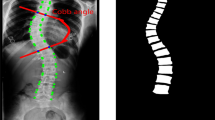
Abstract
The accuracy of the Cobb measurement is essential for the diagnosis and treatment of scoliosis. Manual measurement is however influenced by the observer variability hence affecting progression evaluation. In this paper, we propose a fully automatic Cobb measurement method to address the accuracy issue of manual measurement. We improve the U-shaped network based on the multi-scale feature fusion to segment each vertebra. To enable multi-scale feature extraction, the convolution kernel of the U-shaped network is substituted by the Inception Block. To solve the problem of gradient disappearance caused by the widening of the network structure from the Inception Block, we propose using Res Block. CBAM (Convolutional Block Attention Module) can help the network judges the importance of the feature map to modify learning weight. Also, to further enhance the accuracy of feature extraction, we add the CBAM to the U-shaped network bottleneck. Finally, based on the segmented vertebrae, the efficient automatic Cobb angle measurement method is proposed to estimate the Cobb angle. In the experiments, 75 spinal X-ray images are tested. We compare the proposed U-Shaped network with the state-of-the-art methods including DeepLabV3 + , FCN8S, SegNet, U-Net, U-Net + + , BASNet, and U2Net for vertebra segmentation. Our results show that compared to these methods, the Dice coefficient is improved by 32.03%, 33.58%, 12.42%, 5.65%, 4.55%, 4.42%, and 3.27%, respectively. The CMAE of the calculated Cobb measurement is 2.45°, which is lower than the average error of 5–7° of manual measurement. The experimental results indicate that the improved U-shaped network improves the accuracy of vertebra segmentation. The proposed efficient automatic Cobb measurement method can be used in clinics to reduce observer variability.
Graphical Abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Adolescent scoliosis (AS) is a deformity of the spine that is developed at puberty and before skeletal maturity [1, 2]. Studies show that 2–4% of adolescents suffer from AS. If it is left untreated, progression of the spinal deformity may affect lung and heart functions. It can also compress the spinal cord resulting in paraplegia [3]. The most cost-effective imaging modality for diagnosing scoliosis is X-ray. The Cobb angle measurement [4] is proposed to assess the degree of AS precisely. The Cobb angle measured on the posterior-anterior (PA) X-ray image of the spine is the most common method for assessing AS.
Figure 1 illustrates the manual measurement of the Cobb angle. The accuracy and repeatability of manual measurements however largely depend on the operator’s experience and judgment [5]. Evidence shows that the intra-observer and inter-observer errors are 3 to 5° and 5 to 7°, respectively [6]. Such an error margin however is beyond the 5° threshold for progression assessment. To reduce the manual measurement errors, automatic measurement methods [7] have been proposed, which generally fall into one of the following categories including segmentation-based and direct estimation methods.
In the direct estimation methods, the Cobb angle is obtained using the relationship between PA spinal X-ray images and clinical measurement methods without segmentation results. Finding the landmark corresponding to the vertebra is then used to obtain the deflection angle and calculate the Cobb angle. For instance, Wu et al. proposed the BoostNet [8] to accurately extract the spinal landmarks for Cobb angle measurement. Based on the BoostNet, Wu et al. proposed the MVC-Net [9] to associate the PA and the lateral spinal X-ray images. This method incorporates the global features of the spinal X-ray images to further improve measurement accuracy. Based on the MVC-Net, Wang et al. proposed the MVE-Net [10], this method learns to directly estimate the Cobb angle based on PA and lateral angles. Furthermore, Fu et al. [11] proposed LCE-Net to estimate the Cobb angle. Their proposed method is a multitasking network that combines segmentation and landmark information. The main issue with the direct estimation methods is their low measurement accuracy and their rather high dependency on the corresponding clinical measurement.
Alternative to direct estimation methods, segmentation-based techniques have been shown to address the above issues. Compared with direct estimation methods, segmentation-based methods reduce the measurement error by extracting the outline of the vertebrae accurately. The measurement accuracy of segmentation-based methods is higher than the direct estimation methods. For instance, Zhang et al. [12, 13] proposed an automatic measurement method based on the Hough transform. Also, Sardjono et al. [14] proposed a physics-based model to calculate the Cobb angle. Anitha et al. [15] further proposed an approach based on the custom filter to automatically extract the vertebral endplate. Due to the low quality of the spine X-ray image and the fuzzy contour of the vertebral body, the segmentation accuracy of these methods is low, and the measurement error of the Cobb angle is large. Therefore, the main disadvantages of segmentation-based methods are the existence of larger errors and low segmentation accuracy.
