Abstract
Monitoring of cerebrovascular diseases via carotid ultrasound has started to become a routine. The measurement of image-based lumen diameter (LD) or inter-adventitial diameter (IAD) is a promising approach for quantification of the degree of stenosis. The manual measurements of LD/IAD are not reliable, subjective and slow. The curvature associated with the vessels along with non-uniformity in the plaque growth poses further challenges. This study uses a novel and generalized approach for automated LD and IAD measurement based on a combination of spatial transformation and scale-space. In this iterative procedure, the scale-space is first used to get the lumen axis which is then used with spatial image transformation paradigm to get a transformed image. The scale-space is then reapplied to retrieve the lumen region and boundary in the transformed framework. Then, inverse transformation is applied to display the results in original image framework. Two hundred and two patients’ left and right common carotid artery (404 carotid images) B-mode ultrasound images were retrospectively analyzed. The validation of our algorithm has done against the two manual expert tracings. The coefficient of correlation between the two manual tracings for LD was 0.98 (p < 0.0001) and 0.99 (p < 0.0001), respectively. The precision of merit between the manual expert tracings and the automated system was 97.7 and 98.7%, respectively. The experimental analysis demonstrated superior performance of the proposed method over conventional approaches. Several statistical tests demonstrated the stability and reliability of the automated system.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
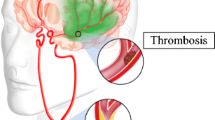
A total of 17.5 million people died from cardiovascular diseases (CVD) in 2012 representing 31% of all global deaths [58]. Arterial atherosclerosis is the primary reason for CVD. Figure 1 shows a representative illustration of atherosclerosis in the common carotid artery (CCA). Lumen narrowing due to plaque growth is currently measured via imaging techniques such as computed tomography (CT), magnetic resonance imaging (MRI) or conventional angiogram [21]. Carotid arterial diameters can be determined by using noninvasive B-mode ultrasound. However, considering the volumes of data available from a sonographic examination, visual analysis may be difficult and prone to error. Therefore, an automated delineation and measurement system would be valuable in standardizing quantitative assessment of luminal characteristics [22, 25, 47, 48].
Illustration of atherosclerosis in the common carotid artery. The plaque buildup in the carotid artery causes the narrowing of the lumen, disturbing proper blood flow to the brain. As the narrowing worsens, pieces of plaque can break free (embolize) and block blood vessels that supply blood to the brain leading to stroke. The enlarged cross-sectional view of plaque buildup inside the carotid artery is shown on the right side
The common carotid artery intima-media thickness (IMT) is currently considered as an early surrogate biomarker for cardiovascular diseases [5, 26, 57]. However, in practice, the presence of ultrasound imaging artifacts and low image quality [25], speckle noise [30], acoustic shadowing [28], echolucency variations [20, 22] and curvature of the vessels [43] makes the automated detection of the boundary interfaces of the intima-media complex (IMC) challenging [31]. Furthermore, evaluation of the IMT becomes harder with an increase in the age due to the increased presence of acoustic shadows (holes or echo dropouts) in the adventitia layer in older individuals [29, 59]. Automated carotid artery segmentation enables real-time measurements of diameters and can support the clinical evaluation of large databases. For example, the carotid arterial diameters have shown an association with myocardial infarction (MI) [6, 14]. The main advantage of using automated carotid lumen diameter (LD) or inter-adventitial diameter (IAD) as an imaging biomarker is to ensure that the measurements are more reliable, accurate and reproducible [34]. Hence, LD and IAD together are considered to be a relevant biomarker for evaluating the risk of atherosclerosis disease and are thus useful in screening of vulnerable patients. Finally, the automatic segmentation technique can be applied to a time series in order to monitor the changes in the carotid LD over the cardiac cycle. This in turn will be helpful in computing the arterial wall stiffness, a risk indicator for cardiovascular disease [17].
Several approaches have been proposed in the literature for the segmentation of the carotid artery lumen contours. This includes deformable contours (snakes) [32], Hough’s transform [19], dynamic programming [46] and classification-based strategies [34]. These approaches are summarized along with a comparative study in the discussion section. Many of the existing techniques proposed so far are either manual or semiautomatic which requires manual intervention such as the selection of a region of interest (ROI) or locating an initial seed point. However, the manual interventions are tedious and subjective. A few other automated studies have reported certain limitations in their techniques in the presence of atherosclerotic plaque [19]. Further, the region-based or boundary-based segmentation techniques developed so far are not fully able to handle the curvature associated with the carotid arteries. This motivated us to introduce an iterative spatial transformation-based technique for the segmentation of carotid artery.
This paper presents a fully automated algorithm for LD and IAD measurements in carotid arteries from B-mode ultrasound images. As in previous work [37], we use the assumption that the far wall of the CCA is the brightest region in the image in order to detect it. Further, we have assumed that the blood inside the lumen region has constant density [17] and can be considered to have a homogeneous region. Our algorithm is comprised of two stages. The stage 1 (global system) constitutes scale-space-based segmentation to identify the lumen axis. The stage 2 is the local processing where a spatial transformation is applied to straighten the curved vessels. Lumen-intima (LI) borders are then extracted using the scale-space approach, and then, the inverse spatial transformation is applied to map these LI borders back on to the original image. Since our empirical formulation of past automated methods on carotid IMT accuracy measurements has been 95% [33, 35], we use the same criteria for establishing the benchmark for LD/IAD precision measurement.
The paper is organized as follows. Section 2 describes the proposed method. Section 3 presents the experimental results and performance evaluation. Discussions are presented in Sect. 4, and the conclusions are drawn in Sect. 5.
