies the lung tissue and an external marker that differentiates outside of the lung region. To do so, the internal marker has been picked out by thresholding the image with value − 400 HU and deleting other remaining parts, and the external marker is defined by morphological dilation of the other extracted marker along with two iterations and after that generating the final result, lastly, the watershed marker is made up by composing the two extracted markers that have dissimilar grayscale level, as shown in Fig. 2.
In the second fraction, after discovering the lung boundaries accurately by performing a watershed marker at the previous fraction, now the Sobel filter is needed and computed, so as to keep corner nodules and to guaranty that these regions and other neighboring areas are included, a “Black Top Hat” filter has been applied. The CT scan original size is well-kept by set − 2000 HU to the filtered locations, as shown in Fig. 3.
The later operation went through the slices that were chosen from every scan. Each of the slices was segmented, normalized, and zero centered. Later, to remove the useless black background as much as possible, each slice was scaled to 350 × 350. Subsequently, to enlarge the dataset, each slice was augmented using random noise, flip up-down, flip up-down with noise, flip left–right, and flip left–right with noise. The samples prepared from this dataset are 83 (scans) × 5(slices) × 6 (times augmentation) = 2490, as shown in Fig. 4.
4 Network architecture
To build our architecture, we reviewed one of the most impressive CNNs, AlexNet [14] to be the baseline for designing the approach of this study. AlexNet is a novel CNN architecture that could win the top 5 test error rate in ILSVRC-2012 challenge. AlexNet consists of 5 convolutional layers, 3 max-pooling, 2 fully connected layers, and the total parameters are more than 60 million. The major issue in AlexNet is the rapid down sampling of the intermediate representations through stride convolutions and max-pooling layers, causing overfitting issues. Medical datasets are commonly stored as scalar fields, with rapid down sampling data implying an inevitable loss of information that provokes the loss of fine details and the modification of the original image; this is not suitable for medical datasets, especially down sampling color data. In the proposed model, to preserve the data and obtain the result more precisely and accurately, we focused on an efficient deep neural network architecture with more simple down sampling.
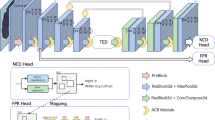
The proposed network architecture is a built-from-scratch CNN (with the benefits of AlexNet) that experiments with several types of layer ordering, hyperparameters, and functions for the various sides of the network. The final resulting model is made up of seven convolutional layers, three pooling layers of the type max-pooling, and only one dense layer. The network is divided into three blocks of layers, as shown in Fig. 5.
To begin with, the first and second blocks consist of three convolutions with one max pool. This is followed by the third block, which consists of one convolution with one max pool, respectively. A batch normalization and activation function layer of type ReLU follow up each of the seven convolution layers. Afterward, to reduce overfitting in the fully connected layers, there is a dropout layer that comes directly after the last pooling layer. The output of the dropout layer is fed into an FC (dense) layer. In this proposed architecture, only one fully connected layer is used to induce better representation learning in the convolutional layers and to create an efficient end-to-end learning algorithm.
The FC layer is followed by an activation function of softmax. Finally, the output layer predicts the output value. The convolution layers are constructed from a stride of 2 and padding of type SAME. The max pooling layer has a kernel size of 2 × 2, a stride of 2, and padding of type SAME, which are the numbers of learnable parameters for the proposed architecture. The total number of parameters is 2,396,674 as shown in Table 1.
5 Hyperparameter tuning and selection
To design an effective architecture that can produce the best outcomes, different hyperparameters and methods were experimented with according to the various aspects of the designed model, such as playing with several dropout rates, for example 0.6, 0.7, and 0.8; trying a variety of learning rates, for instance 0.0001, 0.0003, and 0.0005; and practicing diverse range of filter sizes, like 3 × 3, 5 × 5, and 7 × 7. Moreover, three of the well-known activation functions were also examined: ReLU and a few of its variants, including ELU and Leaky ReLU. Extraparameters were also experimented with, including applying a number of optimization methods, namely Adam, AdaGrad, and Adadelta.
To initialize the weights, a numerous initializer was tested, namely truncated normal and random normal, with different values for standard deviation such as 0.01, 0.03, 0.001, and 0.1. Glorot normal was also examined. Relatedly, the bias was initialized as a constant value, and again multiple values were tested, like 0, 0.1, and 1.
