Abstract
The seasonal surplus and putrefactive property of moist forages inevitably increase the pressure on environmental protection and residual grass disposal. In the current work, the anaerobic fermentation approach was adopted to assist the sustainable recycling of leftover Pennisetum giganteum (LP), and its chemical composition, fermentation performance, bacterial community and functional profiles during anaerobic fermentation were studied. Fresh LP was spontaneously fermented for up to 60 d. At the end of anaerobic fermentation, fermented LP (FLP) displayed homolactic fermentation with low pH value, ethanol, and ammonia nitrogen concentrations but high lactic acid concentration. Weissella was dominant in 3-day FLP, yet Lactobacillus was the overwhelming genus (92.6%) in 60-day FLP. The anaerobic fermentation process promoted (P < 0.05) the metabolism of carbohydrate and nucleotide while suppressing (P < 0.05) that of lipid, cofactors, vitamins, energy, and amino acid. The results showed that the residual grass with LP as an example could be successfully fermented even if no additives were added, without signs of clostridial and fungal contamination.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Feed production has been tightly bound up with the environment, as it is a basic industry that requires resources including energy, land, water, and labor. Considering that the growing world population has increased the demand for animal products, the relationship is likely to get even closer in the future. Forage grass cultivation, a pivotal ring of the feed production industry, has obvious seasonal and regional characteristics all year round (Garcez Neto et al. 2020). According to the feedback of forage grass cultivators, forage yields commonly surpass utilization requirements. Meanwhile, Li et al. (2022) reported that, sometimes, the plants are not used effectively, leading to a considerable amount of residue. The disposal of the residual grass becomes a hot potato because the surplus with high moisture is discarded directly in the fields without any treatments, which easily causes effluent production, thereby inducing groundwater pollution and germ breeding (Mattioli et al. 2017; Yin et al. 2021). It was reported that this type of effluent produces 200 times more pollution than untreated domestic sewage (Woolford 1978). Moreover, severe deamination and protein degradation during plant spoilage cannot be ignored because haze pollution and secondary inorganic aerosols occur when the above degradation products are released and reacted with surrounding acids.
Pennisetum giganteum (P. giganteum Z. X. Lin or giant JUNCAO) is an emerging economic grass that has been cultivated in more than 30 China provinces and more than 80 countries (Li et al. 2020). It is a C4 plant with high water, light and nitrogen use efficiency as well as great biomass yield. Nowadays, P. giganteum is mainly used in the production of fiber, biofuels, and animal feeds. It is reported that P. giganteum can be mowed 6 to 8 times a year, producing 254 tons of fresh grass per hectare (Zhang et al. 2013). With such a high biomass yield, the problem of seasonal surplus and large accumulation of P. giganteum exists in many countries. Although these biomass resources are low-cost, easy-to-obtain and rich-in-nutrient and attracting increasing attention (Li et al. 2022). However, in the evaluation of resource waste, forage waste is often less concerned than food waste, and few studies emphasize the waste of forage. Based on the concept of resource recovery and clean production, long-term conservation of leftover P. giganteum (LP) by anaerobic fermentation may transform the original linear economy into a circular one and ease the burden of waste management and environmental protection (de Mello Santos et al. 2022; Huang et al. 2023).
Forage anaerobic fermentation (ensilage) refers to the fundamentals that epiphytic microbes, especially lactic acid bacteria (LAB), metabolizing substrates of forages to generate organic acids, and quickly reducing the pH of forages, thereby powerfully suppressing the activity of undesirable microbes, to accomplish the goal of long-term conservation (Weinberg and Muck 1996). With the deepening of anaerobic fermentation studies, the traditional approach has been unable to satisfy the need for a clear understanding of microbial ecology during anaerobic fermentation. In recent years, high-throughput sequencing or even single molecule real-time sequencing has assisted scientists to analyze the microbial community structure and diversity involved in different microenvironments (McAllister et al. 2018). Furthermore, microbial functional annotation is increasingly found to be extremely important for microbial ecology studies (Asshauer et al. 2015). The prediction of functionalities involving microbial communities can be recommended as an effective supplement to high-throughput sequencing or single molecule real-time sequencing (Kanehisa and Goto 2000) and will be potentially beneficial for anaerobic fermentation research.
Hence, this work distinguished the fermentation performance, bacterial community and functionality of LP during anaerobic fermentation, and the results can supply a reference for the clean recovery of residual grass.
Materials and methods
Material collection
The LP (P. giganteum outgrown the needs for utilization) was freely supplied by Baima National Agricultural-tech Zone (latitude 31.61°N, longitude 119.18°E, elevation 25.1 m, Jiangsu, China). After collection, LP was cut into approximately 20-mm lengths by a fodder cutter. Subsequently, the cut LP was thoroughly mixed for the subsequent trial.
Anaerobic fermentation preparation
About 0.45 kg of well-mixed LP was loaded into an ultraviolet-light-sterilized laboratory-scale silo (300 × 400-mm polythene plastic bag), vacuum-sealed and kept under surrounding temperature (29 ± 2 ℃). Triplicate silos were opened and sampled on days 1, 3, 7, 15, 30, and 60 of anaerobic fermentation, respectively.