The recent development of deep neural networks resulted in the significant progress of image segmentation methods. For example, Long et al. proposed a fully convolutional network (FCN) [16] to obtain highly accurate pixel-level segmentation results. However, the FCN does not consider helpful global context information. This issue has been addressed using the U-Net [17] using the encoding and decoding structures. Based on the U-net, Zhou et al. proposed U-Net + + [18, 19] to redesign skip connections to aggregate features of varying semantic scales at the decoder sub-networks. Qin et al. proposed BASNet [20] to segment the salient object regions. Furthermore, Qin et al. proposed U2Net [21] to capture more contextual information from different scales thanks to the mixture of receptive fields of different sizes in residual U-blocks. However, the number of model parameters for these methods is too large.
Deep learning methods have been also applied to spine segmentation and Cobb angle estimation. For example, Fang et al. [22] proposed an approach based on the FCN to segment the computed tomography (CT) images of the spine and reconstruct the corresponding three-dimensional module. However, the accuracy of the segmentation in their proposed method is low. Further, Horng et al. [23] developed a method that automatically measures the Cobb angle. Nevertheless, their proposed segmentation processing needs to detect the vertebrae and crop the spine X-ray images into individual vertebral images. The Cobb angle measurement is then obtained by reconstructing the segmentation results of the vertebrae. Tan et al. [24] used U-Net to complete the segmentation of spine X-ray images. Also, [25] reviews the AASCE2019 challenge (i.e., an accurate automated spinal curvature estimation challenge) with spinal anterior–posterior X-Ray images. In this challenge, team XMU segmented the boundary of the spine and used a convolutional neural network to regress angles. Team Tencent regarded the vertebrae and intervertebral space segmentation as an intermediate state and ensembles multiple networks to produce angles. These methods achieved the measurement of the Cobb angle. For the automatic measurement of Cobb angle, the higher accuracy of vertebral segmentation reduces the measurement error.
In this paper, we propose a novel U-shaped network for spine X-ray image segmentation and develop a novel automatic measurement method of the Cobb angle. Our experimental results confirm that the improved segmentation model is efficient and overperforms other improved U-net models.
In summary, we make the following two contributions:
-
We propose a U-shaped segmentation network, which is a semantic segmentation neural network that takes advantage of the Inception Block, Res Block, and CBAM Block to extract multi-scale features, alleviate the gradient explosion and gradient disappearance, and improve segmentation performance.
-
Based on the minimum enclosing rectangle measurement algorithm, we further develop a novel efficient Cobb angle automatic measurement method that finds the upper-end vertebrae and the lower-end vertebrae to achieve highly accurate Cobb angle automatic measurement for spine X-ray images.
2 Materials and methods
The traditional method of manually measuring a Cobb angle has the characteristics of low efficiency and large error. The computer-aided method of measuring the Cobb angle has high efficiency and high accuracy. It can be a good substitute for the manual method of measuring the Cobb angle. There exist some noise and blur in spine X-ray images. To remove the noise and blur of the spine X-ray image and improve the measurement accuracy of the Cobb angle, we adopt an image segmentation method. Traditional image segmentation methods include threshold-based segmentation methods, region-based segmentation methods, edge-based segmentation methods, etc. These methods however have some disadvantages, e.g., low segmentation accuracy and existing segmentation fault. As convolutional neural networks used for feature learning are insensitive to image noise, blur, contrast, etc. [26], they provide excellent segmentation results for medical images. At present, image segmentation methods based on convolutional neural networks have been widely applied in medical image processing.
In this section, firstly, we introduce the framework of the segmentation network and its components. Then, we introduce an efficient automatic Cobb angle measurement method. We then present the implementation details of our experiments.
