Abstract
Automatic and reliable prostate segmentation is an essential prerequisite for assisting the diagnosis and treatment, such as guiding biopsy procedure and radiation therapy. Nonetheless, automatic segmentation is challenging due to the lack of clear prostate boundaries owing to the similar appearance of prostate and surrounding tissues and the wide variation in size and shape among different patients ascribed to pathological changes or different resolutions of images. In this regard, the state-of-the-art includes methods based on a probabilistic atlas, active contour models, and deep learning techniques. However, these techniques have limitations that need to be addressed, such as MRI scans with the same spatial resolution, initialization of the prostate region with well-defined contours and a set of hyperparameters of deep learning techniques determined manually, respectively. Therefore, this paper proposes an automatic and novel coarse-to-fine segmentation method for prostate 3D MRI scans. The coarse segmentation step combines local texture and spatial information using the Intrinsic Manifold Simple Linear Iterative Clustering algorithm and probabilistic atlas in a deep convolutional neural networks model jointly with the particle swarm optimization algorithm to classify prostate and non-prostate tissues. Then, the fine segmentation uses the 3D Chan-Vese active contour model to obtain the final prostate surface. The proposed method has been evaluated on the Prostate 3T and PROMISE12 databases presenting a dice similarity coefficient of 84.86%, relative volume difference of 14.53%, sensitivity of 90.73%, specificity of 99.46%, and accuracy of 99.11%. Experimental results demonstrate the high performance potential of the proposed method compared to those previously published.

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The American Cancer Society estimates about 174,650 new prostate cancer cases, resulting in 31,620 deaths in 2019 only in the USA [37], which in 2018 was considered the second most frequent cancer in men [36]. Moreover, prostate cancer has surpassed lung cancer by turning out to be the most common due to the extensive increase of screening [33]. According to the most recent data, for every 9 men, 1 is bound to develop prostate cancer throughout his lifetime [1].
The prostate is an exocrine gland situated nearby the bladder and the penis that is responsible for the production and storage of a colorless fluid, which along with sperm constitutes the semen [2]. Most prostate cancers grow slowly and cause no symptoms. However, tumors at a later stage are aggressive and can spread quickly [2, 33]. Therefore, in order to diminish prostate cancer cases, early detection is crucial, not only by improving the effectiveness of treatment but also by increasing the survival chance of almost 96% of the patients [1].
The prostate segmentation is a fundamental step to assist the identification, analysis, and treatment of cancer; by instructing medical procedures and therapies, several works [8, 18, 20, 52, 53, 57] propose methods of prostate segmentation with diverse medical imaging techniques, principally magnetic resonance imaging (MRI), for it is more convenient for cancer analysis compared to other approaches such as transrectal ultrasound (TRUS) and computed tomography (CT) [15]. Besides, MRI scans provide high-resolution images with excellent soft-tissue contrast and do not involve ionizing radiation [10].
In current practice, prostate segmentation on MRI scans is carried out by the hand of a professional, mostly a radiologist, visually inspecting slice-by-slice. A manual and time-consuming approach that demands expertise and concentration [20, 57]. Thereby, an accurate and automatic method for prostate segmentation on MRI scans is a meaningful and beneficial contribution to reduce prostate cancer cases [15].
A precise and automatic method to accomplish prostate segmentation on 3D MRI scans would be clinically beneficial, improving efficiency and reducing errors and inconsistencies in the manual segmentation by radiologists [8]. Notwithstanding, the prostate does not have well-defined borders, as it is very similar to its adjacent tissues and may vary in size and shape according to the patient and the resolution of the image. Thus, its automatic segmentation is a challenging task [52, 57].
With the advancement of deep learning techniques, deep convolutional neural networks (DCNN) prove to be a great tool for medical image analysis, presenting encouraging results for segmentation purposes [19, 21, 51]. The DCNN model implicitly extracts features directly from input data, eliminating the explicit feature extraction step [25]. Compared to traditional segmentation methods [14, 31, 32], DCNNs have several advantages, such as the ability to effectively handle extremely diverse data, and are robust to variations in brightness, noise, distortion, and occlusion. Problems that may occur according to how MRI scans were acquired [53, 57].
In addition to the deep learning techniques, the state-of-the-art includes methods based on the probabilistic atlas and active contour models. Nonetheless, these methods present several limitations: (1) the atlas-based approaches need MRI scans with the same spatial resolution [5], (2) the active contour-based approaches need a good contour initialization [16], and (3) the deep learning–based approaches contain a set of hyperparameters that are critical to the training process, making it difficult and costly to manually identify these optimal hyperparameters [38].
Thereby, this paper proposes an automatic and novel coarse-to-fine segmentation method for prostate 3D MRI scans based on a DCNN model, content-sensitive superpixels technique, probabilistic atlas, active contour model, and an optimization algorithm in order to address the limitations mentioned above. As for contributions to this work, we can formulate as follows:
-
1.
Lies in the exploitation of Intrinsic Manifold Simple Linear Iterative Clustering [30] to clustering the pixels in MRI scans. Superpixel analysis guarantees a large amount of data for deep learning techniques. In addition to being more efficient than the pixel approach.
-
2.
Build a probabilistic atlas of any dimension, presenting a wide variety in the prostate anatomy. This process avoids the use of registration techniques, guaranteeing the real information of the MRI scans.
-
3.
Combine two parallel convolutional neural networks, each fed by the superpixel in the original MRI scan and the probabilistic atlas, thus aggregating the texture and spatial features in the learning.
-
4.
Apply the particle swarm optimization (PSO) algorithm [22] to optimize some hyperparameters in the DCNN, eliminating the requirement of a manual search.
-
5.
An efficient coarse segmentation step based on the superpixel classification into prostate and non-prostate tissues. This step guarantees a good and automatic initialization for the 3D Chan-Vese model, without the need for human intervention.