Fermentation product, chemical composition, and microbial population analyses
For fermentation product analysis, approximately 40 g fresh LP or FLP was extracted with 120 mL distilled water at 4 °C for 30 min. After filtering with 4 layers of sterilized cheesecloth and filter paper, the pH of fresh LP or FLP was recorded by a HI 2221 pH/mV/°C bench meter (Hanna Instruments Inc., Rhode Island, USA) immediately. The ammonia nitrogen (NH3-N) concentration of FLP was determined by colorimetry after a reaction with phenol and sodium hypochlorite (Broderick and Kang 1980). The fermentation products (ethanol, 1,2-propanediol, and lactic acid (LA), acetic acid (AA), propionic acid (PA), and butyric acid (BA)) of FLP were determined by a 1260 Infinity II HPLC system (Agilent Technologies Inc., California, USA). The eluent was 2.5 mM H2SO4; the flow rate of eluent was 0.5 mL/min; and the run temperature was 55 °C. The chemical composition and microbial population were analyzed, according to our previous work (Zhao et al. 2018). In brief, dry matter (DM) was determined after oven-drying at 105 °C for 15 min and at 65 °C for 48 h. Water soluble carbohydrate (WSC) content was determined by the anthrone-sulfuric acid method. Neutral detergent fiber (NDF) and acid detergent fiber (ADF) content were analyzed with an Ankom 200 fiber analyzer (Ankom Technology, New York, USA). Total nitrogen (TN) content was analyzed by a Kjeltec 8200 Kjeldahl nitrogen analyzer (Foss Analytics, Höganäs, Sweden). Crude protein (CP) content was calculated by multiplying the TN content by 6.25. After homogenizing with autoclaved saline solution (0.85% NaCl) at a ratio of 1:9 and shaken at 37 °C and 120 rpm for 2 min. Then, 1 mL above solution was serially diluted for microbial counting, and the rest solution was filtered with two layers of sterilized gauze for subsequent bacterial DNA extraction. LAB were counted on de Man, Rogosa and Sharp agar media after anaerobic incubation at 37 °C for 3 days. Aerobic bacteria were counted on nutrient agar medium after aerobic incubation at 37 °C for 2 days. Yeasts and molds were counted on potato dextrose agar medium after aerobic incubation at 30 °C for 2 days. Enterobacteria were counted on violet red bile glucose agar medium after aerobic incubation at 37 °C for 1 day. The microbial count was transformed to a Log10-scale in the form of colony-forming units (CFU) and expressed on a fresh material (FM) basis.
High-throughput sequencing and functional prediction analyses
Preparatory work before sequencing (DNA extraction + PCR amplification) was conducted following the procedures of Zhao et al. (2022a). Concretely, the solution used for microbial DNA extraction was centrifuged at 10,000 g for 15 min at 4 °C to collect microbial precipitates. The FastPrep® Instrument and FastDNA® Spin Kit (MP Biomedicals, California, USA) were used to extract bacterial DNA. The quantity and quality of extracted DNA were detected by 1% sodium borate agarose gel electrophoresis and NanoDrop 2000 ultraviolet–visible spectrophotometer (260/280 nm, Thermo Scientific, Delaware, USA). According to the designated sequencing area, specific primers with barcode were synthesized. The V3-V4 hypervariable region of the bacterial 16S rRNA gene was amplified by ABI GeneAmp® 9700 PCR amplification instrument (Applied Biosystems, California, USA). The primers were 338F (5'-ACTCCTACGGGAGGCAGCAG-3') and 806R (5'-GGACTACHVGGGTWTCTAAT-3'). The PCR products of the same sample were mixed and detected by 2% agarose gel electrophoresis. The PCR amplification products were recovered by AxyPrep® DNA Gel Recovery Kit (Axygen Biotechnology Co., Ltd., California, USA), eluted with Tris–HCl, and purified by 2% agarose gel electrophoresis. According to the preliminary quantitative results of electrophoresis, the purified PCR products were quantitatively detected by QuantiFluor TM-ST blue fluorescence quantitative system (Promega Biotechnology Co., Ltd., Wisconsin, USA), and then mixed in proportion according to the sequencing requirements of each sample. Purified amplicons pooled at equimolar concentrations were paired-end sequenced on a MiSeq PE300 (Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China). The generated paired-end sequencing data were first merged using FLASH (ver. 1.2.11) and quality-filtered by QIIME (ver. 1.9.1) to retain sequences with quality scores > 20. Then, operational taxonomic units (OTUs) were clustered with 97% identities by UPARSE (ver. 7.0.0), and chimeric sequences were eliminated using UCHIME (ver. 4.1). Finally, the taxonomy of OTUs was assigned from phylum to genus using RDP classifier (ver. 2.11) by matching with SILVA database (ver. 132) with 70% confidence threshold. Bacterial alpha diversity reflected by Shannon, Chao1, Ace, Sobs, Simpson, and Coverage indices were visualized using ggplot2 R package (ver. 3.3.6). Bray–Curtis metric beta diversities were calculated using QIIME (ver. 1.9.1), processed by Bray–Curtis distance metric principal coordinates analysis (PCoA), and visualized using vegan R package (ver. 4.1.2). Stream graphs showing the bacterial community successions was constructed using ggplot2 R package (ver. 3.3.6). Spearman’s correlation heatmap between fermentation product and bacterial community was constructed using phearmap R package (ver. 1.0.12). The bacterial Kyoto Encyclopedia of Genes and Genomes (KEGG) functionalities in LP, FLP-3 and FLP-60 were predicted through the Tax4fun developed by Asshauer et al. (2015).