2 Methods
2.1 Image database
Two hundred and two patients’ left and right common carotid artery (404 carotid images) B-mode ultrasound images were retrospectively analyzed (ethics approval with IRB was granted by Toho University, Japan) with mean age 69 ± 15.9 years. In this database of diabetic patients, there were 155 males (76.7%) and 47 females (23.3%) with a mean age of 67 and 75 years, respectively. These patients had a mean HbA1c of 6.28 ± 1.1 (mg/dl), glucose 108 ± 31 (mg/dl), LDL cholesterol 101.27 ± 31.6 (mg/dl), HDL cholesterol of 50.26 ± 14.8 (mg/dl) and total cholesterol of 175.04 ± 38 (mg/dl). The carotid ultrasound images were obtained using Toshiba scanner equipped with a 7.5-MHz linear array transducer by the same sonographer. These patients underwent both (a) B-mode carotid ultrasound using Toshiba scanner and (b) percutaneous coronary interventions using IVUS Boston Scientific® scanner, Marlborough, MA, that used iMAP software. Stable angina pectoris was defined as class I or II angina unchanged for more than 2 months or a positive stress test result. No special inclusion or exclusion criteria were adapted in selecting the 202 patients. The mean pixel resolution of the carotid B-mode scans was 0.05 ± 0.01 mm/pixel. The time period over which these data were acquired is from July 2009 to December 2010.
The manual delineation of the lumen as well as adventitia borders was done by two experienced neuroradiologists (one with 15 years of experience and second about 5 years of experience) using ImgTracer™ (AtheroPoint™, USA), a user-friendly commercial software [38]. The two experts selected 15–25 edge points proximal to the bulb in order to delineate the boundaries of the carotid artery. The numbers of points vary depending upon the length of the carotid artery. The observer had ability to zoom the image in the wall region for visualization of the wall region. The output of the ImgTracer™ was the ordered set of traced (x, y) coordinates.
2.2 Overview of automated lumen segmentation system
We modeled the LD/IAD segmentation system as a two-stage process: a global system which can establish the region of interest (ROI) and a local system which can model the lumen and adventitial borders. The global system combines the scale-space strategy embedded with pixel-classification approach. The power of scale-space can be used to tap the brightest intensity of the far (distal) wall of the common carotid artery, while the power of the pixel-classification approach can be used to tap the constant blood density region of the artery. Thus, the global system is able to extract the approximate lumen region which can be used as a guidance zone for the refined local processing for estimating the lumen borders. Prior to the iterative utilization of the scale-space recursive framework, the transformation concept is built which ensures that the curved vessels are straightened for refined and very accurate measurements for LD/IAD. This transformation framework uses lumen axis computed from the global system and then transformed using spatial transformation to ensure straight vessels.
Figure 2 shows the flow diagram of the automated system which works in an iterative manner. The preprocessing step includes automated cropping and denoising of the input image. Cropping helps in removing the text information from the image, while denoising reduces the effect of speckle which is usually present in ultrasound images [39]. After preprocessing, the image is fed to the global system which constitutes stage 1 of our algorithm. Scale-space-based strategy is employed at this stage in order to capture the bright edges of the adventitia wall which defines the region of interest (ROI). Here we used the assumption that the adventitia region is the brightest in an ultrasound image due to its high tissue density [13, 37]. The ROI covers region between the near and far media-adventitia (MA) walls which encloses the lumen region in it. Our second assumption is that the blood density to be constant over the lumen region and hence the intensity of pixels corresponding to this region must be constant [15]. This hypothesis allows us to use a pixel-based classifier to extract the lumen region. The lumen axis is then approximated as the mean of the near and far LI borders. The ROI containing the lumen axis is the input to the stage 2 (local processing). The spatial image transformation makes the curved vessel straight by using the lumen axis. LI borders are then extracted again using the scale-space approach, and then, inverse image transformation is applied to map these LI borders back on to the original image. The LD is measured as the mean distance between the near and far LI borders, and the IAD is measured as the distance between the near- and the far-wall MA interfaces. Both LD and IAD are measured using polyline distance metric (PDM) [53]. All operations were performed offline.
2.3 Scale-space-based segmentation
Scale-space-based segmentation technique is employed in order to capture the bright edges of the adventitia wall. The adventitial edges were delineated in two steps. The second-order derivative of a 5 × 5 Gaussian kernel [2, 40] is computed and convoluted with the cropped image to enhance the edges corresponding to the near and far adventitia walls. The optimal size of Gaussian kernel is set empirically. The standard deviation of Gaussian kernel (σ) must be chosen as the minimum MA wall thickness in pixels. Using the previous rationale, we have set the value of σ to 12 pixels in our experiments [33, 40]. Convolving an image with derivatives of the Gaussian kernel highlights features of the image with a size larger than the chosen scale σ [10].
We have used vertical spectral analysis of pixel columns to trace the adventitial borders. The proximal and distal adventitial borders can be approximated as the bright dots (peaks) in the spectrum. These bright dots are well separated by the dark lumen region in between. Thus, by using the spectral analysis technique, the adventitial borders are delineated accurately. In spectral analysis, rather than analyzing each column of the image pixels independently, we used a sliding window of size \(W \times L = 2 \times 5\) (in pixels) and estimated the mean intensity. This is to overcome any sudden spurious intensity in the spectrum. This is vital, as it will help in detecting the correct peaks in the spectrum under the scale-space paradigm. On increasing the width, the computation time will increase, but accuracy would not. By moving the aforementioned sliding window from bottom to top and from left to right, we were able to find the pair of peaks corresponding to MA borders. The peaks represent high-intensity value points with a pool of very small intensity values in between. The ROI is then defined as the region covered by these two MA borders.