Briefly, the proposed method has five steps. After the materials step, all MRI scans are preprocessed to reduce the impact of the acquisition of different protocols and equipment. The coarse segmentation step consists of the classification of superpixels in prostate and non-prostate tissue based on the DCNN model in conjunction with the PSO algorithm. Besides the texture features, the probabilistic atlas provides spatial features in the training of the DCNN model. Then, the fine segmentation step is completed using the 3D Chan-Vese model to fine segment the prostate. Lastly, the results obtained are evaluated.
In addition to the introductory section, the paper is structured into five more sections. Section 2 summarizes related works recently published in the literature. Section 3 details the proposed method and materials. Section 4 presents the results obtained, and Section 5 discusses the results. Finally, Section 6 presents final remarks about this research.
2 Related works
Accurate and automatic prostate segmentation on 3D MRI scans is useful for the analysis and treatment of prostate cancer [10, 57]. In this regard, several works have been recently published in the literature [21, 43, 54] in the light of early detection significantly increases the patient’s chance of survival [1]. The literature includes studies based on a probabilistic atlas, active contour models, and deep learning techniques.
In the atlas-based approaches, Li et al. [27] proposed an automatic method based on the random walker algorithm and a probabilistic atlas. The method obtained a dice similarity coefficient (DSC) of 80.7 ± 5.1% using the Prostate 3T database [28]. Stojanov et al. [43] reported an automatic method based on a set of pre-labeled atlas. The method obtained a DSC of 81% using the Prostate MR Image Database [46]. Tian et al. [47] proposed an automatic method divided into two stages based on a multi-atlas framework. The method obtained a DSC of 83.4 ± 4.3% using a private database.
Korsager et al. [23] described an automatic method based on probabilistic atlas in conjunction with a graph cut algorithm. The method obtained a DSC of 88% using a private database. In general, atlas-based approaches can result in poor segmentation if the anatomy and size of the prostate in the MRI scan is very different from the MRI scans used to generate the atlas. Besides, most of the atlas-based approaches are performed in a global registration step, increasing the computational time [5, 34, 45].
In the active contour-based approaches, Al et al. [4] proposed a semi-automatic method based on a multi-resolution level set algorithm with shape prior. The method obtained a DSC of 80% using the Prostate MR Image Database. Tian et al. [48] described a semi-automatic method based on 3D graph cuts and a 3D level set. The method obtained a DSC of 89.3 ± 1.9% using a private database. Yang et al. [53] reported a semi-automatic method based on a level set algorithm with shape prior. The method obtained a DSC of 91.45% using a private database.
Tian et al. [49] proposed a semi-automatic method based on supervoxels in conjunction with a level set algorithm. The method obtained a DSC of 86.9 ± 3.2% using a private database. Yang et al. [54] described a semi-automatic method based on a hierarchical level set, using statistic distance analysis, texture, and shape information. The method obtained a DSC of 92.05% using a private database. Notwithstanding, the active contour-based approaches can result in poor segmentation if the prostate boundaries are not well defined on MRI scan. In addition, this approach needs an initial contour [16].
Apropos of deep learning–based approaches, Yan et al. [52] proposed an automatic method based on geodesic object proposals algorithm and deep convolutional neural networks (DCNN). The method obtained a DSC of 89% using the PROMISE12 database [29]. Guo et al. [18] reported an automatic method based on stacked sparse auto-encoder in conjunction with sparse patch matching. The method obtained a DSC of 87.8% using a private database. Jia et al. [20] suggested an automatic method based on an ensemble DCNN. The method obtained a DSC of 88 ± 0.04% using the PROMISE12 database. Yu et al. [57] proposed an automatic method based on a volumetric convolutional neural network with mixed long and short residual connections. The method obtained a DSC of 89.43% using the PROMISE12 database.
Cheng et al. [8] reported an automatic method based on holistic neural network, exploiting both patch-based and holistic approaches. The method obtained a DSC of 89.77 ± 3.29% using a private database. Jia et al. [21] described an automatic method based on an ensemble DCNN in conjunction with atlas registration. The method obtained a DSC of 91% using the PROMISE12 database. Nevertheless, the deep learning–based approaches can take a long time to be trained using the whole image as input and require a large amount of data. Furthermore, these models contain a set of hyperparameters that are critical to the training process, making it difficult and costly to manually identify these optimal hyperparameters.
Despite the considerable efforts and several methods that have been proposed in recent literature, accurate and automatic prostate segmentation on MRI scans still remains a challenge. Thus, we exploit the effectiveness of the aforementioned approaches to present an automatic superpixel-based deep convolutional neural networks method for prostate segmentation on 3D MRI scans. The proposed method combines local texture and spatial information using superpixels and probabilistic atlas to coarse segment the prostate. Region-based active contour model to fine segment the prostate, producing smooth surfaces. Lastly, an optimization algorithm is used in order to optimize the number of filters capable of discriminating between the prostate and non-prostate tissues in each convolutional layer, eliminating therefore the requirement of a manual search.
3 Materials and method
The proposed method for automatic prostate segmentation consists of five steps as presented in Fig. 1. Briefly, the first step describes the databases used as MRI scans. Secondly, image preprocessing is performed. In the third step, a coarse segmentation is conducted using a probabilistic atlas, superpixels, and deep convolutional neural networks. Afterwards, a fine segmentation is completed using a region-based active contour model to fine segment the prostate. Lastly, the proposed method is evaluated using metrics commonly accepted for performance analysis of image processing based systems.
3.1 Materials
The MRI scans used in our research consist of the Prostate 3T and PROMISE12 databases, both available on the internet for challenges purposes. The Prostate 3T database contains 30 T2-weighted MRI scans along with their ground truth. The whole database is in the DICOM format and has 320 × 320 of dimension with 16 bits per voxel, voxel spacing of 0.6 × 0.6 × 4.0 mm3, and slice thickness of 4 mm [27]. The number of slices varies from 15 to 24. The database was acquired on a 3.0T Siemens scanner using a pelvic phased array coil [28].