Statistical analysis
The effects of storage days on the chemical composition, fermentation characteristics and microbial number of FLP were investigated through the general linear model of SAS software (SAS Institute Inc., North Carolina, USA). Meanwhile, the student’s t-test and analysis of similarity (ANOSIM) with 999 permutations were adopted to statistically compare the difference in alpha diversity and beta diversity of bacterial community, respectively. The statistical differences are significant at P < 0.05.
Results
Characteristics of fresh LP
For chemical composition, the fresh LP had a WSC content of 14.2% DM and a CP of 10.8% DM. The value of BC was 30.8 mEq/kg DM. As for microbial population, the LAB number was higher than 5.0 log10 CFU/g FM. The number of aerobic bacteria, yeast, mold, and enterobacteria was 7.93, 5.60, 5.03, and 7.08 log10 CFU/g FM, respectively (Table 1).
Fermentation products of FLP
Storage days significantly (P < 0.05) affected the pH value, the concentrations of LA, AA, and PA, and the LA/AA of FLP (Table 2). The pH sharply (P < 0.05) declined within the first 7 days of anaerobic fermentation with the lowest value (3.50) on day 30 of anaerobic fermentation, then slightly (P > 0.05) increased. The variation of LA was opposite to that of pH, with a maximum value of 74.1 g/kg DM on day 30. The AA and volatile fatty acid (VFA) concentrations constantly increased with the progress of storage days. During the anaerobic fermentation, the LA/AA showed a trend of rising first and then falling with the maximum on day 30 of anaerobic fermentation. No BA was detected in all FLPs. The WSC content as well as the NH3-N concentration were significantly (P < 0.05) affected by storage days. As anaerobic fermentation proceeded, the NH3-N concentration significantly (P < 0.05) increased, and the DM and WSC contents significantly (P < 0.05) decreased.
As shown in Table 3, storage days significantly (P < 0.05) affected the number of LAB, aerobic bacteria, yeast, molds, and enterobacteria in FLP. The LAB number rapidly increased within the initial 3 days of anaerobic fermentation, peaked on day 15 of anaerobic fermentation and then decreased gradually. Differently, aerobic bacteria, yeast, and mold number gradually reduced to low or undetected levels.
Bacterial community structure of fresh LP and FLP
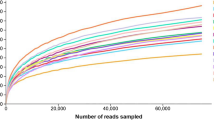
The bacterial alpha and beta diversities of fresh LP and FLP are shown in Fig. 1. The Shannon, Chao1, Ace, and Sobs indices were highest in LP and lowest in FLP-60. The Coverage of each sequenced sample was more than 99% (Fig. 1 A). The PCoA plot revealing the difference of bacterial β-diversity is shown in Fig. 1 B. The symbols of LP, FLP-3, and FLP-60 were well distributed in different quadrants.
At the bacterial phylum level (Fig. 2 A, C), Proteobacteria and Firmicutes were the dominant phyla in fresh LP, accounting for 50.7% and 41.8% of the relative abundance (RA) of bacterial community, respectively. After 3-day anaerobic fermentation, the RA of Proteobacteria, Actinobacteriota, and Bacteroidota decreased while the RA of Firmicutes increased. At the end of anaerobic fermentation (day 60), Firmicutes was the overwhelming phylum (> 90%) in FLP.
Bacterial community composition and succession of fresh LP and FLP. A Relative abundance of bacterial community at the phylum level; B relative abundance of bacterial community at the genus level; C bacterial community successions during anaerobic fermentation of LP are aggregated and colored on a stream-graph by phylum; D bacterial community successions during anaerobic fermentation of LP are aggregated and colored on a stream-graph by genus. The numbered treatments (e.g., LP-1, LP-2, and LP-3) are replicates. LP, leftover P. giganteum; FLP, fermented LP
At the bacterial genus level (Fig. 2 B, D), Paenibacillus (19.0%), Bacillus (15.9%), Enterobacter (15.9%), Acinetobacter (15.3%), and Pantoea (7.92%) were the main genera in fresh LP. With the progress of anaerobic fermentation, the RA of Paenibacillus, Bacillus, Acinetobacter, Exiguobacterium, Sphingomonas, etc. sharply decreased to a low level (< 1%). At the end of anaerobic fermentation (day 60), Lactobacillus predominated in the bacterial community of FLP, with RA accounting for 92.6%.