Using the assumption of constant blood density, we choose a classifier which is suitable for soft tissue characteristics, such as K-mean classifier. K-means classifier with three pre-defined classes is used at this stage for separating the lumen region. These three classes represent low-intensity lumen region, medium-intensity plaque region and high-intensity wall region. The identified lumen region is then converted to binary form with white lumen region on a dark background. The holes in the resultant binary lumen were filled via connected component analysis [51] with a pixel connectivity of eight pixels.
The objective of including a denoising step in the overall pipeline is to reduce the speckle noise, thereby improving the accuracy of the system. Optimized Bayesian non-local means filter (OBNLM) [12] is used for this purpose. This edge-preserved smoothing approach will also help in patching the bleeding of the lumen region into the media region at some places along the lumen border. This bleeding otherwise produces over-estimation when tracing the lumen borders using the classifier.
2.4 Spatial transformation
The heart of the proposed system is a spatial transformation. The whole idea of introducing the spatial transformation is to improve the performance and stability of the system. To address this issue, we followed an approach presented in [62], where a spatial transformation is used to straighten the curved vessels for segmenting the contours of the intima-media complex in the carotid artery wall. Given an approximation of the lumen axis, the aim of the transformation is to generate a sub-image in which the anatomical interfaces become nearly horizontal. The transformed image I t is created by applying the spatial transformation T on the original I W sub-image, such that:
The transformation T consists of selecting L pixels above and below the lumen axis in I W . This approach can overcome the limitations of the existing methods that require the image interfaces to be horizontal at the time of acquisition. The scale-space-based segmentation technique is then again applied to retrieve the lumen region and LD borders in the transformed framework. The inverse transformation is applied to obtain the LD borders back on to the original image. Figure 3 illustrates the flow of stage 2 in detail.
The transformation approach used in [62] is for segmenting the IMT of the carotid artery on a single wall. However, our measurements include both the arterial walls which are more challenging. There are three main reasons for this. First, the near-wall region is low contrast compared to the far-wall region [37]. Second, the curvature plays a role which poses a challenge for an accurate LD estimation process. Third, the validation systems are tedious and expensive to produce, and this further requires increase in costs for tracings. Figure 4 illustrates the algorithmic steps of the proposed technique on a single image having curvature. A visual comparison of the results of both the proposed and the simple scale-space-based techniques is given in the next section.
Illustrating the algorithmic steps on a single image having curvature. a Curved ROI, b lumen axis on ROI, c transformed ROI, d adventitial borders on the transformed ROI, e binary lumen from classifier, f LD borders on transformed ROI, g LD borders on inverse transformed ROI, h LD borders on original ROI
2.5 Evaluation methodology
Two experts supported the manual delineation for computing the inter-observer variability. Statistical analyses were performed using MedCalc software [23]. The overall system’s performance was computed using the first method of precision of merit (PoM1) in percentage as [26]:
where
\({\text{LD}}_{{{\text{Auto}}_{i} }}\) is the measured automated lumen diameter and \({\text{LD}}_{{{\text{Manual}}_{i} }}\) is the measured manual lumen diameter of a particular image. N is the total number of images in the database.
This is a key feature that evaluated the automated traced diameter difference compared to the manual diameter difference. Similarly, the PoM1 for IAD measurements was computed by replacing LD with IAD in the above equation.
where
\({\text{IAD}}_{{{\text{Auto}}_{i} }}\) was the measured automated inter-adventitial diameter and \({\text{IAD}}_{{{\text{Manual}}_{i} }}\) was the measured manual lumen inter-adventitial diameter of a particular image. N is the total number of images in the database.
Second method of precision of merit (PoM2) computation was using error difference between the automated and manual methods for each individual case. This is the PoM per image basis. This is mathematically expressed as [2]:
Similarly, for IAD measurement:
We also used different statistical measures for evaluation of the performance of the automated system. We used regression plots to show the variability between manual tracings which is seen by the deviation from trend line. The Bland–Altman (BA) plot used the Bland–Altman method that demonstrated the level of agreement between two methods when measuring the same variable [4]. Friedman test is used for testing the difference between the sample medians [16, 24]. Further, two-sample Kolmogorov–Smirnov (KS) test [24] was used to verify that the samples follow the same distribution. Student’s t test was used for testing whether the (strong and highly significant) correlation can be regarded as equality or not [7]. In all the above tests, p values <0.05 were considered statistically significant unless otherwise specified. Cumulative frequency plots are shown to illustrate the error distributions. Box plots illustrate the median as a measure of central tendency. Dice similarity (DSC) and Jaccard index [3, 27] were computed to find the similarity between two regions.
3 Experimental results
3.1 Automated carotid LD (Auto LD) measurements
Mean carotid Auto LD measured was 6.58 ± 2.15 mm, while the two manual tracings were 6.43 ± 2.10 and 6.49 ± 2.14 mm, respectively. This indicates that there is no significant difference between the Auto LD measurements and the corresponding manual tracings. The performance evaluation of the Auto LD against the two manual tracings is summarized in Table 1.
3.2 Automated carotid IAD (Auto IAD) measurements
Mean carotid Auto IAD measured was 7.80 ± 0.98 mm, which is close to the two manual tracings, 7.76 ± 0.99 and 7.89 ± 1.00 mm, respectively. Performance of Auto IAD was validated against the two manual tracings and is summarized in Table 2.
Figure 5 illustrates the carotid Auto LD borders against the manual borders for both simple scale-space-based technique and the proposed iterative approach. Here, the simple scale-space-based technique means the same procedure excluding the spatial transformation-based iterative steps. This comparison is to demonstrate the improvement that the spatial transformation step adds. The result of simple scale-space-based technique is given on the left side, and the result of proposed approach is given on the right. It can be observed that the proposed approach improves the performance of the lumen segmentation in curved vessels. We also want to share the fact that 40% images in our database have plaque in it and 23% of them have jugular vein interference. The main criteria adapted for selecting the images which have plaque in it are by visually assessing it on the basis of region covered by plaque in the intimal region. Further, we noticed that these extended regions were hyperechoic (having brighter contrast) compared to the remaining wall region [55, 56, 61]. We also validated our visual description by computing the LD values, and they came out of as low as 3.46 mm. Figure 6 illustrates the LD and IAD borders along with the manual tracings on two sample images to support this fact.