2.1 Overall architecture
An overview of the proposed automatic Cobb measurement is presented in Fig. 2, which illustrates the processing of the measurement of Cobb angle based on the segmentation of the spine X-ray images. Firstly, the spine X-ray images are input to the proposed U-shaped segmentation network for training and testing of the segmentation model. The segmentation results are input to the efficient automatic Cobb angle measurement method to obtain the Cobb measurement data and present a visualization of measurement results.
2.2 The structure of the proposed segmentation method
The structure of our proposed segmentation method is shown in Fig. 3. The size of the input image is 256 × 256, and there is only one channel of the training images. The image passes the Inception Block and the multi-scale feature maps are concatenated to a larger feature map. Feature maps are fused after every skip connection. They are then restored to the original image size by the upsampling layer. The Res Block is concatenated to the inception block to reduce the gradient explosion or gradient disappearance problems. The CBAM Block also improves the performance of the network to extract the features.
2.3 The Inception Block
For a convolutional neural network, the performance of the deep networks is often higher than that of the shallow networks. Nevertheless, increasing the depth of the network may result in issues such as gradient explosion and gradient disappearance. The inception network is a method to extract richer image features using multi-scale convolution kernels and to perform feature fusion. This enables obtaining better feature representation [27]. It also provides higher performance by merging convolution kernels in parallel without increasing the depth of networks. Based on the Inception network, 3 × 3, 5 × 5, and 7 × 7 convolution kernels are used for feature extraction at different scales. However, the 5 × 5 and 7 × 7 convolution kernels are highly complex [28]. To reduce the computational complexity and number of network parameters, two and three 3 × 3 convolution kernels are concatenated to substitute the 5 × 5 and 7 × 7 convolution kernels, respectively. We further use the shortcut of the 1 × 1 convolution kernel to extract the spatial information of the image. This module is called the Inception Block.
Figure 3b shows the structure of the Inception Block. The parallel connection of 3 × 3 convolution kernels is used to replace the 3 × 3, 5 × 5, and 7 × 7 convolution kernels. After this module, the feature map enters the Relu activation and max-pooling layers. Relu activation is proposed to solve the gradient disappearance, while the max-pooling layers compress the feature map.
2.4 Res Block
The Inception Block is proposed to enlarge the depth and width of the convolutional neural network. Increasing the depth and width of the network may result in gradient explosion or gradient disappearance problems during the training process. In ResNet [29], the 1 × 1 shortcut is used to solve the question of gradient explosion or gradient disappearance. With the image passing the inception block, the feature maps of the three convolution kernels of different scales are concatenated, and the size of the feature map is changed. To enable the feature to be fused with the features obtained by upsampling layer, we further added Res Block to adjust the size of the feature map and solve the gradient explosion and gradient disappearance problems. The Res Block is illustrated in Fig. 3c, consisting of 3 × 3 convolution kernels and a 1 × 1 shortcut. The ReLU activation layer and batch normalization are also used to improve the convergence rate and alleviate the gradient disappearance.
2.5 The CBAM Block
The SENet [30] is the first proposed attention module that can automatically learn the importance of channel features to increase the weight of significant features. This step also decreases the weight of useless features during the learning processing of the network. The CBAM Block [31] includes spatial attention and channel attention modules. Considering the importance of the pixels in different channels and their positions in a given channel, we use the CBAM Block to integrate the spatial attention module and channel attention module to improve the segmentation accuracy. Firstly, considering the importance of the spatial and channel based on the SENet, we use the CBAM Block to extract the features from the feature map. Secondly, spatial and channel information is multiplied and fused to the feature map. This step can help the network to learn spatial and channel information. The structure of the CBAM Block is shown in Fig. 3d.
2.6 The loss function
Our objective here is to extract the vertebra in the spine X-ray images. This is a binary segmentation task; hence, we use the binary cross-entropy as the loss function. For an image \(p\), the ground truth of the image is denoted by \(q\), and \(y\) is the ground truth segmentation mask. Also, \(\widehat{y}\) is the predicted value. The binary cross-entropy is defined as
The binary cross-entropy is mainly used for binary classification tasks, and this experiment is mainly to segment the spine. The label has only two types of background and spine, so the binary cross-entropy is used as the loss function.