Correlation analysis of fermentative products and bacterial communities
As shown in Spearman’s correlation heatmap (Fig. 3), Lactobacillus was positively correlated with LA (R = 1.000; P < 0.01) and AA (R = 0.886; P < 0.05) concentration. Negative correlations were found between LA concentration and Enterococcus (R = − 0.943, P < 0.01), Leuconostoc (R = − 0.943, P < 0.01) or Pediococcus (R = − 0.886, P < 0.05). Weissella, Leuconostoc, and Enterobacter were negatively correlated with AA (R = − 0.943, P < 0.01; R = − 0.943, P < 0.01; R = − 0.943, P > 0.05) concentration, while Lactobacillus was positively correlated with AA (R = 0.886, P < 0.05) concentration.
Spearman’s correlation heatmap of fermentation parameters and top 30 genera in FLP. Red squares refer to a positive correlation (0 < r < 1), whereas green squares refer to a negative correlation (− 1 < r < 0). *P < 0.05; **0.001 < P ≤ 0.01; ***P ≤ 0.001. DM, dry matter; WSC, water soluble carbohydrates; PA, propionic acid; NH3-N, ammonia nitrogen; AA, acetic acid; LA, lactic acid; LA/AA, the ratio of lactic to acetic acid
KEGG functional prediction of bacterial communities
In Fig. 4, the abundance of “Metabolism” was much higher than that of other functional categories under pathway level 1. Under pathway level 2, “carbohydrate and amino acid metabolism” were the major functional categories of “Metabolism.” The abundance of “carbohydrate and amino acid metabolism” was significantly (P < 0.05) different among fresh LP, FLP-3, and FLP-60. Concretely, FLP-60 had the highest “carbohydrate metabolism” abundance, followed by FLP-3 and then fresh LP; whereas the “amino acid metabolism” abundance showed an opposite trend, with the maximum in fresh LP and the minimum in FLP-60. “Carbohydrate and amino acid metabolism” were further subdivided in pathway level 3 (Fig. 5). For “carbohydrate metabolism,” the abundance of “amino sugar,” nucleotide sugar, starch, and sucrose,” “galactose, fructose, and mannose metabolism,” “pentose and glucuronate interconversions,” “pentose phosphate pathway,” and “glycolysis/gluconeogenesis” was significantly (P < 0.05) increased in FLP, compared with fresh LP, while the abundance of “C5-branched dibasic acid, butanoate, propanoate, glyoxylate, dicarboxylate, pyruvate, ascorbate, and aldarate metabolism” and “citrate cycle” was significantly (P < 0.05) decreased in FLP in comparison with fresh LP. And for amino acid metabolism, except for the abundance of “lysine biosynthesis,” all the abundance of “amino acid metabolism” categories was significantly (P < 0.05) decreased in FLP, compared with fresh LP.
Discussion
Analysis of fresh material
For anaerobic fermentation, the content of available substrate and the number of key microbes are two critical factors affecting the fermentation quality and efficiency (McDonald et al. 1991; Dong et al. 2020). In the current work, the WSC (14.2% DM) and LAB (5.58 log10 CFU/g FM) levels of fresh LP met the minimum requirement for LA fermentation (Cai et al. 1999). And the low BC (30.8 mEq/kg DM) of fresh LP suggested that an acidic microenvironment is easy to establish via anaerobic fermentation (McDonald et al. 1991). However, the high number (> 7.0 log10 CFU/g FM) of aerobic bacteria and enterobacteria might cause challenges to fermented feed production.
Effect of storage days on the fermentation performance of FLP
The pH decline rate during anaerobic fermentation has long been used as an indicator to determine whether the fermented feed is effectively preserved (Kung and Shaver 2001). It is commonly known that the slower the pH decline rate, the greater the nutrient loss risk. In this work, the low pH value and high LA concentration in FLP could be associated with the sufficient WSC and LAB levels as well as low BC of fresh LP, which effectively suppressed undesirable microbes, thus explaining the insignificant DM difference and relatively high residual WSC content. The AA concentration increased along with anaerobic fermentation, which was similar to most studies, and this might be related to the activity of acetate-producing microbes in the initial stage and the transformation of homolactic fermentation to heterolactic fermentation in the late stage. The butyrate produced by clostridia is easily detected in silage with DM < 30% (van Niekerk et al. 2010). Although the DM content of LP was < 30%, no BA concentration was detected in all FLP, suggesting that no clostridial fermentation occurred in FLP. The VFA are organic acids with 1–6 carbon chains, including acetic acid, propionic acid, isobutyric acid, and n-butyric acid, which are undesirable to silage production (Wang et al. 2020). Considering the undetected or negligible contents of butyric acid and propionic acid, the dynamic change of volatile organic acid content in this study was mainly influenced by acetic acid content. Therefore, VFA content showed the same trend as acetic acid with anaerobic fermentation days.