Carotid Auto LD borders compared against the manual tracings on the grayscale images of four patients for both simple scale-space and transformation-based iterative methods. Carotid Auto LD borders are shown in solid white, while manual LD borders are shown in dashed white (Auto SS—Automated Simple scale-space, Auto SST—Automated scale-space with Transformation, Reader—Manual Reader)
Carotid Auto LD/IAD borders drawn on the grayscale images of patients having jugular vein interference (a) and thick plaque can be seen in (b). a Carotid Auto LD borders (solid white) versus Manual-1 LD borders (dashed white); b Auto LD borders (solid white) versus Manual-2 LD borders (dashed white); c Auto IAD borders (solid black) versus Manual-1 IAD borders (dashed black); d Auto IAD borders (solid black) versus Manual-2 IAD borders (dashed black)
3.3 Performance evaluation
Figure 6 compares the automated LD and IAD tracings against the two corresponding manual tracings on two sample images. Figure 6a, b shows the Auto LD against the two manual tracings, while Fig. 6c, d shows the corresponding Auto IAD against the two manual tracings.
3.3.1 Inter-observer variability
Inter-observer differences were estimated by calculating the correlation coefficient (CC) between the measurements on the same subject. The correlation coefficient (CC) between carotid Auto LD and the two manual tracings was 0.98 (p < 0.0001) and 0.99 (p < 0.0001), respectively. The scatter diagrams of Auto LD with respect to the two manual tracings are given in Fig. 7a, b. These diagrams show the closeness of the automated measurements with the manual one. The Bland–Altman plots of Auto LD with respect to the two manual tracings are given in Fig. 7c, d. Using two-sample KS test, the null hypothesis that the data samples are drawn from the same distribution is retained (p = 0.7533, p = 0.9659). Similarly, based on the result of paired t test (p < 0.0001, p < 0.0003), the null hypothesis is that the means of Auto LD and manual tracings are equal cannot be retained. As a consequence, the relation between carotid Auto LD and two manual tracings cannot be regarded as equality (see Table 5 in “Appendix”).
The correlation coefficient (CC) between carotid Auto IAD and the two manual tracings was 0.93 (p < 0.0001) and 0.94 (p < 0.0001), respectively. The scatter diagrams of Auto IAD with respect to the two manual tracings are given in Fig. 8a, b, respectively. The Bland–Altman plots of Auto IAD with respect to the two manual tracings are given in Fig. 8c, d, respectively. Using two-sample KS test, the null hypothesis that the data samples are drawn from the same distribution is retained (p = 0.9659, p = 0.4104). Similarly, based on the result of paired t test (p < 0.0093, p < 0.0001), the null hypothesis is that the means of Auto IAD and the two manual tracing are equal cannot be retained. As a consequence, the relation between carotid Auto IAD and the two manual tracings cannot be regarded as equality (see Table 6 in “Appendix”).
3.3.2 Friedman test
The Friedman test is a nonparametric test for testing the difference between several related samples. The null hypothesis for the Friedman test is that there are no differences between the sample medians. If the calculated p value is less than the selected significance level, then the null hypothesis will get rejected and it can be concluded that at least 2 of the sample medians are significantly different from each other. The detailed analysis of Friedman test is given in the “Appendix” (Tables 7, 8). It is observed that the null hypothesis got rejected for both LD and IAD samples with a very low p value (p < 0.00001). Further analysis has carried out after dividing the entire population into 10 equal parts (percentiles) based on mean absolute bias with respect to the manual tracings. It is observed that in the case of LD, the null hypothesis retained at the 50th percentile (with 50% of population) (p = 0.463). However, in the case of IAD, the null hypothesis got rejected even with 10% of the population.
This can be justified in the following way. With a large set of data (404 images) in hand, there is always a fair chance that, at least in a few cases, either of the manual or the auto-measurement goes wrong. This will reflect in the result of Friedman test which may not produce a high p value in this case. There are many factors which influence the measurement variability such as the presence of speckle noise and poor contrast on some images. Thus, there can be some pairs of values (Auto and Manual) which significantly differ. Another important point in this regard is that the consistency (regularity) should be maintained for the Friedman test in order to retain the null hypothesis. Here the consistency means the distribution of errors (or bias) of auto-measurement compared to manual as well as between the manual measurements. The signed error distribution for individual measurements can be seen directly from the Bland–Altman plots in Figs. 7 and 8 which is not in equal proportion in both positive and negative directions. There are several reasons for this such as the observer’s training (novice vs. experienced). We cannot always expect the observer’s tracings to be perfect because of lack of proper training.
There are some studies in the literature which used Friedman test to show the difference between the sample medians [8, 60]. These studies reported a low p value for the Friedman test. Further, the test is shown to be very sensitive to the regularity in the observations. If one of the readers consistently underestimates or overestimates the variable, irrespective of the magnitude of bias between them, the Friedman test rejects the null hypothesis. Along the same lines, our strategy has obtained consistent results for the Friedman test.
3.3.3 Cumulative frequency plots of signed and unsigned LD/IAD errors
Cumulative frequency plots of unsigned and signed Auto LD errors against manual tracings are given in Fig. 9a, b. The unsigned cumulative frequency plot shows the total number of measurements with less than a particular error value irrespective of the sign (positive or negative). From Fig. 9, it can be seen that above 90% of the LD measurements are within 1 mm error compared to the manual tracings. Similarly above 90% of the IAD measurements were taken with <0.5 mm error with respect to the two manual tracings. Further, it can be seen that the maximum error value not exceeds 2 mm in any of the cases. Figure 9c, d shows the cumulative frequency error plots for IAD.