2.7 Performance evaluation metric
The dice coefficient, precision, and recall are used as the performance evaluation metrics of the experiments. The dice coefficient is used to compare the similarity of the segmentation results and labels and is defined as
where TP, TN, FP, and FN are the set of true positive, true negative, false positive, and false negative. These parameters are used to evaluate the reliability and degree of accuracy. The dice coefficient is used to compare the similarity of the segmentation results and labels.
For Cobb angle estimation, we adopt circular MAE (CMAE) and symmetric mean absolute error (SMAPE) to evaluate the relative error. Given a list of N angles \(\left[{\alpha }_{0},{\alpha }_{1},\dots ,{\alpha }_{N}\right]\), the circular mean is defined as
The circular MAE is defined as
The SMAPE metric is defined as
where Ma is the automatic measurement value of Cobb angle and Lm is the manual measurement value of Cobb angle. SUM is the element-wise summation of a vector.
The Euclidean distance is defined as in Eq. 10:
where \(({\alpha }_{i1}, {\alpha }_{i2},{\alpha }_{i3})\) is the estimated Cobb angles, (\({\beta }_{i1}, {\beta }_{i2},{\beta }_{i3})\) is the ground truth, and N is the number of images.
The Manhattan distance is defined as in Eq. 11:
where \(({\alpha }_{i1}, {\alpha }_{i2},{\alpha }_{i3})\) is the estimated Cobb angles, \({(\beta }_{i1}, {\beta }_{i2},{\beta }_{i3})\) is the ground truth, and N is the number of images.
The Chebyshev distance is defined as in Eq. 12:
where \(({\alpha }_{i1}, {\alpha }_{i2},{\alpha }_{i3})\) is the estimated Cobb angles, \({(\beta }_{i1}, {\beta }_{i2},{\beta }_{i3})\) is the ground truth, and N is the number of images.
2.8 Efficient automatic Cobb angle measurement method
The efficient automatic Cobb angle measurement method is proposed to calculate the Cobb angle in AS patients. It is developed from the “minAreaRect()” function in the python OpenCV package. The “minAreaRect()” function obtains the rotated rectangle that encloses the smallest area of the input 2D point set. Figure 4 is shown the processing of the minimum enclosing rectangle algorithm. This processing then extracts four vertex coordinates and deflection angle to the horizontal plane of the minimum enclosing rectangle.
Based on this function, we design an efficient automatic Cobb measurement method. The processing of this method is illustrated in Fig. 5. Our efficient automatic Cobb angle measurement is divided into four following steps:
-
1.
Traversing the contour information of each segmented vertebrae and recording the contour information.
-
2.
Automatic creation of the minimum enclosing rectangle and recording the coordinate of the minimum enclosing rectangle.
-
3.
Extract the deflection corner between the minimum enclosing rectangle and the horizontal plane.
-
4.
Traversing all of the twisted corners, finding the upper-end vertebrae and the lower-end vertebrae.
2.9 Dataset
The dataset contains 185 PA spine X-ray images. The images from the dataset included the spine of the normal and the spine of idiopathic scoliosis patients. The PA spine X-ray images of the dataset are provided by the orthopedic surgeon at the First People’s Hospital of Yunnan Province. All images are taken using the equipment manufactured by the same manufacturer.
Additional dataset: The testing dataset of AASCE2019 [25] contains 98 PA spine X-ray images. This testing dataset is used to verify the robustness and generalization of our proposed method.
2.10 Image preprocessing
The ground truth of the images is approved by the orthopedic surgeon at the First People’s Hospital of Yunnan Province. Before the annotation of the dataset, the image is preprocessed and resized to 256 × 256 and a bit depth of 8. The label of the vertebrae is also transformed into the binarization image.
2.11 Implementation details
The software package used in the experiment is implemented using TensorFlow 1.12 and Keras. An RTX 2080 GPU with 8 GB of memory is used in the experimental hardware configuration. We train the model using Adaptive Moment Estimation (Adam) with batch size 11 and a learning rate of 0.0001. The dataset is contained 185 images where 110 images are used as the training set, and the remaining 75 images are used as the testing set. We have enhanced the training data by data enhancement setting in Keras. The data of the training set is enhanced by flipping, cropping, and zooming the images. The rotation range and the cropping range are also 0.2 and 0.05, respectively. The flipping model is horizontal. For measuring the performance of the trained model, the Dice coefficient is used as the metric.