The generation of NH3-N is the result of protein degradation and amino acid deamination. The concentration of NH3-N should be no more than 10% of TN in high-quality silage (McDonald et al. 1991). While the final NH3-N concentration of FLP (10.0% TN) was just at the maximum allowed, it indicates that the proteolysis of FLP was barely acceptable. The protein degradation, amino acid decarboxylation, and ammonia generation during anaerobic fermentation are a complex biochemical process, involving the activities of plant protease, enterobacteria, clostridia, etc. The degradation extent of forage protein and amino acids relies on the pH decline rate during anaerobic fermentation (van Niekerk et al. 2010). Thus, the acceptable NH3-N concentration of FLP could ascribe to the effective pH decline in the initial stage (0–7 days) of anaerobic fermentation.
The high number of LAB in FLP was because the sufficient WSC content of fresh LP (142 g/kg DM) accelerated the reproduction of LAB. As expected, the strict aerobe (aerobic bacteria and molds) number sharply decreased to an undetected level under the oxygen-free microenvironment. As for facultative anaerobic yeast and enterobacteria, their negligible number in FLP could be related to its low pH value. Furthermore, the reduction of yeasts in FLP was also associated with fast LAB proliferation because yeasts are less competitive in acidic and anaerobic habitats (Zhao et al. 2021).
Effect of storage days on the bacterial community structure of FLP
All the Coverage (> 99%) indicated that most of bacterial community had been fully captured. The anaerobic fermentation process reduced the bacterial alpha diversities of fresh LP, which was associated with the inactivation of acid-intolerant aerobes. Mendez-Garcia et al. (2015) reported that the decline of pH can reduce microbial diversity in the habitat. The sufficient WSC and LAB levels in fresh LP ensured rapid reproduction of LAB and quick inhibition of undesirable microbes, thus decreasing the bacterial alpha diversities of FLP, which was reflected by the lowest indices of Shannon, Chao1, Ace, and Sobs in FLP-60 with the lowest pH value.
In the PCoA plot, the differences of microbial community composition are directly visualized as distances between symbols. The symbols of LP, FLP-3, and FLP-60 were clearly distributed in various quadrants showing the distinct bacterial community compositions of fresh LP before and after anaerobic fermentation, which also ascribed to the inactivation of acid-intolerant aerobes after anaerobic fermentation as abovementioned.
The distinct succession of Proteobacteria by Firmicutes during anaerobic fermentation resulted from the inhibition of aerobes (Sphingomonas, Pseudomonas, etc.) and the thriving of LAB (Weissella, Lactococcus, Lactobacillus, etc.). The anaerobic fermentation environment benefited Firmicutes because this phylum is representative bacteria under anaerobic and acidic conditions (Zhao et al. 2017). Similar to Muck et al. (2018) and Zhao et al. (2021), Lactococcus, Weissella and Lactobacillus were the 3 most common genera during anaerobic fermentation in this work. Lactococcus and Weissella are widely accepted as initial colonists during anaerobic fermentation (Muck 2013; Graf et al. 2016) because the initial acid environment created by Lactococcus and Weissella lends themselves to the subsequent growth of Lactobacillus (Cai et al. 1998). Thus, with the progress of anaerobic fermentation, the FLP bacterial community was initially dominated by Weissella (d 3) and eventually dominated by Lactobacillus (day 60).
Effect of storage days on the potential functionalities of bacterial community in FLP
High-throughput sequencing is a potent tool for assessing microbial ecology but does not directly provide insights into microbial metabolic potential, even though functional diversity is critical for microbial community analysis (Asshauer et al. 2015). Among PICRUSt and Tax4fun tools, the superior Tax4fun was adopted to reveal the potential bacterial functionalities in fresh LP and FLP. Anaerobic fermentation can effectively inhibit the metabolism of harmful microbes by establishing an oxygen-free microenvironment and acidifying effect. Unsurprisingly, the anaerobic fermentation process effectively suppressed the bacterial metabolism of fresh LP. Previous studies found that the functional categories of bacterial metabolism involved in ensilage were dominated by carbohydrate, amino acid, cofactors, vitamins, energy, and nucleotide metabolism (Guan et al. 2020; Bai et al. 2021; Guo et al. 2023). Similarly, the results of the present work presented that those metabolisms were more abundant than other metabolisms in the microbiota of fresh LP and FLP. Ensilage or anaerobic fermentation is based on the principle that available carbohydrate in forage is metabolized to mainly lactate by LAB in anaerobic environments, which could interpret the increase of carbohydrate metabolism abundance in FLP. Amino acids as essential substrates for organisms (microorganisms are no exception) play a significant role in protein synthesis and primary metabolism. The complex and diverse epiphytic microorganisms might explain the higher amino acid metabolism abundance in fresh LP. And the lower amino acid metabolism abundance in FLP was due to its low pH. It was previously reported that the acidic condition can suppress the amino acid metabolism of harmful microorganisms (Flythe and Russell 2004). The cofactor and vitamin metabolism abundance was decreased along the anaerobic fermentation process, which was in line with the results of Zhao et al. (2022b). Bai et al. (2021) stated that the inoculation of functional LAB increased the cofactor and vitamin metabolism abundance and compensated for vitamin loss during ensiling. Hence, LAB with vitamin-producing functions can be recommended in practice to further improve the feeding value of FLP. Different from the study of Xu et al. (2021), the anaerobic fermentation process did not increase the energy metabolism abundance of LP. The energy metabolism of LAB is essential for LA fermentation during anaerobic fermentation (Pessione et al. 2010) and should be promoted in satisfactory anaerobic fermentation theoretically (Xu et al. 2021). These inconsistent results are difficult to explain and need to be further studied. Nucleotides are known for their ability to supply energies for cellular processes and synthesize nucleic acid (Kilstrup et al. 2005). Strangely, the variation of energy metabolism abundance during anaerobic fermentation was opposite to that of nucleotide metabolism abundance. Thus, it is necessary to exactly clarify the bacterial metabolic pathways during anaerobic fermentation by means of emerging omics approaches.