Cumulative frequency analysis for Auto LD errors against manual tracings. a Unsigned LD error wrt Manual-1 (shown in dashed line with empty circles) and Manual-2 (shown in dotted line with filled circles); b signed LD error wrt Manual-1 (shown in dashed line with empty circles) and Manual-2 (shown in dotted line with filled circles). c Unsigned IAD error wrt Manual-1 (shown in dashed line with empty circles) and Manual-2 (shown in dotted line with filled circles); d Signed IAD error wrt Manual-1 (shown in dashed line with empty circles) and Manual-2 (shown in dotted line with filled circles)
3.3.4 Box plot analysis of Auto LD versus two manual tracings
Figure 10 shows box plots that illustrate the median as a measure of central tendency. As can be seen from the figure, there is little difference in the median values of Auto LD and manual LD tracings. The median is 6.21 mm for Auto LD and 6.16 mm for both manual LD tracings. Similarly, the median is 7.60 mm for both Auto IAD and Manual IAD tracings.
3.3.5 Dice similarity and Jaccard index
Two similarity coefficients, namely Dice similarity and Jaccard index, have been computed between automatically and manually extracted binary lumen regions (LR). Similarly, these similarity coefficients have been computed for binary inter-adventitial regions (IAR) and are given in Table 3. If both regions are equal, then the dice similarity or Jaccard index will be 100%.
4 Discussion
The objective of this work was the development of a fully automated system that is able to segment the carotid lumen and inter-adventitial borders accurately from B-mode ultrasound images. The automated system has been designed as a two-stage process. Stage 1 is the global system which combines scale-space and pixel-classification approach for the identification of lumen axis, whereas the stage 2 is designed as a local processing system where we used spatial transformation combined with iterative scale-space-based procedure to identify the lumen interfaces. Our automated system gives accurate segmentation results for both near and far LI borders, irrespective of the presence of LD, which are normal, moderately low and medium low representing varying plaque thicknesses. The main advantage of this strategy is that it prevents interference of vessel-like structures such as jugular vein and other muscles whose interfering intensities can affect the automated process. Further, we use a non-local mean-based denoising [12] to provide robustness to our system. The heart of the system is its ability to handle the curved vessels. This is due to the fact that spatial transformation was embedded in the scale-space framework in stage II of the system. Another secondary advantage of our system is that no matter how irregular the plaque is or how bright (hyperechoic) or less bright (hypoechoic) the plaque is, it circumvents these challenges to estimate the LD/IAD automatically. Finally, we demonstrated the comparison of the automated system (without and without spatial transformation) against the two manual expert tracings and showed the stability of the system.
A detailed overview and comparison of algorithms proposed so far for the carotid lumen segmentation is given in Table 4. Although similar works have been reported in the literature, a direct comparison to previously published results is not easy since different authors may have evaluated their algorithm on different image databases using different performance metrics [11, 19, 34, 46]. Second, as discussed below, some of the other methods make use of semi-automated techniques rather than fully automated and most of them are not measuring the LD and IAD. Hence, we have not included the performance criteria for these techniques in Table 4.
4.1 Brief survey and benchmarking
We selected the most recent research articles while benchmarking our proposed method against the previous published literature which can be seen in Table 4. Since we have considered only the recent publications, we suggest the readers to look into some of the older publications by Golemati et al. [19] and Cinthio et al. [11] which were based on Hough transform and threshold-based criteria. Further, we suggest readers to check on previous literature review in [35, 41, 42]. A combination of model-based line fitting approach was attempted by Molinari et al. [34]. This automated technique adapted an integrated approach for the automated tracing of CCA. The main components of this approach included geometric feature extraction, line fitting and classification. This produced the tracings of the proximal and distal adventitia layers. The system was not fully geared for LD measurement, but does, however, suggest computer vision-based model for automated segmentation of the lumen region. Further, the study highlights that the model faces challenge if the carotid walls are very close to jugular vein.
Rocha et al. [44] proposed a semiautomatic technique to segment the CCA in ultrasound images. The segmentation of the far and near adventitia boundaries adapted a RANSAC and cubic splines model on 50 longitudinal B-mode images. Further, Rocha et al. [45] applied a fuzzy classification-based approach for the automated segmentation of carotid arteries to detect the lumen axis. The edges corresponding to the intima and adventitia boundaries were detected and classified by extracting various features of the carotid wall interfaces and integrating it into a fuzzy score map. The results were validated using manual tracings. Even though there was an attempt to segment the CCA, there were no results on LD measurement. In continuation, Rocha et al. [46] attempted an automated detection of lumen axis in CCA based on dynamic programming. This method was validated on a total of 199 US scans. Even though the study reported a success rate of 99.5%, the near wall was not detected, and hence, the study did not report results on LD measurement.
The deformable model consisting of parametric and geometric snakes has dominated the imaging industry [54] and its applications [33, 36, 38, 52]. Using these foundational strategies, two authors prominently did parametric and geometric snakes for lumen segmentation. Loizou et al. [32] showed a snake-based segmentation approach for automatic segmentation of the carotid lumen on 50 longitudinal US images and measured the LD. The study reported an automated mean LD of 5.77 ± 0.99 mm against 5.59 ± 0.84 mm for manual tracings. The CC between the automated and the manual measurement was 0.63 (p = 0.001). The segmentation of carotid lumen region has been carried out by Santos et al. [49] based on Chan-Vese level set segmentation model. An anisotropic diffusion filter was applied for speckle removal followed by detection of the carotid artery using morphologic operator. Threshold-based region detection was based on the hypoechogenic (bright) characteristics of the lumen. The Chan-Vese model required an initialization of the wall contours. LD/IAD measurements were not reported in this study.