3 Results
3.1 Effectiveness of the CBAM Block
In this section, we examine the effectiveness of the CBAM Block. The different training strategies are compared in our experiments. SE Block or CBAM Block is added in various locations in the network to verify the effectiveness of the CBAM Block. The Dice coefficient % is used to evaluate the effectiveness of the CBAM Block.
Table 1 shows the compared segmentation results using different training strategies. As it is seen in the case where the CBAM Block is added to the bottleneck, the highest performance is achieved compared with other training strategies. The location of the CBAM Block is the bottleneck of the network that can reach a value of (81.46 ± 0.41)% for Dice coefficient %. Compared with the model without the CBAM Block or SE Block, the Dice coefficient is improved by 3.39%. Regardless of the number of added SE Block to different locations in the network, the segmentation remains inferior to adding the CBAM Block. The location of the SE Block is the bottleneck of the network that can reach a value of 78.07% for a Dice coefficient %. As it is seen, compared with the model with the added CBAM Block, the Dice coefficient is lower. The experimental results confirm that the CBAM Block can efficiently supervise the network to extract the underlying features of the image.
3.2 Performance evaluation of the proposed method
To confirm the effectiveness of the network, we evaluate the performance of different blocks in the network. The results are shown in Table 2. Inception block, Res Block, and CBAM Block are added to the network, respectively. The Dice coefficient is used to evaluate the effectiveness of these blocks. Different training parameters are set to confirm the effectiveness of the various blocks. The model of experiment 1 is the original U-Net. Convolutional kernels are replaced by the Inception Block in experiment 2, experiment 3, and experiment 4. The difference between experiment 2 and experiment 3 is whether to use the CBAM Block. The similarity between experiment 2 and experiment 3 is that one layer of the Res Block is added to the network. We use the same comparison strategy as in experiment 2 and experiment 3 between experiment 4 and our proposed method. A four-layer Res Block is added to the network. The experiments also verify that adding four Res Block and CBAM Block is effective. Compared with the original U-Net, the Dice coefficient of our method is also improved by 5.65%.
The performance of the segmentation result is shown in Fig. 6. The difference between various models is marked on the images using red boxes. From Fig. 6, it is seen that the segmentation result of the proposed model has less adhesion and lower segmentation error. The segmentation result is close to the ground truth of the testing set. It is seen that compared with the original network, the segmentation result is significantly improved.
3.3 Comparison with other methods
Here, we apply other methods to our developed dataset and compare the results with the proposed method in this paper. The dice coefficient results presented in Table 3 confirm that our proposed method is higher than that of the other methods. It is also seen in Table 3 that compared with DeepLabV3 + , FCN8s, SegNet, U-Net, U-Net + + , BASNet, and U2Net using the proposed U-Shaped network, the Dice coefficient is improved by 32.03%, 33.58%, 12.42%, 5.65%, 4.55%, 6.22%, and 3.04%, respectively. The number of the model parameter is an essential factor affecting the cost of that model. Compared with other models, the number of parameters of our proposed model is an order of magnitude smaller.
The segmentation results using different methods are shown in Fig. 7. It is seen that the performance of our model is higher than that of the other models.
3.4 Measuring results of the Cobb angle
To evaluate the Cobb measurement, we compare the Cobb measurement performed by the orthopedist and that obtained by automatic computer measurement. The segmentation results of 75 test images from the test set are used to calculate the measurement results. We assess the stability of the Cobb angle calculation result by calculating the CMAE between the manual and automatic measurements. Two orthopedists performed manual Cobb angle measurements on the test set.
As shown in Table 4, our method achieves CMAE of 2.45° and 2.50° from different orthopedists. We assess the stability of the Cobb angle calculation result by calculating the CMAE and SMAPE between the manual and automatic measurements. The results in Table 4 indicate that the efficient automatic Cobb angle measurement method is stable.