To further trace the key bacterial functionalities in fresh LP and FLP, the important carbohydrate and amino acid metabolism were specifically explored. The metabolism abundance of most sugar components was increased with anaerobic fermentation, confirming that LAB can utilize multiple carbon sources for growth and reproduction. Of which, the higher fructose, mannose, and galactose metabolism abundance in 60-day fermented samples could be mainly associated with Lactobacillus. Differently, the TCA cycle abundance was increased by anaerobic fermentation. The inhibition of the TCA cycle after anaerobic fermentation could be due to the consumption of O2 as this pathway should be conducted under aerobic conditions (Banfalvi 1991). Glycolysis/gluconeogenesis is essential for LAB to carry out homolactic and heterolactic fermentations; while the pentose phosphate (PPP) pathway is a unique metabolic pathway of heterofermentative LAB, compared with homofermentative LAB (Abdel-Rahman et al. 2011). Thus, the higher abundance of glycolysis/gluconeogenesis in the late stage of anaerobic fermentation (day 60) might be associated with the abundant homofermentative Lactobacillus, and the high relative abundance of heterofermentative Weissella (day 3) might interpret the high abundance of PPP pathway. Almost all amino acid metabolism abundance was decreased by anaerobic fermentation and the decreased abundance of valine, leucine, isoleucine, and lysine degradation might primarily account for the acceptable NH3-N concentration (< 100 g/kg TN) in FLP.
It should be noted that high-throughput sequencing has a very limited taxonomic resolution; these functional profiles are highly speculative and should be examined with caution. The predicted functional profiles should be further validated by transcriptomics, proteomics, and metabolomics studies to provide clear indications.
Correlation of fermentative products and bacterial communities of FLP
In FLP, AA concentration was positively related to Lactobacillus but negatively related to Weissella, Leuconostoc, and Enterobacter, implying that the AA production in FLP was associated with the heterofermentative strains of Lactobacillus. While the distinct negative correlations between LA concentration and Enterococcus or Pediococcus were unexpected because it is inconsistent with the conventional understanding of cocci-type LAB promoting LA fermentation. This is probably because Enterococcus or Pediococcus was succeeded by Lactobacillus as LA accumulated and pH decreased, resulting in a low RA of Enterococcus or Pediococcus detected in FLP-60 with high LA concentration. Normally, forage protease, enterobacteria, and/or clostridia are involved in NH3-N generation during anaerobic fermentation (McDonald et al. 1991). However, no clostridia were detected in FLP, and there was a slightly negative relationship between NH3-N and Enterobacter. The above results implied that enterobacteria and clostridia could not explain the generation of NH3-N in FLP. The NH3-N generation during anaerobic fermentation of LP might ascribe to forage protease, but it needs further study.
Conclusions
Although LP is regarded as “waste grass,” it still has good characteristics with high content of WSC and CP. The enthusiasm of growers for the management of residual grass may depend on the value of reusing grass. The anaerobic fermentation technique showed great potential to improve the added value of LP reflected by the fermentation performance, bacterial communities, and functionalities and could be a sustainable solution for residual grass. It is thus important to encourage participation in this way to reduce the waste of resources, and this can be achieved by publishing publicity information and making instructions.
Data availability
The sequencing data underlying this article are available in the SRA Database of NCBI and can be accessed with the accession number of PRJNA938912.
