Abstract
An understanding of athlete ground reaction forces and moments (GRF/Ms) facilitates the biomechanist’s downstream calculation of net joint forces and moments, and associated injury risk. Historically, force platforms used to collect kinetic data are housed within laboratory settings and are not suitable for field-based installation. Given that Newton’s Second Law clearly describes the relationship between a body’s mass, acceleration, and resultant force, is it possible that marker-based motion capture can represent these parameters sufficiently enough to estimate GRF/Ms, and thereby minimize our reliance on surface embedded force platforms? Specifically, can we successfully use partial least squares (PLS) regression to learn the relationship between motion capture and GRF/Ms data? In total, we analyzed 11 PLS methods and achieved average correlation coefficients of 0.9804 for GRFs and 0.9143 for GRMs. Our results demonstrate the feasibility of predicting accurate GRF/Ms from raw motion capture trajectories in real-time, overcoming what has been a significant barrier to non-invasive collection of such data. In applied biomechanics research, this outcome has the potential to revolutionize athlete performance enhancement and injury prevention.

Using data science to model high-fidelity motion and force plate data frees biomechanists from the laboratory
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
One of the strongest criticisms of sports biomechanics is that measurements of GRF/Ms, necessary for the estimation of internal and external musculoskeletal loads and associated injury risk, can only be collected in controlled research laboratory environments using external force transducers. Subsequently, the sport biomechanist is forced to trade ecological validity of the more desirable field-based data collection for laboratory-based methods in order to record higher fidelity data outputs (Fig. 1) [4, 10, 19, 30].
Knee anterior cruciate ligament (ACL) injury can be a season or career-ending event for a professional athlete and increases the risk of later osteoarthritis pathology [13, 20]. The majority of ACL injuries (51 to 80%) occurring in team sports such as Australian Rules Football, basketball, and hockey are non-contact in nature, with more than 80% suffered during a sidestep maneuver or single-leg landing [18, 32]. In-silico, in vitro and laboratory studies have identified an increase in knee joint moments as indicators of ACL injury risk [15, 17, 21] and an understanding of on-field GRF/Ms constitutes the first step towards the development of a monitoring system that estimates knee joint moments, thereby providing an early warning system for ACL injury risk. The ability to monitor real-time ACL injury risk enables the development of counter-measure preventative strategies including new biofeedback measures.
Previous studies have attempted to improve the ecological validity of laboratory-based GRF/Ms data collections, with Müller et al [29] investigating properties of artificial turf using varying shoe stud configurations. Samples of turf were mounted to the surface of a force plate and 50 m2 of the surrounding area. Similarly, Jones et al. [22] tested the effects of different artificial turf types on landing and knee biomechanics by mounting samples in a tray fixed above the force plate. Others have attempted to measure GRF/Ms in the field through a variety of in-shoe pressure-sensitive sensors or attachments [24, 26, 33, 38], however, such devices suffer from being cumbersome to the athlete and measure points of contact or pressure distributions (rather than center of pressure). Importantly, the reported values differ significantly from those derived directly from force plates, although Sim et al. [33] did cite improvements via the use of neural networks (NNs). Researchers have derived GRF/Ms from kinematics using linear statistics, or again from NNs [23, 31], with these studies conducted indoors using gait trials. Jung et al. [23] tested ten participants at speeds up to 3.0 m/s while Oh et al. [31] trained a single hidden layer NN using 48 participants (one trial per participant) each walking at a self-selected pace. Efforts to predict GRF/Ms using non-invasive computer vision techniques show promise but either lack validation to a gold standard or criterion reference [34, 37] or relevance to sporting tasks [9]. This paper proposes a novel approach, where the scale of historically collected big data is used to predict GRF/Ms using the input variables: (1) eight marker motion capture trajectories, and (2) participant mass, sex, and height [1].
The School of Human Sciences at The University of Western Australia (UWA) was one of the first to establish a Sport Science/Human Movement university degree in the southern hemisphere and houses one of the largest sports-related marker-based movement data repositories in the world [6]. This study capitalizes on this data by employing PLS [27] and its kernel variants to learn linear and nonlinear models whereby, given a new sample of motion capture data (marker-based data) we can estimate a participant’s GRF/Ms in the absence of a force plate. The accuracy and validity of this approach is confirmed by reporting the mean correlations between GRF/Ms traditionally derived, and those predicted by the PLS methods. We aim first to test the hypothesis that our interpretation of mass and acceleration (via motion capture marker data) and force (recorded from a force plate) is complete enough that PLS can establish a strong relationship between these variables.