To verify the effectiveness of the efficient automatic Cobb angle measurement method, we compare the CMAE and SMAPE between our proposed method and other recently proposed methods in Table 5. The Tencent and XMU experiment data is derived from the AASCE2019 challenge [25]. They are the top 2 in this challenge. The test set of this challenge is used to estimate the Cobb angle. Compared with the method from Tencent and XMU, the CMAE of our method is reduced, and the SMAPE of our method is lower. The experiment results are illustrated that the proposed efficient automatic Cobb angle measurement method avoids the effects of noise and blur in spine X-ray images.
4 Discussion
In this paper, the U-shaped network is proposed to extract the outline of the vertebra of the spine PA’s X-ray images. After the segmentation, the efficient automatic measurement algorithm of the Cobb angle is used to obtain the minimum enclosing rectangle and calculate the deflection angle of the vertebra.
Figure 8 shows a boxplot of the average metrics values for the spine X-ray image compared with the six different segmentation algorithms. It illustrates the dispersion of a set of data. The green dotted line is the mean value of the dice coefficient, and the orange line is the median of the dice coefficient in the boxplot. The figure indicates that the performance of our model is higher than other models. It illustrates that our proposed segmentation network is more stable compared with other existing models.
To verify the effectiveness of the Cobb angle measurement using our proposed method, we compare the Cobb measurement result based on different segmentation methods in Table 6. The measurement results are obtained by the proposed efficient automatic Cobb angle measurement method. From Table 6, we can find that the measurement result of our proposed method has a lower CMAE and SMPE compared with U-Net and U-Net + + . From Table 3, the Dice coefficient of our proposed method is higher than U-Net and U-Net + + . It illustrates that our proposed model can reduce the measurement error of the Cobb angle compared to U-Net and U-net + + . And it also illustrates that the measurement error decreases with the increase of the segmentation accuracy.
For our proposed efficient automatic Cobb angle measurement method, the segmentation accuracy of vertebrae is significant. Figure 9 is the visualization of the efficient automatic Cobb angle measurement method with terrible and fine segmentation results. The yellow line is parallel to the centerline of the lower-end vertebrae. The green line is the parallel of the centerline of the upper-end vertebrae.
From Fig. 9a, the efficient automatic measurement method can extract the minimum enclosing rectangle of every vertebra. The wrong segmentation results cause errors in the Cobb angle calculation. This mistake affects the measurement results due to the large deflection angle of the wrong bodies. The good performance of our proposed method is shown in Fig. 9b, which illustrates that a good segmentation result is significant for Cobb angle measurement. The visualization also confirms high accuracy segmentation of vertebrae in our proposed automatic measurement method.
5 Conclusion
Based on vertebra segmentation, an automatic approach for the measurement of the Cobb angle is proposed. Our model successfully acquires a high precision segmentation in the PA X-ray images. Compared with other segmentation methods, our model has better performance. Based on the segmentation results, the deflection angles of the vertebrae are obtained to calculate the Cobb angle. The CMAE of measurement results is 2.45°. The results presented in this paper confirm that the measurement results of our model are highly reliable.
Soon, we will explore applying transformer layers to our model and use transfer learning approaches to improve segmentation accuracy. Future studies will also explore applying our methods to estimate other clinical parameters based on spinal curvature. In addition, to further strengthen the robustness of the model and reduce measurement errors, experimental data from different devices and different manufacturers will be added to the training set in the future.