Other than the above-named changes, more and more modifications were made to the model to get to a better structure that can be relied on involving a number of dense layers and the number of nodes for the dense layers. Likewise, two mini-batch sizes of 30 and 44 were tested. Additionally, the network was trained to adopt 10, 12, and 15 epochs. Also, we employed the L2 regularization technique to penalize the weights to reduce over-fitting and the loss rate.
Eventually, the best-resulting model used a filter size of 7 × 7 for every convolution except the 1 × 1 convolution layers; a learning rate of 0.0001 with a dropout ratio of 0.6; and the weights were initialized utilizing truncated normal with the value 0.01 for standard deviation. To eliminate biases, a constant value of 0 was used. The main nonlinearity function was ReLU along with Adam as a primary optimization function. The dense layers, based on the outcomes, decided to be only one dense layer together with 256 nodes and a mini-batch size of 30 and 12 epochs of training.
6 Training procedure
As the function of the last (i.e., last dense layer) layer is to turn the coming inputs from the previous layer into outputs, we used the softmax activation function in this layer to interpret the inputs as output probabilities. Similarly, the loss rate was measured using a cross-entropy function, as it is the preferred technique for classification and working on the probabilities produced by softmax in the output layer.
To monitor the network performance, each epoch’s training accuracy and loss rate were printed to the screen. Subsequently, after every epoch of training, the network was validated using a specific validation set. Validation accuracy and loss rate were also determined. Finally, the model was tested utilizing the test set. The testing confusion matrix, accuracy, sensitivity, specificity, precision, and area under the curve (AUC) were measured for all different hyperparameters and methods that were experimented with.
All different experiments were executed on a laptop computer with an Intel core i7 CPU, 16 GB of memory, NVIDIA GeForce GTX 960 M -4 GB VRAM GPU, and Windows 10 operating system. The algorithm was performed using a GPU version of the Keras deep learning library. Each experiment took around 1 h to complete.
7 Discussion and results
To test the capability of the model directly after the training and before the program execution terminated, a particular portion of the dataset specifically separated for testing purposes was fed to the network. This set consisted of 2200 samples (1106 malignant and 1094 benign) from the entire dataset. The testing confusion matrix was computed and from that, the four components of binary classification confusion matrix were generated, which are TN, FP, FN, and TP. Consequently, accuracy, sensitivity, specificity, and precision were calculated. Table 2 presents a detailed description of a variety of our experimented results (including all metrics which are accuracy, sensitivity, specificity, precision, and AUC) that were achieved using a wide range of hyperparameters. The highest result is shown in bold and the lowest shown in italics. The best set of hyperparameters was able to obtain an accuracy of 98.7%, sensitivity of 98.6%, and specificity of 98.9%. The too-high value of accuracy, sensitivity, and specificity demonstrates the effectiveness of our model. Note that this result was chosen to be the top outcome of our CNN model because it could gain the highest score in all used metrics, except for the sensitivity in the one with similar hyperparameters, but a dropout of 0.7 could have a better false negative by 0.5%.
In addition to that, among every employed parameter noticed that the filter size has more impact on the results, and the larger one could be more powerful by achieving higher scores. While the other parameters and hyperparameters each had its own influence on the consequence, they were not as noteworthy as the filter size. Figure 6(a) and (b) visualize a ROC curve of the highest and lowest accuracy result. From that, we can realize remarkably how the curve rose and how efficient the tuning we added to the architecture of the model was. The last thing to clarify is that the two 1 × 1 filter convolution layers were added to the architecture as the final configuration that we made on the CNN method, which could increase the scores significantly. So, this configuration was chosen as our final architecture.
The evaluation is the process of measuring the ability of the model after saving and loading it again. To do so, we split 100 samples from the Lungx dataset, 50 benign and 50 malignant. Likewise, 390 samples were separated from the DSB dataset evenly for the two classes. In total, 490 samples were ready to be applied for the evaluation procedure.