2 Background
For over 30 years, Vicon (Oxford Metrics, Oxford, UK) has been developing motion capture technology, and the company is considered the world leading gold standard manufacturer of passive marker-based motion analysis systems. High-speed video cameras together with near-infrared (IR) light strobes are used to illuminate small spherical retro-reflective markers attached to the body [10, 25], with Carse et al. [8] citing the reconstruction error of such optical systems at less than 0.3 mm [19].
Often captured concurrently with motion data, force platforms/plates are used to measure the forces and moments applied to its top surface as a participant stands, steps (walk/run), jumps from, or lands on it. Three orthogonal force (axes) and three moment components are measured when a participant is in contact with the plate including Fx and Fy representing the horizontal (shear) forces and Fz the vertical force, and Mx, My, and Mz the three rotation moments around the corresponding x, y, and z force axes, respectively. Force platforms used to record this data may utilize a wide variety of force transducer types (e.g., piezo-resistive, piezo-electric) which are generally located in each of the four corners of the platform. Installation of force plates must be carried out in such a manner as to minimize vibration, and with regard to the frequency and absolute force of the intended movement to be captured. For this reason, specialized force plate mounting, directly inside a concrete pad during laboratory construction, produces the best ongoing results [2] but which makes the platform difficult to move or install in sporting environments. GRF/Ms are fundamental to the calculation of joint kinetics, the forces that lead to movement [38], and consequently, this information is critical for all research that seeks to gain an understanding of the mechanism behind performance, injury, and disease.
PLS is a class of supervised multivariate regression techniques which projects data to a lower dimensional space where the covariance between predictor and response variables is maximized [14]. This generally leads to a more accurate regression model compared with, for example, principle component regression (PCR) which maximizes the variance of the predictor variables without taking into account the response variables. PLS is generally referred to as a multilinear regression (MLR) technique; however, it is able to perform nonlinear regression by projecting the data to a higher dimensional nonlinear space where the relationship between the two variable types is linear [7]. First developed in the 1960s, the characteristic of PLS to perform well with many predictor variables, but few examples was found to be a good fit for statistical problems in the natural sciences [12, 27]. More recently, sparse PLS techniques have emerged which can better deal with multivariate responses when some of the predictor variables are noisy. Because of the economic nature of marker-based motion capture representation (compared with video for example), a secondary hypothesis for this study is that sparse PLS will return the strongest predictor (motion capture plus mass, sex, and height) to output (GRF/Ms) response.
3 Methods
3.1 Design and setup
The methodological design schematic of this study is shown in Fig. 2. Original setup and data capture was carried out at one of the two UWA Sports Biomechanics Laboratories (Fig. 4) over a15-year period (2000–2015). All participants used in the archive studies were from ayoung healthy athletic population (male and female, amateur to professional) as opposed to any medical or clinical cohort. Dynamic movement trials included awide variety of generic movement patterns such as walking and running, but also sport-specific movements such as football kicking and baseball pitching. UWA employs acustom, repeatable and well-published upper and lower limb marker set comprising 67 full body retro-reflective markers [5, 11, 15]. This includes markers placed arbitrarily on body segments and markers positioned on anatomically relevant landmarks used to define the joint centers and axes required for anatomical coordinate system definition (e.g., pelvis anterior superior iliac spines, lateral ankle malleoli). Given that the marker set has evolved considerably over the 15-year period, asubset of markers was identified that were consistently and reliably present across all static and dynamic trials of the motion data repository. With the goal of describing movement completely enough that PLS can establish the motion–force relationship, and following earlier pilot testing with larger and smaller marker subsets, the following eight anatomically relevant markers were selected for inclusion in the present study (Fig. 3):
C7, SACR sacrum (automatically constructed between LPSI and RPSI – posterior superior iliac spine left and right), LMT1 left hallux (big toe), LCAL left calcaneus (heel), LLMAL left lateral ankle malleolus (outer ankle), and likewise for the right foot RMT1, RCAL, and RLMAL.