Sifakis et al. [50] proposed a fully automated real-time algorithm for carotid artery localization. This technique exploited basic statistics along with anatomical knowledge of the carotid artery. A statistics-based procedure was used for the identification of individual lumen center points. Then, a subset of the resulting lumen center points, designated as the “backbone,” is further processed to accurately estimate the CCA lumen position. A procedure based on threefold cross-validation was performed to validate the algorithm. The study did not discuss the LD measurement. This algorithm failed in the following cases: (a) non-uniform luminal intensity caused by high speckle content, (b) an abruptly curved arterial shape (e.g., due to a relatively large plaque) and (c) presence of a mimicking structure covering a significant part of the entire image width.
Recently, Carvalho et al. [9] proposed a method for lumen segmentation which included motion estimation from image sequences and image registration. The method is fully automatic, provided that the image contains just a single branch of the carotid artery. Further, the method quantifies carotid lumen geometry in subjects with atherosclerotic plaque from simultaneously acquired B-mode US and contrast-enhanced ultrasound (CEUS) image sequences. In order to compensate for the motion in the B-mode US and CEUS image sequence, the method averaged image intensities pixelwise over the complete sequence. This leads to a single integrated image which is used in the automated lumen segmentation process.
The work by Araki et al. [1] developed a model which combines the scale-space paradigm with the pixel-classification paradigm as global and local models, respectively. They benchmarked their approach against a boundary-based approach where the global shape is extracted first followed by lumen edge capturing using a level set paradigm. The method proposed in our current study is an improved version of the study presented in [1]. The proposed method here is different in its iterative procedure with the inclusion of a spatial transformation to handle the curved vessels, thereby adding robustness to the system. Further, the current study measures both LD and IAD. This is important since both LD and IAD together can become a relevant marker to evaluate the atherosclerosis disease unlike LD alone. We showed a significant improvement in CC values as depicted in Fig. 5. We benchmarked our LD measurement results as shown in Table 4 by comparing our strategy against [1] by using the same patient pool. Unlike other existing techniques, the proposed system has only one parameter σ which was set to 12 pixels (approximate size of minimum MA wall thickness in pixels). This σ needs to change only if there is a significant variation in the resolution of the image dataset. Hence, the proposed system can be easily adapted to a real-time clinical environment. We believe that it can support LD and IAD measurements in clinical practice and can be adapted for stroke monitoring.
4.2 Clinical interpretation
We believe that the LD and the IAD together might represent a biomarker of stroke risk. In patients with significantly high volume of plaque which causes luminal narrowing, LD will be smaller. Hence, LD is inversely related to stenosis severity. As a result, as LD decreases, it seems reasonable to believe that there may be a resultant increased risk of ischemic stroke. However, the plaque growth causes the adventitia region to bulge up according to the Glagov phenomenon [18], which causes the IAD to increase. This adaptive response is directly linked to the atherosclerotic process. Because of this positive remodeling, the relationship between LD and IAD is important to consider and together may be useful imaging biomarkers for stroke risk stratification. Future clinical investigations are warranted to further test the value of LD and IAD in stroke risk assessment in junction with the automated ultrasound measurement technologies that we and others have proposed.
4.3 Strengths and weakness
Utilizing our hypothesis on adventitia brightness, a fully automated method is used for measurement of the LD and IAD with a Gaussian derivative filter to trace the edges. In doing so, many challenges were addressed such as limits of resolution, data size, structural variations and inherent speckle noise in ultrasound scanning. To the best of our knowledge, this study is the first fully automated measurement method for LD and IAD which combines spatial transformation with iterative scale-space paradigm. However, we believe that multiresolution technique can be adapted to improve the automated system. More validation needs to be done on other ethnic groups from different countries. One future scope is to acquire temporal images from each patient to look at the cardiac cycle variations affecting the diameter estimations.
Our lumen region segmentation model is based on the assumption that the blood density is constant. It is very unlikely that this assumption can be disqualified. But there can be cases in which there is no blood in the arterial region during the image acquisition. This can be due to several reasons such (1) as blockage of the artery causing no blood in the other side of the stenosis or (2) non-uniform pumping of the blood from the heart to brain. In cases like these, there can be multiple grayscale contrasts. Such challenges can be considered by modeling it as a multi-class problem. Then, these multiple classes can be merged using the region-growing methods [55, 56].
5 Conclusions
We have developed a fully automated system for the accurate segmentation of carotid lumen and adventitial borders. The system uses iterative scale-space strategy embedded with spatial transformation. Stage one was a global processing system based on scale-space embedded with classifier to extract the region of interest. The spatial transformation was designed during the local processing to delineate the lumen and adventitial borders while handling the curved vessels which are otherwise difficult to compute using conventional methods. We validated our system against the two expert readers and demonstrated high correlations, accuracies and stability tests. Even though the current results are very promising, more multi-center evaluations need to be performed for establishing the automated lumen and IADs for adaption in clinical settings.