Additionally, to do this operation, a saved model was loaded, and each sample of the evaluation data was fed to the model. The model predicts every sample to be benign or malignant. Based on the actual labels of every single image, we computed the percentage of recognized samples that will be the final result of the evaluation process. However, in a total of 490 supplied samples, the model can identify 484 images with only 6 misclassified ones. This means the model can recognize 98.77% of the fed data samples exactly as the achieved accuracy by the test set. In turn, this shows the great success of our CNN method.
So as to show the capability of the model, the architecture has been experimented with 30 different parameters; for each parameter, the architecture was trained 3 times, totalling 90 train pieces, the results of every train piece have been stored with all the used metrics, and then the average (mean) and the standard deviation (std dev) of each 3 trains that were experimented with the same parameters have been computed and shown as a final obtained results.
However, the comparison of the results found in this work with those obtained in related works and published in high-ranking journals is summarized in Table 3. Nevertheless, the comparison uses three of each work’s most recommended metrics. Frankly speaking, it is difficult to compare with the works that have been trained on identical datasets, since the studies either employed a DSB or Lungx dataset, and moreover, the Lungx dataset has only been used to train traditional machine learning models that could not achieve a good enough result to compare. To this end, we decided to make a differentiation within the mentioned works, as the whole idea is to be able to construct an architecture that precisely distinguishes benign nodules from malignant ones. Our method has the highest accuracy, sensitivity, and specificity among all the presented results, which indicates the huge success of the proposed classifier in determining cancer/no-cancer nodules in CT scan images.
The typical CADx system should demonstrate a well-balanced rate between the three dependable metrics: accuracy, sensitivity, and specificity. Therefore, a well-designed model must show its capability in recognizing both benign and malignant nodules with approximately equal ratios. Figure 7 shows changes in accuracy and loss rate per step of the highest and lowest result.
Finally, to make a comparison between the proposed model and the original AlexNet architecture, the prepared data were fed into the architecture to check its capability within our data; however, along with several training times, it could not achieve more than 50% of the accuracy, as shown in Table 4. In addition to that, in one training time, it could recognize 100% TN with 0% TN, while in another time the obtained result was completely the opposite of that, which is a dangerous case in medical diagnosis. Nonetheless, this comparison shows a major improvement over the AlexNet for the task. After training and validation are completed, in order to measure the obtained results on a test set, a confusion matrix is generated, shown in Fig. 8. From the matrix, the concept of true negative (TN, the situation that is actually negative and predicted as negative), false positive (FP, the situation that is incorrectly classified as positive), false negative (FN, the situation that is incorrectly classified as negative), and true positive (TP, the situation that is actually positive and predicted as positive) can be calculated respectively.
8 Conclusion
Lung cancer is one of the most dangerous diseases in today’s human life; it is recorded as the deadliest type of cancer in both men and women together, so, detecting and diagnosing lung cancer in its early stage is considered the key that opens the door of treatment and survival time enhancement. One of the most effective and advanced approaches is to use a computer system for detecting and analyzing a lung nodule as it is more accurate than human and requires much less time. In this study, we proposed a system for diagnosing lung nodules into benign or malignant by using the most effective classification algorithm, which is CNN. The system was built on top of AlexNet, one of the most cited and successful CNNs. The modification in the original AlexNet architecture is done in order to get a reasonable structure that has high nodule analysis sensitivity. The changes include two different sides of the original architecture. The initial change was modifying the layered architecture, for instance, removing one of the dense layers and adding two 1 × 1 convolution layers. The later change was altering the parameters and hyperparameters of the architecture; this causes reduced training time from several days to several hours. The dataset utilized to train our model was a mixture of 2490 samples from the Lungx dataset and 20,000 samples from the DSB dataset. The first dataset is a small well-annotated dataset that was used to improve the quality of the data, while the second one was large and used to enlarge the data size. Additionally, the dataset required many preprocessing and preparation stages which makes the data samples more adequate to train a CNN. Also, the distinct elements of the base network were worked on, for instance, layer ordering, filter size, dropout rate, weight and bias initialization methods and values, learning rate, and optimization methods to build an appropriate architecture for the task and toward a novel outcome. Among the whole set of the modified elements, the filter size has the biggest impact on the obtained consequences.
Furthermore, the developed CADx for this study is capable of gaining top scores, with an accuracy of 98.7%, sensitivity of 98.6%, and specificity of 98.9%, which are, according to our knowledge and search for scores from other developed methods in the same domain, the best state-of-the-art result.