Between 12–20 Vicon near-infrared cameras across a combination of model types (MCam2, MX13, and T40S) were mounted on tripods and wall-brackets and aimed at the desired reconstruction volume space (Fig. 4). Camera calibration (static and dynamic) for all data collection sessions was conducted in accordance with manufacturer recommendations. An AMTI force plate (Advanced Mechanical Technology Inc, Watertown, MA, USA) measuring 1,200 × 1,200 mm, operating at 2,000 Hz and installed flush with the floor was used to record the six GRF/Ms: Fx, Fy, Fz, Mx, My, and Mz. The biomechanics laboratory is a controlled space which utilizes lights and wall paint with reduced IR properties. The floor surface coverings have varied over the 15-year data collection period ranging from short-pile wool carpet squares to artificial turf, both laid on the force plate surface and the wood parquetry surrounding the platform. The relevant proprietary motion capture software that was distributed by the system hardware manufacturer at the time of data collection was used to record and reconstruct the marker trajectories. Irrespective of hardware and software configuration at the time of data collection all reconstructed marker data was compiled and stored in the industry standard c3d file format for motion trajectory and analog data (‘coordinate 3D,’ Motion Lab Systems, Baton Rouge, LA).
3.2 Data mining phase
Over the past two decades, much attention has been paid to identifying the biomechanical precursors to ACL injury, and consequently, the analysis of change of direction (sidestep) maneuvers has been a strong research theme of the biomechanics group at UWA and their collaborators. Given this long data collection history and the subsequent likelihood of a large number of sidestepping motion trials within the legacy motion capture repository, this paper focuses on establishing the motion–force relationship of a single motion trial type: sidestep maneuvers to the left that are performed off the right limb (i.e., right foot plant, Figs. 5 and 6). Data mining of the department’s motion/force plate capture repository was carried out under UWA ethics exemption RA/4/1/8415. Contrary to the traditional scientific method approach of the sport sciences, the philosophy of this study was one of scale, with a mandate to use data capture from as many different sessions as possible (intra-laboratory, multiple testers), and to avoid manual editing of source c3d files. Data mining was conducted using MATLAB R2016b (MathWorks, Natick, MA) in conjunction with the Biomechanical ToolKit v0.3 [3] both running on Ubuntu v14.04 (Canonical, London, UK), a development environment being well-suited to the prototype nature of the study. Hardware employed was a desktop PC, Core i7 4GHz CPU, with 32GB RAM.
From a given top-level folder, the file-system was scanned for motion capture standard c3d files, to which several pre-processing steps were applied to confirm the integrity of the marker trajectories and force plate data before a trial was deemed acceptable and added to the overall data-set. First, the data mining relied only on trials with contiguous markers being labeled and present in the trial and was agnostic to any post-processing artifact associated with filtering or biomechanical modeling (i.e., we only utilized the labeled trajectories of eight real markers). Mass was considered a mandatory input feature, but it was theorized that sex (female = 1, male = 0) and height may also have an important contribution, so they were added to the predictor (input) variable set. These participant specific values (mass, sex, and height) were retrieved from the c3d file or the associated mp file (mp is a proprietary extensible markup language XML file format used by Vicon for session and anthropometric data). At this time, children were excluded by rejecting trials where the participant height was less than 1,500 mm (two standard deviations below the average Australian adult female height 1,644 ± 72 mm, age 19–25 years [36]).
The foot-strike event was automatically determined by detecting vertical force Fz greater than a threshold (20 N) over a defined period (0.025 s) [28]. Compared with trials where the foot-strike event was previously visually identified by the biomechanist undertaking the original data collection, the mean correspondence of the automatic method was ± 0.0054 s. Analog force plate data sampled at frequencies lower than 2,000 Hz and motion capture lower than 250 Hz were time normalized using piecewise cubic spline interpolation. The lead-in period before the foot-strike was deemed to be more important for the predictor movement, and therefore, the marker data was trimmed around the foot-strike event from -0.20 to + 0.30 s (125 frames f ), and force plate data from -0.05 to + 0.30 s (700 frames f ).