The PROMISE12 database contains 50 T2-weighted MRI scans along with the respective ground truth. The whole database presents a large variation in the voxel spacing, dynamic range, and anatomic appearance into MRI scans [57]. This strain is due to different acquisition protocols and different types of equipment, such as 1.5T and 3.0T Siemens scanners using a pelvic phased array coil (PAC) or an endorectal coil (ERC) [29].
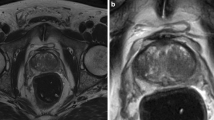
Notwithstanding, there are several reasons to use but the MRI scans without ERC in our method. (1) ERC deforms the prostate anatomy due to the introduction of the specific coil, compressing the peripheral zone, (2) patients with a historic of rectal therapy or rectal stenosis can not use ERC to obtain MRI scans, and (3) ERC causes discomfort and pain to patients [11, 55]. Therefore, in current practice, ERC is still not commonly used. Figure 2 shows examples of PAC and ERC MRI scans.
3.2 Preprocessing
The preprocessing step consists of applying two techniques to normalize the intensity values of the MRI scans. As described in the Section 3.1, the databases were obtained with different protocols using different types of equipment. Hence, prostate tissues may present different texture patterns inter-scans. To overcome such kind of problem, the first technique applied was the histogram matching [17] algorithm, using a single MRI scan of the database as a reference for all others. Then, the second technique applied was the uniform quantization [17] algorithm to normalize the voxel values at 8 bits intra-scans. Figure 3 illustrates its applications.
3.3 Coarse segmentation
The prostate coarse segmentation step combines local texture and spatial information using a content-sensitive superpixels technique and probabilistic atlas in a deep convolutional neural networks (DCNN) model, as shown in Fig. 4. The coarse segmentation is divided into three steps: (1) probabilistic atlas construction, (2) superpixel clustering, and (3) a DCNN combined with the particle swarm optimization algorithm for superpixel classification.
3.3.1 Probabilistic atlas construction
A probabilistic atlas is a rough approximation of the position of the prostate on MRI scans derived from aligning ground truth, in which each voxel indicates the probability that the voxel belongs to the prostate [27, 52]. In the proposed method, we used the probabilistic atlas to incorporate spatial information of the prostate in the DCNN model. Thereon, in the probabilistic atlas construction, the images must be aligned presenting the same dimensions [27]. However, as described in Section 3.1, the databases present variation in some dimension (z-axis). To overcome this problem, we propose an algorithm that creates a probabilistic atlas with any dimension based on an initial probabilistic atlas, as shown in Fig. 5. The algorithm is detailed in the following steps:
-
1.
Group the images of the training subset based on the dimensions (x, y, z).
-
2.
Select the cluster that has the highest number of MRI scans. The goal of this step is to ensure a greater diversity in the variation of prostate anatomy.
-
3.
Align all the ground truth of the selected cluster in step 2 to the center of the image. The idea is to create a probabilistic atlas in the form of a Gaussian distribution, guaranteeing a high probability of prostate in the central region of the MRI scan.
-
4.
Create the initial probabilistic atlas, defined by Eq. 1, using all aligned ground truth in step 3,
$$ Atlas = \frac{1}{N}\sum\limits_{i=1}^{N}{G_{i} * 100<percent>} $$(1)where Gi represents the i th aligned ground truth and N is the number of MRI scans in the cluster selected in step 2.
-
5.
Finally, interpolate the initial probabilistic atlas voxels to any dimension using the trilinear interpolation method. At this step, we can create probabilistic atlas of any dimension presenting a wide variety in the prostate anatomy.
Figure 5 presents the construction of the probabilistic atlas. The higher the value of voxel in the atlas the more likely it is to belong to the prostate and vice versa. Hence, the probabilistic atlas provides prior information of the prostate localization in the MRI scan. After the probabilistic atlas construction step, each MRI scan in the database has a corresponding probabilistic atlas with the same dimensions.
3.3.2 Superpixel clustering
The coarse segmentation uses superpixels for extracting the prostate surface in both the MRI scan and the probabilistic atlas. Considering the fact that spacing between transverse slices in the databases is much larger than the spacing between two voxels within the slice, as described in the Section 3.1, the superpixel clustering and classification are performed in the two-dimensional approach. This would degrade the formation of supervoxels due to high slice thickness. Furthermore, it increases the number of samples in the network training.
The superpixel technique significantly reduces the computational cost while potentially increasing the detection accuracy, as it is more robust to noise than pixel-based method [48, 49]. In our method, the superpixel clustering is generated by applying the Intrinsic Manifold Simple Linear Iterative Clustering (IMSLIC) algorithm. The IMSLIC [30] algorithm extends SLIC [3] to compute content-sensitive superpixels; meanwhile, it inherits all the favorable features of SLIC, such as simplicity and high performance. The algorithm requires two parameters, such as the mean superpixel size and the compactness factor. Furthermore, it can effectively capture non-homogenous features in an image, presenting small superpixels in content-dense regions and large superpixels in content-sparse regions [30].
The IMSLIC algorithm clusterizes each MRI slice to obtain superpixels that describe local texture features. To aggregate spatial features, the same region of the superpixel has been extracted into the corresponding probabilistic atlas. Figure 6 presents this process. Both superpixels are centered on a 64 × 64 patch image, and so, they are used simultaneously for the classification of prostate and non-prostate tissues, as illustrated in Fig. 4.
3.3.3 Deep convolutional neural network-based PSO model
The next step consists of classifying both superpixels into prostate and non-prostate tissues using a deep convolutional neural networks (DCNN) model. Therefore, it is necessary to label the superpixels in two classes: prostate and non-prostate. To define which superpixels are prostate tissues, ground truths are used. A superpixel is considered a prostate tissue if it has at least 90% of its pixels found in the ground truth. If a superpixel touches the ground truth in a proportion of less than 90%, they are ignored from DCNN model training to avoid underfitting. Finally, all the others are labeled as non-prostate tissues. Figure 7 details the process of superpixels labelling.