References
American Cancer Society (2018) Key statistics for lung cancer. Am Cancer Soc. https://www.cancer.org/cancer/lungcancer/about/key-statistics.html. Accessed Feb 2019
Lakshmanaprabu SK, Mohanty SN, Shankar K, Arunkumar N, Ramirez G (2019) Optimal deep learning model for classification of lung cancer on CT images. Futur Gener Comput Syst 92:374–382. https://doi.org/10.1016/j.future.2018.10.009
da Silva GLF, Valente TLA, Silva AC, de Paiva AC, Gattass M (2018) Convolutional neural network-based PSO for lung nodule false positive reduction on CT images. Comput Methods Programs Biomed 162:109–118. https://doi.org/10.1016/j.cmpb.2018.05.006
da Nóbrega RVM, Rebouças Filho PP, Rodrigues MB, da Silva SPP, Dourado Júnior CMJM, de Albuquerque VHC (2018) Lung nodule malignancy classification in chest computed tomography images using transfer learning and convolutional neural networks. Neural Comput. https://springerlink.bibliotecabuap.elogim.com/article/. https://doi.org/10.1007/s00521-018-3895-1. Accessed Feb 2019
Winkels M, Cohen TS (2019) Pulmonary nodule detection in CT scans with equivariant CNNs. Med Image Anal 55:15–26. https://doi.org/10.1016/j.media.2019.03.010
de Pinho Pinheiro CA, Nedjah N, de MacedoMourelle L (2019) Detection and classification of pulmonary nodules using deep learning and swarm intelligence. Multimed Tools Appl. https://doi.org/10.1007/s11042-019-7473-z
Xie Y, Xia Y, Zhang J, Song Y, Feng D, Fulham M, Cai W (2018) Knowledge-based collaborative deep learning for benign-malignant lung nodule classification on chest CT. IEEE Trans Med Imaging 1–1. https://pubmed.ncbi.nlm.nih.gov/30334786/. Accessed Feb 2019
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88. https://doi.org/10.1016/j.media.2017.07.005
Naqi SM, Sharif M, Jaffar A (2018) Lung nodule detection and classification based on geometric fit in parametric form and deep learning. Neural Comput Appl. https://springerlink.bibliotecabuap.elogim.com/article/. https://doi.org/10.1007/s00521-018-3773-x . Accessed Feb 2019
Tran GS, Nghiem TP, Nguyen VT, Luong CM, Burie JC, Levin-Schwartz Y, Tran Thi Phuong AO, Nguyen AO, Thi V, Chi Mai L, Burie AO, Jean-Christophe AO-N (2019) Improving accuracy of lung nodule classification using deep learning with focal loss. J Healthc Eng. https://doi.org/10.1155/2019/5156416
Xie Y, Zhang J, Xia Y (2019) Semi-supervised adversarial model for benign-malignant lung nodule classification on chest CT. Med Image Anal. https://doi.org/10.1016/j.media.2019.07.004
Data Science Bowl | Kaggle (2017) https://www.kaggle.com/c/data-science-bowl-2019. Accessed Feb 2019
Armato SG, Drukker K, Li F, Hadjiiski L, Tourassi GD, Engelmann RM, Giger ML, Redmond G, Farahani K, Kirby JS, Clarke LP (2016) LUNGx challenge for computerized lung nodule classification. J Med Imaging 3:044506. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5166709/. Accessed Feb 2019
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 1097–1105. https://doi.org/10.1145/3065386
Acknowledgements
The authors gratefully acknowledge the financial support for this study from the Ministry of Higher Education and Scientific Research-Kurdistan Regional Government, Department of Computer, College of Science, University of Sulaimani, Sulaimani, Iraq, and we wish to give our full thanks and gratitude to the Kaggle Team and SPIE—with the support of the American Association of Physicists in Medicine (AAPM) and the National Cancer Institute (NCI), who prepared this useful dataset with its annotations and gave everyone the accessibility to download it.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mahmood, S.A., Ahmed, H.A. An improved CNN-based architecture for automatic lung nodule classification. Med Biol Eng Comput 60, 1977–1986 (2022). https://doi.org/10.1007/s11517-022-02578-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-022-02578-0