A number of consistency checks were performed to consider the overall integrity of the laboratory equipment setup and calibration. Trials where the participant appeared to move backward, where the vertical height of markers was unexpected, where all marker coordinates dropped to zero (i.e., missing data), where the start and end vertical force value was unexpected, or the foot-strike was incomplete, were rejected. Templates were used to automatically classify the range of indoor movements found into one of six types:
Static (still), walk, run, run and jump, sidestep left, and sidestep right (regardless of whether the sidestep was planned or unplanned, crossover or regular, or foot-strike technique).
If the motion capture and force plate data passed these checks for quality, it was reassembled into the data-set arrays X (predictor samples × input features) and y (response samples × output features) typical of the format used by multiple regression [27], Fig. 7. Trials with duplicate X data were rejected, therefore avoiding the situation where the same motion capture input referred to multiple pre- and post-filtered analog force plate data.
Ethics approval was based on the only personal information collected (that of mass, sex, and height) being de-identified and acknowledged that the new data science techniques being employed by the current investigation are within the scope of the original studies and would have been included had they existed at the time. In terms of intellectual property of the motion capture pipeline, only the first step of labeling and gap-fill is required by this study, later analysis including modeling, filtering, and classification by meta-data is disregarded.
3.3 Training phase
We performed tenfold cross-validation using a number of PLS methods to test whether our description of movement and force was sufficient, the goal being a strong correlation coefficient. The data-set was randomly shuffled and split into ten training sets (353 samples = 80%, illustrated for each of the eight markers in Fig. 8) and corresponding test-sets (88 samples = 20%), then for each PLS method, the predicted GRF/Ms were compared with the actual recorded force plate analog output. The use of tenfold experiments decreased the risk of overfitting [16]. A total of eleven PLS methods were compared, three from PLS_Toolbox v8.1.1 (EVRI Eigenvector Research, Inc., Manson, WA, USA), four from the R-pls package [27], and four from the R-spls Sparse PLS package [12]. PLS_Toolbox runs directly in MATLAB, while pls and spls functions were executed using system calls from MATLAB to R [35]. Handshake protocols were used between MATLAB and R to ensure success/fail conditions were exchanged. Within this mix of three proprietary and open source PLS packages, different fit algorithms were investigated for their prediction power, performance, and in the case of sparse implementations, variable selection, for the given multivariate data-set where the number of predictor variables (3003) was much greater than the number of training samples (353). Model training and prediction times were used to illuminate differences between methods such as Kernel and Orthogonal Scores PLS which produce the same results. Overall, PLS methods were selected for relevance to (a) perceived state of the art, (b) anticipated benefits of including nonlinear kernel methods to match nonlinearity in the source data, and (c) sparse methods to capitalize on the ranking importance of predictor input markers rather than the traditional PLS approach of simply maximizing the covariance between predictors and response.
Sidestep left eight marker trajectories shown by MATLAB, for one training-set (353 examples = 80%). The physical location of the markers is given in Fig. 3
The primary tuning parameter for PLS is the number of hidden internal components, nc. For every sample in the test-set, the mean correlation coefficient r was calculated by comparing the six vectors Fx, Fy, Fz, Mx, My, and Mz of ground truth force plate data with that predicted by the specified PLS method. A range of nc from 1 to 81 (in steps of 5) was used to select nc via the corresponding maximum r by GRF/Ms for the subsequent tenfold experiments. This range was arrived at empirically using the root mean squared error of prediction (RMSEP) function in R-pls; use of the mean squared prediction error (MSPE) in R-spls; and by noting the maximum value at which MATLAB exhausted system memory. MSPE was also used to determine the sparsity tuning parameter eta of 0.9. Although this granular approach increased the risk of missing the precise optimal value of nc, meaningful results were observed. The average nc over all GRF/Ms for each PLS method gave a range of training times from 00:00:10.534 (hh:mm:ss.sss) for R-pls Wide Kernel PLS to 00:18:28.552 R-spls Orthogonal Scores PLS (mean timing over ten iterations).
4 Results and discussion
A high-potential subset of the entire historical archive containing 20,066 c3d files was scanned, and after quality assurance and automatic categorization of movement type, a total of 441 sidestep left-directed motion trials were identified. The original data capture for these trials was carried out between February 7, 2007 and November 12, 2013 using a range of Vicon proprietary software (from Workstation v5.2 to Nexus v2.2).