References
Araki T, Kumar KP, Suri HS et al (2016) Two automated techniques for carotid lumen diameter measurement: regional versus boundary approaches. J Med Syst 40(7):1–19
Babaud J, Witkin AP, Baudin M et al (1986) Uniqueness of the Gaussian kernel for scale-space filtering. IEEE Trans Pattern Anal Mach Intell 8(1):26–33
Bharatha A, Hirose M, Hata N et al (2001) Evaluation of three-dimensional finite element-based deformable registration of pre-and intraoperative prostate imaging. Med Phys 28(12):2551–2560
Bland JM, Altman DG (1986) Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 327(8476):307–310
Bots ML, Hoes AW, Koudstaal PJ et al (1997) Common carotid intima-media thickness and risk of stroke and myocardial infarction: the Rotterdam Study. Circulation 96(5):1432–1437
Bots ML, Grobbee DE, Hofman A et al (2005) Common carotid intima-media thickness and risk of acute myocardial infarction: the role of lumen diameter. Stroke 36(4):762–767
Box JF (1987) Guinness, Gosset, Fisher, and small samples. Stat Sci 2(1):45–52
Bray R, Derpapas A, Fernando R et al (2016) Does the vaginal wall become thinner as prolapse grade increases? Int Urogynecol J. doi:10.1007/s00192-016-3150-1
Carvalho DD, Akkus Z, van den Oord SC et al (2015) Lumen segmentation and motion estimation in B-mode and contrast-enhanced ultrasound images of the carotid artery in patients with atherosclerotic plaque. IEEE Trans Med Imaging 34(4):983–993
Cheng HD, Li J (2003) Fuzzy homogeneity and scale-space approach to color image segmentation. Pattern Recognit 36(7):1545–1562
Cinthio M, Jansson T, Eriksson A et al (2010) Evaluation of an algorithm for arterial lumen diameter measurements by means of ultrasound. Med Biol Eng Comput 48(11):1133–1140
Coupé P, Hellier P, Kervrann C et al (2009) Nonlocal means-based speckle filtering for ultrasound images. IEEE Trans Image Process 18(10):2221–2229
Delsanto S, Molinari F, Giustetto P et al (2007) Characterization of a completely user-independent algorithm for carotid artery segmentation in 2-D ultrasound images. IEEE Trans Instrum Meas 56(4):1265–1274
Eigenbrodt ML, Sukhija R, Rose KM et al (2007) Common carotid artery wall thickness and external diameter as predictors of prevalent and incident cardiac events in a large population study. Cardiovasc Ultrasound 5(1):1–11
Filardi V (2013) Carotid artery stenosis near a bifurcation investigated by fluid dynamic analyses. Neuroradiol J 26(4):439–453
Friedman M (1940) A comparison of alternative tests of significance for the problem of m rankings. Ann Math Stat 11(1):86–92
Gamble G, Zorn J, Sanders G et al (1994) Estimation of arterial stiffness, compliance, and distensibility from M-mode ultrasound measurements of the common carotid artery. Stroke 25(1):11–16
Glagov S, Weisenberg E, Zarins CK et al (1987) Compensatory enlargement of human atherosclerotic coronary arteries. N Engl J Med 316(22):1371–1375
Golemati S, Stoitsis J, Sifakis EG et al (2007) Using the Hough transform to segment ultrasound images of longitudinal and transverse sections of the carotid artery. Ultrasound Med Biol 33(12):1918–1932
Grønholdt MLM, Nordestgaard BG, Schroeder TV et al (2001) Ultrasonic echolucent carotid plaques predict future strokes. Circulation 104(1):68–73
Gupta A, Nair S, Schweitzer AD et al (2012) Neuroimaging of cerebrovascular disease in the aging brain. Aging Dis 3(5):414–425
Gupta A, Kesavabhotla K, Baradaran H et al (2015) Plaque echolucency and stroke risk in asymptomatic carotid stenosis a systematic review and meta-analysis. Stroke 46(1):91–97
Hollander M, Wolfe DA, Chicken E (2013) Non-parametric statistical methods. Wiley, Hoboken
Ikeda N, Araki T, Dey N et al (2014) Automated and accurate carotid bulb detection, its verification and validation in low quality frozen frames and motion video. Int Angiol 33(6):573–589
Ikeda N, Gupta A, Dey N et al (2015) Improved correlation between carotid and coronary atherosclerosis SYNTAX score using automated ultrasound carotid bulb plaque IMT measurement. Ultrasound Med Biol 41(5):1247–1262
Jaccard P (1912) The distribution of the flora in the alpine zone. New Phytol 11(2):37–50
Jodas DS, Pereira AS, Tavares JMR (2016) A review of computational methods applied for identification and quantification of atherosclerotic plaques in images. Expert Syst Appl 46(2016):1–14
Lamont D, Parker L, White M et al (2000) Risk of cardiovascular disease measured by carotid intima-media thickness at age 49–51: life course study. Br Med J 320(7230):273–278
Loizou CP, Pattichis CS, Christodoulou CI et al (2005) Comparative evaluation of despeckle filtering in ultrasound imaging of the carotid artery. IEEE Trans Ultrason Ferroelectr Freq Control 52(10):1653–1669
Loizou CP, Pattichis CS, Pantziaris M et al (2007) Snakes based segmentation of the common carotid artery intima media. Med Biol Eng Comput 45(1):35–49
Loizou CP, Kasparis T, Spyrou C et al (2013) Integrated system for the complete segmentation of the common carotid artery bifurcation in ultrasound images. Artif Intell Appl Innov 412(1):292–301
Molinar F, Meiburger KM, Saba L et al (2012) Constrained snake vs. conventional snake for carotid ultrasound automated IMT measurements on multi-center data sets. Ultrasonics 52(7):949–961
Molinari F, Zeng G, Suri JS (2010) An integrated approach to computer based automated tracing and its validation for 200 common carotid arterial wall ultrasound images. J Ultrasound Med 29(3):399–418
Molinari F, Zeng G, Suri JS (2010) A state of the art review on intima–media thickness (IMT) measurement and wall segmentation techniques for carotid ultrasound. Comput Methods Programs Biomed 100(3):201–221