There are certain reasons for using the DCNN in our research: (1) the DCNN model implicitly extracts features using the convolutional layers and classifies them using the fully connected layers at the top of the network [25, 26], thus eliminating the explicit feature extraction step, and (2) the pooling layers guarantees the invariance of the extracted features as the geometric transformations. Problems that may occur with varying the MRI scans acquisition protocol [53, 57].
Each layer within the DCNN model contains a set of hyperparameters that are critical to the training process [21, 38, 51]. Setting these hyperparameters manually, however, is a costly task, as each application can perform well with totally different sets of hyperparameters [38, 39]. Therefore, the swarm intelligence technique of particle swarm optimization (PSO) [12] was used in conjunction with the DCNN model to automatically define some set of hyperparameters. Thus, the PSO algorithm was used to optimize the number of filters capable of discriminating between prostate and non-prostate tissues in each convolutional layer, so eliminating the requirement of a manual search [39].
The proposed architecture of the DCNN model is shown in Fig. 4. The DCNN model contains five convolutional layers with two max pooling layers in parallel, followed by two fully connected layers with dropout regularization of 0.5, and an output layer with softmax activation for binary classification. The kernel size of all convolutional layers is defined as 5 × 5 and all pooling layer is defined as 2 × 2. Finally, the number of neurons in both fully connected layers is determined as 4096.
The particles of the PSO algorithm are encoded as a list of ten coordinates, corresponding to the hyperparameters to be optimized, as described in Table 1. Each coordinate corresponds to a number of filters in a given convolution layer. Once these hyperparameters are automatically obtained, the DCNN model may prioritize one of the information given as input, texture, or spatial features, in order to obtain a model with a greater discrimination capacity between prostate and non-prostate tissues.
Every single particle of the initial swarm is randomly initialized with integer values between 96 and 384. The range was based on the smallest and largest number of filters found in the well-known architecture proposed by Krizhevsky et al. [24]. The fitness of each particle in the swarm is defined by the performance obtained from the DCNN model in the validation subset, as described in Eq. 2. The metric sensitivity has been given a higher weight as it represents the ability of the DCNN model to correctly classify prostate tissue.
At each PSO iteration, the swarm particles are trained and their fitness evaluated. After that, all particles are updated based on the best-known position (Pbest) in the search-space as well as the best-known position in the whole swarm (Gbest). The hyperparameters optimization is performed until the maximum number of iterations. In the end, the particle represented by Gbest is the optimal hyperparameters for the superpixel classification into prostate and non-prostate. Figure 8 illustrates the proposed DCNN-based PSO flowchart.
In summary, the prostate coarse segmentation consists of 5 steps, with the image of the preprocessing step as input: (1) construction of the corresponding probabilistic atlas, (2) applying the IMSLIC algorithm for superpixels clustering, (3) extraction of superpixels with texture and spatial features, (4) classification of superpixels using the best DCNN-based PSO model, and finally, (5) output image reconstruction. Figure 9 presents this process.
3.4 Fine segmentation
The superpixel-based deep convolutional neural network model in conjunction with probabilistic atlas to incorporate spatial information is an efficient step. Nevertheless, the resulting image may have an irregular boundary if superpixels do not find the edge efficiently. Therefore, a 3D active contour in conjunction with a cubic Bézier spline was used in our method to fine segmentation of the prostate surface.
Chan et al. [7] proposed one of the most efficient active contour models without edges information. The algorithm optimizes the initial segmentation based on information from internal and external regions the curve, respectively the prostate and the adjacent tissues. Only a few iterations of the algorithm are sufficient to refine the prostate edge. Since the coarse segmentation step ensures a good initialization for the 3D Chan-Vese model.
The prostate fine segmentation consists of three steps: (1) get the largest volume in the resulting image of the coarse segmentation step, (2) fit a cubic Bézier spline in some points to produce a smoother prostate surface, and (3) apply a 3D Chan-Vese model to obtain the final prostate surface. Figure 10 shows this process.
3.5 Evaluation metrics
The last step of the proposed method is the evaluation of results using metrics that are used in 3D segmentation analysis [19, 21, 44]. The evaluation metrics used in our research were the dice similarity coefficient (DSC), relative volume difference (RVD), sensitivity (SEN), specificity (SPE), and accuracy (ACC) [44].
The dice similarity coefficient (DSC) measures the spatial overlap between the ground truth of the prostate and the final segmentation.
The relative volume difference (RVD) measures the absolute size difference between the final segmentation (X) and the ground truth (Y ).
The sensitivity (SEN) measures the proportion of prostate voxels correctly classified in the final segmentation.
The specificity (SPE) measures the proportion of non-prostate voxels correctly classified in the final segmentation.
The accuracy (ACC) measures the proportion of voxels correctly classified (both prostate voxels and non-prostate voxels) in the final segmentation.
where TP is true positive, FP is false positive, TN is true negative, and FN is false negative.
4 Results
The proposed method was developed using ITK [56] and Keras libraries [9] on a computer with an Intel Core i7-6700HQ processor, 16GB RAM, and GeForce GTX 1070 of 8GB. The strategy for result analysis is detailed as follows: (1) database separation, (2) preprocessing evaluation, (3) coarse segmentation results, (4) fine segmentation result, and (5) case study.
4.1 Database separation
Both databases, as described in Section 3.1, contain 56 T2-weighted MRI scans along with their ground truth. In order to evaluate the proposed method, the databases were randomly partitioned into three subsets: training, validation, and test. The proportions of database separation used in this research were 60%, 20%, and 20%, respectively. Therefore, the training subset contains 34 MRI scans, the validation subset contains 11 MRI scans, and the test subset contains 11 MRI scans.