The mean correlation coefficient r between the estimated and actual GRF/Ms was calculated using the nc derived by the earlier cost analysis, for which the prediction times ranged from 00:00:00.064 (hh:mm:ss.sss, mean timing over ten iterations) for EVRI-pls Direct Scores PLS to 00:00:00.403 R-pls Kernel PLS. The mean ± SD between each of the tenfolds, and prediction times, by PLS method and by GRF/Ms are given in Table 1 (and illustrated by animation Online Resource 1), in which the best values of r by GRF/Ms are shown in bold, as are r(Fmean) and r(Mmean) for the strongest package overall. The highest correlation was seen in the vertical r(Fz), explained by the influence of mass in this axis and the corresponding greater variation for PLS to associate with. R-spls SIMPLS was identified as the strongest method overall, with average r of 0.9804 for GRFs and 0.9143 for GRMs. These high correlation coefficients proved the hypothesis, that our interpreted force, mass, and acceleration by the abstract methods of marker-based motion capture were sufficient enough to establish a strong relationship with the analog force plate output.The combined mean ± SD results r(Fmean) 0.9796 ± 0.0004 and r(Mmean) 0.9113 ± 0.0036 illustrate the proximity of all the PLS methods investigated.
Figure 9 illustrates the performance of R-spls SIMPLS for r(Fmean) and r(Mmean) over the range of nc from 1 to 81. Ahead of nc 55 selected by the cost analysis for this PLS method, the high r(Fmean) offsets the gradual decline in r(Mmean). At greater nc, this relationship breaks down as r(Mmean) is increasingly affected by noise.
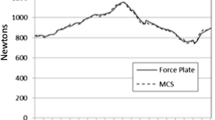
With R-spls SIMPLS outperforming other methods, the second hypothesis that a sparse PLS method would prevail was also proven. The individual sample with the highest r(Fmean) was identified for R-spls SIMPLS, and Fig. 10 shows the predictions for this sample by the SIMPLS implementation by each of the three packages.
Ground truth GRF/Ms (blue ticks) and predicted (red), plotted as Fx, Fy, Fz, Mx, My and Mz versus force plate frame for the same sample using each of the strongest PLS methods by package: EVRI-pls SIMPLS, R-pls SIMPLS and R-spls SIMPLS. The sample was selected for having the highest r(Fmean) with R-spls SIMPLS
The mean R-spls SIMPLS results exceed the maximum correlation coefficients r for the six vectors as reported by Oh et al. [31] and shown in Table 2. Using PLS, rather than a single hidden layer NN, with a data-set an order of magnitude greater (441 versus 48 samples), our study demonstrated greater correlations for a more complex movement pattern (sidestep versus walking gait), and the importance of data scale for NNs.
Sparse PLS methods by nature retain the input features useful for prediction, and therefore, R-spls SIMPLS can be used to illustrate the relative influence of markers and mass/sex/height. Using fold one of the training-set/test-set split, the movement type is confirmed as sidestep left by virtue of the greater emphasis on the markers of the right stance foot (RMT1, RCAL, and RLMAL) at the expense of those on the swing limb on the left (Table 3).
5 Conclusions
To the best of our knowledge, this is the first study which mines big data to predict GRF/Ms of a complex movement pattern from marker-based motion capture (and using a reduced marker set). We investigated the connection between PLS and the relationship of marker-based motion capture to force plate output. Using historical movement and force data (441 sidestep samples), and 11 PLS methods, we observed average correlation coefficients between ground truth and predicted of 0.9804 for GRFs and 0.9143 for GRMs thus proving our first hypothesis. This strongest response was predicted by the R-spls SIMPLS sparse method in support of our second hypothesis.
Our results using PLS methods against a complex sidestep movement pattern improved on those reported using a single hidden layer NN and a simple gait pattern by Oh et al. [31] illustrating the relevance of big data. We intend to extend this work through greater intra and inter-laboratory historical data, to analyze other movement patterns, validate in real-time with a dual data capture in the laboratory, then ultimately test in the field of play with outdoor cameras and less invasive methods of motion capture. The information provided by R-spls allows for fine-tuning of motion and force temporal input parameters, and an investigation of the relative importance of markers and the discrete features mass/sex/height. The success of PLS methods suggests this data is a candidate for deep learning. This study begins to address the significant barrier to non-invasive collection of real-time on-field kinetic data to inform athlete performance enhancement and injury prevention.