References
Abdel-Rahman MA, Tashiro Y, Sonomoto K (2011) Lactic acid production from lignocellulose-derived sugars using lactic acid bacteria: overview and limits. J Biotechnol 156:286–301. https://doi.org/10.1016/j.jbiotec.2011.06.017
Asshauer KP, Wemheuer B, Daniel R, Meinicke P (2015) Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31:2882–2884. https://doi.org/10.1093/bioinformatics/btv287
Bai J, Ding Z, Ke W et al (2021) Different lactic acid bacteria and their combinations regulated the fermentation process of ensiled alfalfa: ensiling characteristics, dynamics of bacterial community and their functional shifts. Microb Biotechnol 14:1171–1182. https://doi.org/10.1111/1751-7915.13785
Banfalvi G (1991) Conversion of oxidative energy to reductive power in the citrate cycle. Biochem Educ 19:24–26. https://doi.org/10.1016/0307-4412(91)90138-x
Broderick GA, Kang JH (1980) Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J Dairy Sci 63:64–75. https://doi.org/10.3168/jds.S0022-0302(80)82888-8
Cai Y, Benno Y, Ogawa M, Ohmomo S, Kumai S, Nakase T (1998) Influence of Lactobacillus spp. from an inoculant and of Weissella and Leuconostoc spp. from forage crops on silage fermentation. Appl Environ Microbiol 64:2982–2987. https://doi.org/10.1128/AEM.64.8.2982-2987.1998
Cai Y, Benno Y, Ogawa M, Kumai S (1999) Effect of applying lactic acid bacteria isolated from forage crops on fermentation characteristics and aerobic deterioration of silage. J Dairy Sci 82:520–526. https://doi.org/10.3168/jds.S0022-0302(99)75263-X
de Mello Santos VH, Campos TLR, Espuny M, de Oliveira OJ (2022) Towards a green industry through cleaner production development. Environ Sci Pollut Res Int 29:349–370
Dong Z, Shao T, Li J, Yang L, Yuan X (2020) Effect of alfalfa microbiota on fermentation quality and bacterial community succession in fresh or sterile Napier grass silages. J Dairy Sci 103:4288–4301. https://doi.org/10.3168/jds.2019-16961
Flythe MD, Russell JB (2004) The effect of pH and a bacteriocin (bovicin HC5) on Clostridium sporogenes MD1, a bacterium that has the ability to degrade amino acids in ensiled plant materials. FEMS Microbiol Ecol 47:215–222. https://doi.org/10.1016/s0168-6496(03)00259-9
Garcez Neto AF, dos Santos TM, da Silva J, Fernandes SR (2020) Effect of whey permeate and Lactobacillus buchneri on biomass conservation, chemical characteristics and aerobic stability of elephant grass silage. Waste Biomass Valori 12:879–893. https://doi.org/10.1007/s12649-020-01035-z
Graf K, Ulrich A, Idler C, Klocke M (2016) Bacterial community dynamics during ensiling of perennial ryegrass at two compaction levels monitored by terminal restriction fragment length polymorphism. J Appl Microbiol 120:1479–1491. https://doi.org/10.1111/jam.13114
Guan H, Shuai Y, Ran Q et al (2020) The microbiome and metabolome of Napier grass silages prepared with screened lactic acid bacteria during ensiling and aerobic exposure. Anim Feed Sci Technol 269:114673. https://doi.org/10.1016/j.anifeedsci.2020.114673
Guo X, Xu D, Li F, Bai J, Su R (2023) Current approaches on the roles of lactic acid bacteria in crop silage. Microb Biotechnol 16:67–87. https://doi.org/10.1111/1751-7915.14184
Huang F, Wang T, Zhang J et al (2023) Exploring the bacterial community succession and metabolic profiles of Lonicera japonica Thunb. residues during anaerobic fermentation. Bioresour Technol 367:128264. https://doi.org/10.1016/j.biortech.2022.128264
Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. https://doi.org/10.1093/nar/28.1.27
Kilstrup M, Hammer K, Ruhdal Jensen P, Martinussen J (2005) Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol Rev 29:555–590. https://doi.org/10.1016/j.femsre.2005.04.006
Kung L Jr, Shaver R (2001) Interpretation and use of silage fermentation analysis reports. Focus Forage 3:1–5
Li Q, Xiang C, Xu L et al (2020) SMRT sequencing of a full-length transcriptome reveals transcript variants involved in C18 unsaturated fatty acid biosynthesis and metabolism pathways at chilling temperature in Pennisetum giganteum. BMC Genomics 21:52. https://doi.org/10.1186/s12864-019-6441-3
Li X, Chen F, Wang X et al (2022) Innovative utilization of herbal residues: exploring the diversity of mechanisms beneficial to regulate anaerobic fermentation of alfalfa. Bioresour Technol 360:127429. https://doi.org/10.1016/j.biortech.2022.127429
Mattioli A, Boscaro D, Dalla Venezia F et al (2017) Biogas from residual grass: a territorial approach for sustainable bioenergy production. Waste Biomass Valor 8:2747–2756
McAllister TA, Duniere L, Drouin P et al (2018) Silage review: using molecular approaches to define the microbial ecology of silage. J Dairy Sci 101:4060–4074. https://doi.org/10.3168/jds.2017-13704
McDonald P, Henderson AR, Heron SJE (1991) The biochemistry of silage, 2nd edn. Chalcombe Publication, Marlow, Bucks, UK