4.2 Preprocessing evaluation
All the MRI scans in the databases were submitted to the preprocessing step to normalize the intensity values. Since the histogram matching requires a reference image, an exhaustive search was carried out to find the best MRI scan in the training subset. The idea is to extract the superpixels using the IMSLIC algorithm in all MRI scans on the validation subset. Then, check which superpixels touch the ground truth and create a segmented volume. Finally, compute the dice similarity coefficient (DSC) based on the ground truth. In the end, the MRI scan of the training subset with the highest mean DSC is the reference image. Moreover, the uniform quantization algorithm was applied at 8 bits. Experimental results proved to be better than using the original image (16 bits). Figure 11 illustrates the qualitative results of the preprocessing step application in some MRI scans.
In order to evaluate the quantitative results of the preprocessing step, only the validation subset with 11 MRI scans was used in the experiment. An approach similar to that used to find the reference image was used to verify each technique applied in the preprocessing step. As can be seen in Table 2, the preprocessing step using histogram matching and uniform quantization obtained the best result with a mean DSC of 80.79%, being higher than the result if used only one technique separately. This demonstrates the importance of both techniques in the proposed method.
4.3 Coarse segmentation results
In order to identify the optimal hyperparameters in the convolutional layers for the DCNN model using the PSO algorithm, the IMSLIC algorithm clusterized all MRI slices into superpixels. The IMSLIC algorithm parameters used in our research were: 25 × 25 of mean superpixel size and 10 of compactness factor, in which it balances the proximity of the color space with the spatial regularization, that is, the smaller this parameter more adjusted to the color space will be the superpixel [3].
Then, two procedures were performed to decrease the unbalance of the non-prostate class in both training and validation subsets. The first procedure is to use only the bounding box of the correspondent probabilistic atlas with a 30% slack on each side for superpixel extraction. Secondly, rotational transformations at the 90, 180, and 270 angles and flip in the axial plane were applied to the prostate class superpixels.
After the two procedures, the training subset contains 100,817 superpixels (63,040 prostate and 37,777 non-prostate) extracted on 34 MRI scans, the validation subset contains 34,170 superpixels (23,240 prostate and 10,930 non-prostate) extracted on 11 MRI scans. At last, the test contains 82,521 superpixels (3,250 prostate and 79,271 non-prostate) extracted on 11 MRI scans. The data augmentation was performed only in the prostate superpixels training and validation subsets in order to improve DCNN model performance in correctly classifying prostate tissues.
The DCNN model training configuration is defined as follows. The number of epochs was defined as 100 with batches size of 128. The loss function was the cross entropy [35], and the weights update was based on the standard backpropagation algorithm [26] with a learning rate of 0.01. To reduce the training process, the image patches were resized from 64 × 64 to 28 × 28. As for the PSO algorithm, the swarm size was defined as 10 particles with the maximum number of iterations equal to 5. The cognitive and social parameters were determined to 2.0 [6, 13]. Lastly, the inertia weight was equal to 0.7 [13].
Table 3 presents the results obtained in both validation and test subsets for the Gbest particle at the end of the PSO algorithm. The results include the sensitivity (SEN), specificity (SPE), and accuracy (ACC). The Gbest particle consists of the following coordinates: 180, 384, 252, 121, 216, 96, 193, 96, 305, and 384. The particle coordinates have been described in Table 1.
Analyzing the Gbest particle, we verified that the DCNN model prioritized the texture features in the first three convolutional layers (180, 384, 252) more than the spatial features (96, 193, 96), and only in the last two convolutional layers the model used more spatial features (305 and 384) than (121 and 216) texture features. Thus, the texture features are more relevant when the image has a higher resolution since the max pooling layers reduce the dimensions of the image while maintaining the maximum values, the spatial features become more important to discriminate the superpixels in the last layers.
To evaluate the DCNN-based PSO performance, the hyperparameters proposed by Krizhevsky et al. [24] and Cheng et al. [8] were used in the same test subset. To produce a fair comparison between the hyperparameters, all experiments were performed with identical subsets and the same training configuration. Notwithstanding, the hyperparameters obtained by Gbest particle presented better results for superpixel classification into prostate and non-prostate than the others, as detailed in Table 4.
The Gbest particle took approximately 54 min to train the DCNN model, obtaining a sensitivity of 98.43%, a specificity of 98.48%, an accuracy of 98.65%, a fitness of 3.914, and an error of 0.045 in the validation subset with 34,170 superpixels (23,240 prostate and 10,930 non-prostate). As for the other particles, the mean training time was about 45 min. Finally, the total execution time of the PSO algorithm was 93 h. Figure 12 presents the training curves.
In addition to the experiment using superpixel, we extracted overlapping patches for pixel-wise classification. This experiment analyzes the importance of the superpixel clustering step, Section 3.3.2, in the proposed method. In summary, the same configuration of the experiment based on superpixel was used, such as the size of the patch, number of epochs, and parameters of the PSO. The training subset contains 96,110 superpixels (35,754 prostate and 60,356 non-prostate) extracted on 34 MRI scans and the validation subset contains 69.115 superpixels (34,296 prostate and 34,819 non-prostate) extracted on 11 MRI scans.
The Gbest particle obtained a sensitivity of 91.39%, a specificity of 98.87%, and an accuracy of 95.16% with the fitness of 3.768 on the validation subset. Notwithstanding, the results were worse and the pixel-based experiment is slower compared to the experiment based on superpixels. The delay is due to the patch extraction being proportional to the size of the MRI scans. Table 5 presents results obtained on the test subset by both experiments in the coarse segmentation step.
4.4 Fine segmentation results
Since the superpixel clustering and classification are performed in the two-dimensional approach, the resulting image in 3D was generated to compare with corresponding ground truth. Then, the prostate fine segmentation step, described in Section 3.4, was performed to obtain the final prostate surface. As the resulting image in 3D of the coarse segmentation step is a satisfactory initialization for the 3D Chan-Vese active contour model, few iterations are needed to achieve convergence. In this research, the number of iterations was defined equal to 20, as it was noticed in experiments that reached the best results.