References
Alderson J, Johnson W (2016) The personalized ’digital athlete’: an evolving vision for the capture, modeling and simulation, of on-field athletic performance. In: 34th International conference on biomechanics in sports
AMTI (2004) Biomechanics platform instruction manual. Watertown, Advanced Mechanical Technology Inc. ISBN 1134298730
Barre A, Armand S (2014) Biomechanical toolkit: open-source framework to visualize and process biomechanical data. Comput Methods Programs Biomed 114(1):80–87. ISSN 0169-2607
Bartlett R (2007) Introduction to sports biomechanics: analysing human movement patterns. Routledge, New York. ISBN 1134298730
Besier T F, Sturnieks D L, Alderson J A, Lloyd D G (2003) Repeatability of gait data using a functional hip joint centre and a mean helical knee axis. J Biomech 36(8):1159–1168. ISSN 0021-9290
Bloomfield J (2012) From ugly duckling to soaring swan: the story of sport science at The University of Western Australia. Round House Press, South Fremantle
Boulesteix A-L (2004) PLS dimension reduction for classification with microarray data. Statist Appl Gene Molecul Biol 3(1):1–30. ISSN 1544-6115
Carse B, Meadows B, Bowers R, Rowe P (2013) Affordable clinical gait analysis: an assessment of the marker tracking accuracy of a new low-cost optical 3d motion analysis system. Physiotherapy 99(4):347–351. ISSN 0031-9406
Chen N, Urban S, Osendorfer C, Bayer J, Van Der Smagt P (2014) Estimating finger grip force from an image of the hand using convolutional neural networks and gaussian processes. In: 2014 IEEE International conference on robotics and automation (ICRA). IEEE, pp 3137–3142. ISBN 1479936855
Chiari L, Croce U D, Leardini A, Cappozzo A (2005) Human movement analysis using stereophotogrammetry: part 2 instrumental errors. Gait & Posture 21(2):197–211. ISSN 0966-6362
Chin A, Lloyd D, Alderson J, Elliott B, Mills P (2010) A marker-based mean finite helical axis model to determine elbow rotation axes and kinematics in vivo. J Appl Biomech 26(3):305–315. ISSN 1065–8483
Chun H, Keleş S (2010) Sparse partial least squares regression for simultaneous dimension reduction and variable selection. J R Statist Soc Series B (Statist Methodol) 72(1):3–25. ISSN 1467-9868
Dallalana R J, Brooks J H, Kemp S P, Williams A M (2007) The epidemiology of knee injuries in English professional rugby union. Amer J Sports Med 35(5):818–30. https://doi.org/10.1177/0363546506296738. ISSN 0363-5465 (Print) 0363-5465 (Linking)
De Bie T, Cristianini N , Rosipal R (2005) Eigenproblems in pattern recognition. Springer-Verlag, Berlin, pp 129–167. ISBN 978-3-540-28247-1
Dempsey AR, Lloyd D G, Elliott B C, Steele J R, Munro B J, Russo K A (2007) The effect of technique change on knee loads during sidestep cutting. Med Sci Sports Exer 39(10):1765–1773. ISSN 0195–9131
Domingos P (2012) A few useful things to know about machine learning. Commun ACM 55(10):78–87. ISSN 0001-0782
Donnelly C J, Lloyd D G, Elliott B C, Reinbolt J A (2012) Optimizing whole-body kinematics to minimize valgus knee loading during sidestepping: implications for acl injury risk. J Biomech 45(8):1491–1497. ISSN 0021–9290
Donnelly C J, Chinnasee C, Weir G, Sasimontonkul S, Alderson J (2016) Joint dynamics of rear-and fore-foot unplanned sidestepping. J Sci Med Sport, 1–6 (in press). ISSN 1440-2440
Elliott B, Alderson J (2007) Laboratory versus field testing in cricket bowling :a review of current and past practice in modeling techniques. Sports Biomech 6(1):99–108. ISSN 1476-3141
Filbay S R, Ackerman I N, Russell T G, Macri E M, Crossley K M (2014) Health-related quality of life after anterior cruciate ligament reconstruction: a systematic review. Amer J Sports Med 42(5):1247–1255. ISSN 0363–5465