References
Hefti F et al (2013) Pathogenesis and biomechanics of adolescent idiopathic scoliosis (AIS). J Children’s Orthop 7(1):17–24
Little JP, Izatt MT, Labrom RD et al (2013) An FE investigation simulating intra-operative corrective forces applied to correct scoliosis deformity. Scoliosis 8(1):9
Cobb JR (1947) Outline for the study of scoliosis. Instruct Course Lect 5
Asher MA, Burton DC (2006) Adolescent idiopathic scoliosis: natural history and long term treatment effects. Scoliosis 1(1):2–2
Weinstein SL, Dolan LA, Cheng JCY et al (2008) Adolescent idiopathic scoliosis. Lancet 371(9623):1527–1537
Vrtovec T, Pernu F, Likar B (2009) A review of methods for quantitative evaluation of spinal curvature. Eur Spine J 18(5):593–607
Pruijs JEH, Hageman MAPE, Keessen W et al (1994) Variation in Cobb angle measurements in scoliosis. Skelet Radiol 23(7):517–520
Wu H et al (2017) Automatic landmark estimation for adolescent idiopathic scoliosis assessment using BoostNet. International Conference on Medical Image Computing and Computer-Assisted Intervention Springer, Cham
Wu H, Bailey C, Rasoulinejad P et al (2018) Automated comprehensive adolescent idiopathic scoliosis assessment using MVC-Net. Med Image Anal 48:1–11
Lw A et al (2019) Accurate automated Cobb angles estimation using multi-view extrapolation net. Med Image Anal 58:101542
Fu X et al (2020) An automated estimator for Cobb angle measurement using multi-task networks. Neural Comput Appl 1–7
Zhang J, Lou E, Hill DL et al (2010) Computer-aided assessment of scoliosis on posteroanterior radiographs. Med Biol Eng Comput 48(2):185–195
Zhang J, Lou E, Le LH et al (2009) Automatic Cobb measurement of scoliosis based on fuzzy hough transform with vertebral shape prior. J Digit Imaging 22(5):463–472
Sardjono TA, Wilkinson MHF, Veldhuizen AG et al (2013) Automatic Cobb angle determination from radiographic images. Spine 38(20):1256–1262
Anitha H, Karunakar AK, Dinesh KVN (2014) Automatic extraction of vertebral endplates from scoliotic radiographs using customized filter. Biomed Eng Lett 4(2):158–165
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition. 3431–3440
Ronneberger O, Fischer P, Brox T (2015) U-Net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 234–241
Zhou Z, Siddiquee MMR, Tajbakhsh N et al (2018) Unet++: a nested u-net architecture for medical image segmentation. Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, Cham, p 3–11
Zhou Z et al (2020) UNet++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans Med Imaging 39(6):1856–1867
Qin X et al (2019) BASNet: boundary-aware salient object detection. 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), IEEE
Qin X et al (2020) U2-Net: going deeper with nested U-structure for salient object detection. Pattern Recognit 106:107404
Fang L, Liu J, Liu J, et al (2018) Automatic segmentation and 3D reconstruction of spine based on FCN and marching cubes in CT volumes. 2018 10th International Conference on Modelling, Identification and Control (ICMIC), IEEE. 1–5
Horng MH, Kuok CP, Fu MJ et al (2019) Cobb angle measurement of spine from X-ray images using convolutional neural network. Comput Math Methods Med
Tan Z, Yang K, Sun Y et al (2018) An automatic scoliosis diagnosis and measurement system based on deep learning. 2018 IEEE International Conference on Robotics and Biomimetics (ROBIO), IEEE, p 439–443
Wang L et al (2021) Evaluation and comparison of accurate automated spinal curvature estimation algorithms with spinal anterior-posterior X-ray images: the AASCE2019 challenge. Med Image Anal 72(1):1
Lei T et al (2020) Medical image segmentation using deep learning: a survey
Szegedy C, Ioffe S, Vanhoucke V et al (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. Thirty-First AAAI Conference on Artificial Intelligence
He K, Zhang X, Ren S, et al (2016) Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition, p 770–778
He K, Zhang X, Ren S et al (2016) Identity mappings in deep residual networks. European conference on computer vision, Springer, Cham, p 630–645
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. Proceedings of the IEEE conference on computer vision and pattern recognition, p 7132–7141
Woo S, Park J, Lee J Y, et al (2018) CBAM: convolutional block attention module. Proceedings of the European Conference on Computer Vision (ECCV). 3–19
Chen L C, Zhu Y, Papandreou G, et al., “Encoder-decoder with atrous separable convolution for semantic image segmentation,” Proceedings of the European conference on computer vision (ECCV). 801–818 (2018).
Badrinarayanan V, Kendall A, Cipolla R (2017) SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Transactions on Pattern Analysis & Machine Intelligence. 1–1
Acknowledgements
This work was funded by the National Natural Science Foundation of China (Grant No. 62063034). The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn/) for the expert linguistic services provided.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, Y., Zhang, J., Li, H. et al. Automatic Cobb angle measurement method based on vertebra segmentation by deep learning. Med Biol Eng Comput 60, 2257–2269 (2022). https://doi.org/10.1007/s11517-022-02563-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-022-02563-7