Table 6 describes the results obtained in each step by the proposed method for automatic prostate segmentation on 3D MRI scans of the test subset. The results include the dice similarity coefficient (DSC), relative volume difference (RVD), sensitivity (SEN), specificity (SPE), and accuracy (ACC).
Experimental results shown in Table 6 demonstrate the importance of the fine segmentation step since all evaluation metrics have improved. The mean DSC of 84.86% with a minimum DSC of 78.56% and a maximum DSC of 93.46% represents an accurate segmentation result. Positive mean RVD of 14.53% reflects an over-segmentation of the prostate. This is due to the model training ignoring the superpixels of the border. Finally, the mean SEN, mean SPE, and mean ACC present a good precision of voxels classified correctly. All results exceeded 90%.
4.5 Case study
Figure 13 illustrates a success case obtained by the proposed method on the test subset. In this MRI scan, the evaluation metrics were as follows: DSC of 93.46%, RVD of 6.3%, SEN of 96.40%, SPE of 98.78%, and ACC of 98.52%. The final prostate surface is highlighted in green and the ground truth is outlined in red. Moreover, for the same success case, Fig. 14 displays the results reached in each step of the method and its corresponding ground truth in a series visualization and Fig. 15 presents the final result in a 3D view. It is noticed that the volume obtained by the coarse segmentation step is very close to the final volume, demonstrating to be a good initialization for the fine segmentation step.
Nevertheless, Fig. 16 exhibits a fail case obtained by the proposed method on the test subset. The evaluation metrics for this MRI scan were as follows: DSC of 78.56%, RVD of 42.73%, SEN of 92.91%, SPE of 99.27%, and ACC of 99.17%. The final prostate surface is highlighted in green, and the ground truth is outlined in red. Although a mismatch was found in some MRI slices, the final prostate surface is alike the ground truth in the middle region. Additionally, for the same fail case, Fig. 17 shows the results achieved in each step of the method and its corresponding ground truth in series visualization and Fig. 18 illustrates the final result in a 3D view. In this case, the fine segmentation step removed regions that were erroneously segmented and it incorporated regions that had been lost by the coarse segmentation step.
5 Discussion
Considering the fact that none of the related work described in Section 2 made available the database divided into the three subsets (training, validation, and testing), an accurate and trustworthy comparison becomes difficult to perform. Since the only piece of information available in the works is the database used and the number of MRI scans. Table 7 synthesizes the results recently published in the literature with the result obtained by the proposed method based on the DSC metric.
Analyzing these works, the proposed method obtained better results than some works [4, 27, 43, 47]. However, only Li et al. [27] used the Prostate 3T database. Besides, our method does not impose limitations on the use of the probabilistic atlas in the light of the fact that it generates a probabilistic atlas of any dimension from an initial probabilistic atlas. Some works [8, 18, 23, 48, 49, 53, 54] presented DSC higher than that of our method. Nonetheless, these works used private databases. Generally, these databases have MRI scans acquired using the same equipment with well-defined acquisition protocols. Furthermore, the works of [48, 49, 53, 54] require human intervention to achieve final segmentation.
Although the works of [20, 21, 52, 57] had results superior to ours, some of these works had to modify the spatial resolution of MRI scans using registration techniques, thus losing real information from the images. Also, only the PROMISE12 database was used in the evaluation of these works, generally with a number of MRI scans in the test subset less than ours (11 MRI scans). Analysis of published works revealed that the proposed method obtained results as good as to the most reliable previously reported works, as shown in Table 7, though using two completely different MRI databases. The findings obtained indicated the following:
-
1.
The use of the IMSLIC algorithm was promising for the clustering of the pixels in MRI scans. It reduced the computational and memory costs while potentially increasing the detection accuracy, as it is more robust to noise than pixels-based method.
-
2.
The construction of a probabilistic atlas of any dimension presented a wide variety of the prostate anatomy. This made it possible to use MRI scans with different resolutions, making the method more robust and generic.
-
3.
The combination of two parallel convolutional neural networks, each fed by the superpixel in the original MRI scan and the probabilistic atlas, aggregated the texture and spatial features in the model learning.
-
4.
The application of the PSO algorithm to optimize some hyperparameters in the DCNN model eliminated the requirement of a manual search. Besides, it obtained better results than the hyperparameters found in the literature.
-
5.
The use of Chan-Vese active contour model refined the prostate surface when the superpixel failed to find the boundary of the prostate accurately.
-
6.
Lastly, it is important to emphasize that the databases used in our research are extremely complex and diverse, containing countless different cases of MRI scans.
It is also worth mentioning the performance of the proposed method in noise images, a common problem in medical imaging [40,41,42]. The databases contain MRI scans acquired by 1.5T and 3.0T scanners. Although the 3.0T provides better quality images, they have a higher likelihood of artifacts appearing in the image due to the movement of blood or fluid and produces more noise than the 1.5T scanner [50]. Nevertheless, the proposed method obtained good results, because the fine segmentation step used a region-based active contour method that exploits texture features inside the curve.
As for limitations, the proposed method has two drawbacks: (1) the superpixels are treated individually in the classification step, described in Section 3.3.2, and thus the explored deep features do not reflect the holistic information ie the prostate anatomy, and (2) the high computational cost in processing (clustering and classification) of non-prostate superpixels away from the prostate region, since the prostate is usually located in the center of the MRI slice.
6 Conclusion
The present research proposed an automatic coarse-to-fine segmentation method for prostate on 3D MRI scans. As for the contributions hereby, the coarse segmentation step combining local texture and spatial information using the IMSLIC algorithm and probabilistic atlas in a DCNN model jointly with the PSO algorithm to classify the prostate and non-prostate superpixels is presented. Thereupon, the fine segmentation uses the 3D Chan-Vese active contour model to obtain the final prostate surface. To the best of our knowledge, this is the first method that applies the IMSLIC algorithm to the problem of MRI scan segmentation.