Hashemi J, Breighner R, Chandrashekar N, Hardy DM, Chaudhari A M, Shultz S J, Slauterbeck J R, Beynnon B D (2011) Hip extension, knee flexion paradox: a new mechanism for non-contact acl injury. J Biomech 44(4):577–585. ISSN 0021–9290
Jones P L, Kerwin D G, Irwin G, Nokes LDM (2009) Three dimensional analysis of knee biomechanics when landing on natural turf and football turf. J Med Biol Eng 29(4):184–188. ISSN 1609-0985
Jung Y, Jung M, Ryu J, Yoon S, Park S-K, Koo Seungbum (2016) Dynamically adjustable foot-ground contact model to estimate ground reaction force during walking and running. Gait & Posture 45:62–68. ISSN 0966-6362
Liu T, Inoue Y, Shibata K (2010) A wearable ground reaction force sensor system and its application to the measurement of extrinsic gait variability. Sensors 10(11):10240–10255
Lloyd DG, Alderson J, Elliott BC (2000) An upper limb kinematic model for the examination of cricket bowling: a case study of Mutiah Muralitharan. J Sports Sci 18(12):975–982. ISSN 0264-0414
Manal KT, Thomas S (2002) Buchanan. Biomechanics of human movement. McGraw-Hill Professional, New York, pp 5.1–5.27. ISBN 0071356371
Mevik B-H, Wehrens R (2007) The pls package: principal component and partial least squares regression in R. J Statist Softw 18(2):1–24
Milner C E, Paquette M R (2015) A kinematic method to detect foot contact during running for all foot strike patterns. J Biomech 48(12):3502–3505. ISSN 0021-9290
Müller C, Sterzing T, Lange J, Milani T L (2010) Comprehensive evaluation of player-surface interaction on artificial soccer turf. Sports Biomech 9(3):193–205. ISSN 1476-3141
Mündermann L, Corazza S, Andriacchi T P (2006) The evolution of methods for the capture of human movement leading to markerless motion capture for biomechanical applications. J NeuroEng Rehab 3(6):j1—11. ISSN 1743-0003
Oh S E, Choi A, Mun J H (2013) Prediction of ground reaction forces during gait based on kinematics and a neural network model. J Biomech 46(14):2372–2380. ISSN 0021-9290
Shimokochi Y, Shultz S J (2008) Mechanisms of non-contact anterior cruciate ligament injury. J Athletic Train 43(4):396–408. ISSN 1062-6050
Sim T, Kwon H, Oh S E, Joo S-B, Choi A, Heo H M, Kim K, Mun J H (2015) Predicting complete ground reaction forces and moments during gait with insole plantar pressure information using a wavelet neural network. J Biomech Eng 137(9):091001:1–9. ISSN 0148-0731
Park HS, Shi J (2016) Force from motion: Decoding physical sensation in a first person video. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 3834–3842
R Core Team (2016) A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna. http://www.R-project.org
Ward S (2011) Anthropometric data and australian populations – do they fit? In: HFESA 47th Annual Conference 2011
Wei X, Chai J (2010) Videomocap: modeling physically realistic human motion from monocular video sequences. In: ACM Transactions on graphics (TOG), vol 29. ACM, pp 42. ISBN 1450302106
Winter D A (2009) Biomechanics and motor control of human movement. Wiley, Hoboken. ISBN 0470398183
Acknowledgements
This project was partially supported by the ARC Discovery Grant DP160101458 and an Australian Government Research Training Program Scholarship. We gratefully acknowledge NVIDIA for providing a GPU through its Hardware Grant Program, and Eigenvector Research for the loan licence of PLS_Toolbox. Portions of data included in this study have been funded by NHMRC grant 400937.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Johnson, W.R., Mian, A., Donnelly, C.J. et al. Predicting athlete ground reaction forces and moments from motion capture. Med Biol Eng Comput 56, 1781–1792 (2018). https://doi.org/10.1007/s11517-018-1802-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-018-1802-7