Molinari F, Zeng G, Suri JS (2011) Inter-greedy technique for fusion of different segmentation strategies leading to high-performance carotid IMT measurement in ultrasound images. J Med Syst 35(5):905–919
Molinari F, Krishnamurthi G, Acharya UR et al (2012) Hypothesis validation of far-wall brightness in carotid-artery ultrasound for feature-based IMT measurement using a combination of level-set segmentation and registration. IEEE Trans Instrum Meas 61(4):1054–1063
Molinari F, Meiburger KM, Saba L et al (2012) Fully automated dual-snake formulation for carotid intima-media thickness measurement a new approach. J Ultrasound Med 31(7):1123–1136
Molinari F, Meiburger KM, Zeng G et al (2012) Automated carotid IMT measurement and its validation in low contrast ultrasound database of 885 patient Indian population epidemiological study: results of AtheroEdge™ Software. Int Angiol 31(1):42–53
Molinari F, Pattichis CS, Zeng G et al (2012) Completely automated multiresolution edge snapper—a new technique for an accurate carotid ultrasound IMT measurement: clinical validation and benchmarking on a multi-institutional database. IEEE Trans Image Process 21(3):1211–1222
Molinari F, Meiburger KM, Saba L et al (2012) Ultrasound IMT measurement on a multi-ethnic and multi-institutional database: our review and experience using four fully automated and one semi-automated methods. Comput Methods Programs Biomed 108(3):946–960
Molinari F, Meiburger KM, Zeng G et al (2012) Carotid artery recognition system: a comparison of three automated paradigms for ultrasound images. Med Phys 39(1):378–391
Rocha R, Campilho A, Silva J et al (2010) Segmentation of the carotid intima-media region in B-mode ultrasound images. Image Vis Comput 28(4):614–625
Rocha R, Campilho A, Silva J et al (2011) Segmentation of ultrasound images of the carotid using RANSAC and cubic splines. Comput Methods Programs Biomed 101(1):94–106
Rocha R, Silva J, Campilho A (2012) Automatic segmentation of carotid B-mode images using fuzzy classification. Med Biol Eng Comput 50(5):533–545
Rocha R, Silva J, Campilho A (2014) Automatic detection of the carotid lumen axis in B-mode ultrasound images. Comput Methods Programs Biomed 115(3):110–118
Saba L, Raz E, di Martino M et al (2015) Is there an association between asymmetry of carotid artery wall thickness (ACAWT) and cerebrovascular symptoms? Int J Neurosci 125(6):456–463
Saba L, Araki T, Kumar KP et al (2016) Carotid inter-adventitial diameter is more strongly related to plaque score than lumen diameter: an automated tool for stroke analysis. J Clin Ultrasound 44(4):210–220
Santos AMF, Tavares JMRS, Sousa L et al (2013) Automatic segmentation of the lumen of the carotid artery in ultrasound B-mode images. Expert Syst Appl 40(16):6570–6579
Sifakis EG, Golemati S (2014) Robust carotid artery recognition in longitudinal B-mode ultrasound images. IEEE Trans Image Process 23(9):3762–3772
Soille P (2013) Morphological image analysis: principles and applications. Springer Science & Business Media, Berlin
Suri JS, Laxminarayan S (2002) PDE and level sets. Springer Science & Business Media, Berlin
Suri JS, Haralick RM, Sheehan FH (2000) Greedy algorithm for error correction in automatically produced boundaries from low contrast ventriculograms. Pattern Anal Appl 3(1):39–60
Suri JS, Liu K, Singh S et al (2002) Shape recovery algorithms using level sets in 2-D/3-D medical imagery: a state-of-the-art review. IEEE Trans Inf Technol Biomed 6(1):8–28
Suri JS, Yuan C, Wilson DL (eds) (2005) Plaque imaging: pixel to molecular level, vol 113. IOS Press, Amsterdam
Suri JS, Kathuria C, Molinari F (2010) Atherosclerosis disease management. Springer Science & Business Media, Berlin
Suri JS, Ikeda N, Gupta A et al (2015) 2083060 A new technique for compartmental-IMT estimation in presence of bulb in carotid ultrasound scans: a stroke risk assessment system. Ultrasound Med Biol 41(4):S74
WHO factsheet on cardiovascular disease. (Last updated on January 2015). http://www.who.int/mediacentre/factsheets/fs317/en/
Wilhjelm JE, Gronholdt ML, Wiebe B et al (1998) Quantitative analysis of ultrasound B-mode images of carotid atherosclerotic plaque: correlation with visual classification and histological examination. IEEE Trans Med Imaging 17(6):910–922
Wu Q, Yang B, Cao C et al (2016) Age-dependent impact of inferior alveolar nerve transection on mandibular bone metabolism and the underlying mechanisms. J Mol Histol. doi:10.1007/s10735-016-9697-9
Xiao G, Brady M, Noble JA et al (2002) Segmentation of ultrasound B-mode images with intensity inhomogeneity correction. IEEE Trans Med Imaging 21(1):48–57
Zahnd G, Orkisz M, Sérusclat A et al (2014) Simultaneous extraction of carotid artery intima-media interfaces in ultrasound images: assessment of wall thickness temporal variation during the cardiac cycle. Int J Comput Assist Radiol Surg 9(4):645–658
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Dr. Jasjit S. Suri has a relationship with AtheroPoint™, Roseville, CA, USA, which is dedicated to Atherosclerosis Disease Management including Stroke and Cardiovascular imaging.
Rights and permissions
About this article
Cite this article
Krishna Kumar, P., Araki, T., Rajan, J. et al. Accurate lumen diameter measurement in curved vessels in carotid ultrasound: an iterative scale-space and spatial transformation approach. Med Biol Eng Comput 55, 1415–1434 (2017). https://doi.org/10.1007/s11517-016-1601-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-016-1601-y