The results obtained indicate the high performance-potential of the proposed method compared to those previously published. Besides, the proposed DCNN model in conjunction with the PSO algorithm eliminates the explicit feature extraction step and the manual definition of optimal hyperparameters. Although the databases used in this method assure a high diversity in the variation of prostate anatomy, further experiments with other databases are needed to improve the proposed method and make it more robust and generic.
Future works include analyzing the supervoxels extraction using the IMSLIC algorithm. Apart from the aforementioned, the application of a swarm intelligence technique to find the best parameters of the IMSLIC algorithm to segment the prostate on 3D MRI scans. Also, using the PSO algorithm to build the DCNN architecture instead of optimizing only the hyperparameters.
References
American cancer society: key statistics for prostate cancer (2018). https://www.cancer.org/cancer/prostate-cancer/about/key-statistics.html. Accessed: 2018-04-15
Instituto nacional do câncer, tipos de câncer: Próstata (2018). http://www2.inca.gov.br/wps/wcm/connect/tiposdecancer/site/home/prostata. Accessed: 2018-04-15
Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S (2012) Slic superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell 34(11):2274–2282
Al-Qunaieer FS, Tizhoosh HR, Rahnamayan S (2014) Multi-resolution level sets with shape priors: a validation report for 2d segmentation of prostate gland in t2w mr images. J Digit Imaging 27(6):833–847
Alves RS, Tavares JMR (2015) Computer image registration techniques applied to nuclear medicine images. In: Computational and experimental biomedical sciences: methods and applications. Springer, pp 173–191
Cai X, Cui Z, Zeng J, Tan Y (2009) Individual parameter selection strategy for particle swarm optimization. In: Particle swarm optimization. Intechopen, pp 89–112
Chan TF, Vese LA (2001) Active contours without edges. IEEE Trans Image Process 10(2):266–277
Cheng R, Roth HR, Lay NS, Lu L, Turkbey B, Gandler W, McCreedy ES, Pohida TJ, Pinto PA, Choyke PL et al (2017) Automatic magnetic resonance prostate segmentation by deep learning with holistically nested networks. J Med Imaging 4(4):041,302
Chollet F et al (2015) Keras. https://github.com/fchollet/keras
Damião R, Figueiredo RT, Dornas MC, Lima DS, Koschorke MA (2015) Câncer de próstata Revista Hospital universitário Pedro Ernesto, pp 14
De Visschere P (2018) Improving the diagnosis of clinically significant prostate cancer with magnetic resonance imaging. Journal of the Belgian Society of Radiology 102(1)
Eberhart R, Kennedy J (1995) A new optimizer using particle swarm theory In: , 1995. MHS’95., Proceedings of the Sixth International Symposium on Micro Machine and Human Science. IEEE, pp 39–43
Eberhart RC, Shi Y (2000) Comparing inertia weights and constriction factors in particle swarm optimization. In: 2000. Proceedings of the 2000 Congress on Evolutionary Computation, vol 1. IEEE, pp 84–88
Ferreira A, Gentil F, Tavares JMR (2014) Segmentation algorithms for ear image data towards biomechanical studies. Comput Methods Biomech Biomed Eng 17(8):888–904
Ghose S, Oliver A, Martí R, Lladó X, Vilanova JC, Freixenet J, Mitra J, Sidibé D, Meriaudeau F (2012) A survey of prostate segmentation methodologies in ultrasound, magnetic resonance and computed tomography images. Comput Methods Programs Biomed 108(1):262–287
Gonċalves PC, Tavares JMR, Jorge RN (2008) Segmentation and simulation of objects represented in images using physical principles. Comput Model Eng Sci 32(1):45–55
Gonzalez RC, Wintz P (1977) Digital image processing(book). Reading, Mass., Addison-Wesley Publishing Co., Inc. Applied Mathematics and Computation (13), pp 451
Guo Y, Gao Y, Shen D (2017) Deformable mr prostate segmentation via deep feature learning and sparse patch matching. In: Deep learning for medical image analysis. Elsevier, pp 197–222
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin PM, Larochelle H (2017) Brain tumor segmentation with deep neural networks. Med Image Anal 35:18–31
Jia H, Xia Y, Cai W, Fulham M, Feng D (2017) Prostate segmentation in mr images using ensemble deep convolutional neural networks. In: 2017 IEEE 14th international symposium on Biomedical imaging (ISBI 2017). IEEE, pp 762–765
Jia H, Xia Y, Song Y, Cai W, Fulham M, Feng D (2018) Atlas registration and ensemble deep convolutional neural network-based prostate segmentation using magnetic resonance imaging. Neurocomputing 275:1358–1369
Kennedy J (2011) Particle swarm optimization. In: Encyclopedia of machine learning. Springer, pp 760–766
Korsager AS, Fortunati V, Lijn F, Carl J, Niessen W, ØStergaard LR, Walsum T (2015) The use of atlas registration and graph cuts for prostate segmentation in magnetic resonance images. Med Phys 42 (4):1614–1624
Krizhevsky A, Sutskever I, Hinton G (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp 1097–1105
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436
LeCun Y, Kavukcuoglu K, Farabet C (2010) Convolutional networks and applications in vision. In: Proceedings of 2010 IEEE International Symposium on Circuits and Systems (ISCAS). IEEE, pp 253–256
Li A, Li C, Wang X, Eberl S, Feng D, Fulham M (2013) Automated segmentation of prostate mr images using prior knowledge enhanced random walker. In: International conference on digital image computing: Techniques and applications (DICTA). IEEE, pp 1–7
Litjens G, Futterer J, Huisman H (2015) Data from prostate-3t: the cancer imaging archive. https://doi.org/10.7937/K9/TCIA.2015.QJTV5IL5. Accessed: 2018-01-15