Mendez-Garcia C, Pelaez AI, Mesa V, Sanchez J, Golyshina OV, Ferrer M (2015) Microbial diversity and metabolic networks in acid mine drainage habitats. Front Microbiol 6:475. https://doi.org/10.3389/fmicb.2015.00475
Muck RE (2013) Recent advances in silage microbiology. Agr Food Sci 22:3–15. https://doi.org/10.23986/afsci.6718
Muck RE, Nadeau EMG, McAllister TA, Contreras-Govea FE, Santos MC, Kung L Jr (2018) Silage review: recent advances and future uses of silage additives. J Dairy Sci 101:3980–4000. https://doi.org/10.3168/jds.2017-13839
Pessione A, Lamberti C, Pessione E (2010) Proteomics as a tool for studying energy metabolism in lactic acid bacteria. Mol Biosyst 6:1419–1430. https://doi.org/10.1039/c001948h
van Niekerk WA, Hassen A, Bechaz FM (2010) Influence of growth stage at harvest on fermentative characteristics of Panicum maximum silage. S Afr J Anim Sci 40:334–341
Wang M, Franco M, Cai Y, Yu Z (2020) Dynamics of fermentation profile and bacterial community of silage prepared with alfalfa, whole-plant corn and their mixture. Anim Feed Sci Technol 270:114702. https://doi.org/10.1016/j.anifeedsci.2020.114702
Weinberg ZG, Muck RE (1996) New trends and opportunities in the development and use of inoculants for silage. FEMS Microbiol Rev 19:53–68. https://doi.org/10.1111/j.1574-6976.1996.tb00253.x
Woolford MK (1978) The problem of silage effluent. Herbage Abstr 48:397–403
Xu D, Wang N, Rinne M et al (2021) The bacterial community and metabolome dynamics and their interactions modulate fermentation process of whole crop corn silage prepared with or without inoculants. Microb Biotechnol 14:561–576. https://doi.org/10.1111/1751-7915.13623
Yin X, Wu J, Tian J, Wang X, Zhang J (2021) Dried soybean curd residue: a promising absorbent for cleaner production of high-quality silage. J Clean Prod 324:129300. https://doi.org/10.1016/j.jclepro.2021.129300
Zhang J, Lei H, Li J et al (2013) Growth performance of Pennisetum sp. at different elevation. Guizhou Agric Sci 41:112–115 (in Chinese with English abstract)
Zhao X, Liu J, Liu J et al (2017) Effect of ensiling and silage additives on biogas production and microbial community dynamics during anaerobic digestion of switchgrass. Bioresour Technol 241:349–359. https://doi.org/10.1016/j.biortech.2017.03.183
Zhao J, Dong Z, Li J et al (2018) Ensiling as pretreatment of rice straw: the effect of hemicellulase and Lactobacillus plantarum on hemicellulose degradation and cellulose conversion. Bioresour Technol 266:158–165. https://doi.org/10.1016/j.biortech.2018.06.058
Zhao J, Yin XJ, Wang SR, Li JF, Shao T (2021) Separating the effects of chemical and microbial factors on fermentation quality and bacterial community of Napier grass silage by using gamma-ray irradiation and epiphytic microbiota transplantation. Anim Feed Sci Technol 280:115082. https://doi.org/10.1016/j.anifeedsci.2021.115082
Zhao J, Yin X, Dong Z et al (2022a) Using gamma-ray irradiation and epiphytic microbiota inoculation to separate the effects of chemical and microbial factors on fermentation quality and bacterial community of ensiled Pennisetum giganteum. J Appl Microbiol 132:1675–1686. https://doi.org/10.1111/jam.15323
Zhao J, Yin XJ, Wang SR, Li JF, Dong ZH, Shao T (2022b) Changes in the fermentation products, taxonomic and functional profiles of microbiota during high-moisture sweet sorghum silage fermentation. Front Microbiol 13:967624. https://doi.org/10.3389/fmicb.2022.967624
Funding
This work was financially supported by the National Natural Science Foundation of China (NSFC, Grant No. 32201464), Jiangsu Funding Program for Excellent Postdoctoral Talent (Grant No. 2022ZB319), and United Fund for Regional Inno & Dev of NSFC (Grant No. U20A2003).
Author information
Authors and Affiliations
Contributions
Investigation, data curation, Visualization, and writing — original draft preparation: J. Z.; methodology, software, formal analysis, and visualization: Z. J. and X. Y.; methodology, formal analysis, data curation, and project administration: J. L; funding acquisition and investigation: Z. D. and S. W.; resources, supervision, data curation, funding acquisition, and writing — review and editing: T. S.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Informed consent was obtained from all the individual participants included in the work.
Conflict of interest
The authors declare no competing interests..
Additional information
Responsible Editor: Diane Purchase
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, J., Jing, ZD., Yin, XJ. et al. Sustainable utilization of residual grass: effect of anaerobic storage days on chemical composition, fermentation performance, microbial community, and functional profiles of Pennisetum giganteum. Environ Sci Pollut Res 31, 38866–38877 (2024). https://doi.org/10.1007/s11356-023-26524-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-023-26524-1