Litjens G, Toth R, van de Ven W, Hoeks C, Kerkstra S, van Ginneken B, Vincent G, Guillard G, Birbeck N, Zhang J et al (2014) Evaluation of prostate segmentation algorithms for mri: the promise12 challenge. Med Image Anal 18(2):359–373
Liu YJ, Yu M, Li BJ, He Y (2017) Intrinsic manifold slic: a simple and efficient method for computing content-sensitive superpixels. IEEE transactions on pattern analysis and machine intelligence
Ma Z, Tavares JMR, Jorge RN (2009) A review on the current segmentation algorithms for medical images. In: Proceedings of the 1st International Conference on Imaging Theory and Applications (IMAGAPP), pp 135–140
Ma Z, Tavares JMR, Jorge RN, Mascarenhas T (2010) A review of algorithms for medical image segmentation and their applications to the female pelvic cavity. Comput Methods Biomech Biomed Eng 13(2):235–246
Mohler JL, Armstrong AJ, Bahnson RR, D’Amico AV, Davis BJ, Eastham JA, Enke CA, Farrington TA, Higano CS, Horwitz EM et al (2016) Prostate cancer, version 1.2016. J Natl Compr Cancer Netw 14(1):19–30
Oliveira FP, Tavares JMR (2014) Medical image registration: a review. Comput Methods Biomech Biomed Eng 17(2):73–93
Rubinstein RY, Kroese DP (2013) The cross-entropy method: a unified approach to combinatorial optimization, Monte-Carlo simulation and machine learning. Springer Science & Business Media
Siegel R, Miller K, Jemal A (2018) Cancer statistics, 2018. CA: Cancer J Clin 68:7–30
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA: a Cancer J Clin 69(1):7–34
da Silva GL, da Silva Neto OP, Silva AC, de Paiva AC, Gattass M (2017) Lung nodules diagnosis based on evolutionary convolutional neural network. Multimed Tools Appl 76(18):19,039–19,055
da Silva GLF, Valente TLA, Silva AC, de Paiva AC, Gattass M (2018) Convolutional neural network-based pso for lung nodule false positive reduction on ct images. Comput Methods Prog Biomed 162:109–118
Sołtysiński T (2008) Bayesian constrained spectral method for segmentation of noisy medical images. theory and applications. In: Advanced computational intelligence paradigms in healthcare-3. Springer, pp 181–206
Sołtysiński T (2008) Novel quantitative method for spleen’s morphometry in splenomegally. In: International conference on artificial intelligence and soft computing. Springer, pp 981–991
Sołtysiński T, Kałużynski K, Pałko T (2006) Cardiac ventricle contour reconstruction in ultrasonographic images using bayesian constrained spectral method. In: International conference on artificial intelligence and soft computing. Springer, pp 988–997
Stojanov D, Koceski S (2014) Topological mri prostate segmentation method. In: Federated conference on computer science and information systems (fedCSIS). IEEE, pp 219–225
Taha AA, Hanbury A (2015) Metrics for evaluating 3d medical image segmentation: analysis, selection, and tool. BMC Med Imaging 15(1):29
Tavares JMR (2014) Analysis of biomedical images based on automated methods of image registration. In: International symposium on visual computing. Springer, pp 21–30
for Therapy Guided Image, N.C.: Prostate mr image database (2008). http://prostatemrimagedatabase.com/. Accessed: 2017-08-19
Tian Z, Liu L, Fei B (2015) A fully automatic multi-atlas based segmentation method for prostate mr images. In: Medical imaging 2015: Image processing, vol 9413. International society for optics and photonics, pp 941340
Tian Z, Liu L, Zhang Z, Fei B (2016) Superpixel-based segmentation for 3d prostate mr images. IEEE Trans Med Imaging 35(3):791–801
Tian Z, Liu L, Zhang Z, Xue J, Fei B (2017) A supervoxel-based segmentation method for prostate mr images. Med Phys 44(2):558–569
Ullrich T, Quentin M, Oelers C, Dietzel F, Sawicki L, Arsov C, Rabenalt R, Albers P, Antoch G, Blondin D et al (2017) Magnetic resonance imaging of the prostate at 1.5 versus 3.0 t: a prospective comparison study of image quality. Eur J Radiol 90:192–197
Xu J, Luo X, Wang G, Gilmore H, Madabhushi A (2016) A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing 191:214–223
Yan K, Li C, Wang X, Li A, Yuan Y, Feng D, Khadra M, Kim J (2016) Automatic prostate segmentation on mr images with deep network and graph model. In: IEEE 38Th annual international conference of the engineering in medicine and biology society (EMBC). IEEE, pp 635–638
Yang X, Zhan S, Xie D (2016) Landmark based prostate mri segmentation via improved level set method. In: IEEE 13Th international conference on signal processing (ICSP). IEEE, pp 29–34
Yang X, Zhan S, Xie D, Zhao H, Kurihara T (2017) Hierarchical prostate mri segmentation via level set clustering with shape prior. Neurocomputing 257:154–163
Ye X, Oyoyo U, Lu A, Dixon J, Rojas H, Randolph S, Kelly T (2016) Comparison of 3.0 t pac versus 1.5 t erc mri in detecting local prostate carcinoma. bioRxiv, pp 058123
Yoo TS, Ackerman MJ, Lorensen WE, Schroeder W, Chalana V, Aylward S, Metaxas D, Whitaker R (2002) Engineering and algorithm design for an image processing api: a technical report on itk-the insight toolkit. Studies in health technology and informatics, pp 586–592
Yu L, Yang X, Chen H, Qin J, Heng PA (2017) Volumetric convnets with mixed residual connections for automated prostate segmentation from 3d mr images. In: AAAI, pp 66–72
Funding
The authors acknowledge CAPES and CNPq for their financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
da Silva, G.L.F., Diniz, P.S., Ferreira, J.L. et al. Superpixel-based deep convolutional neural networks and active contour model for automatic prostate segmentation on 3D MRI scans. Med Biol Eng Comput 58, 1947–1964 (2020). https://doi.org/10.1007/s11517-020-02199-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-020-02199-5