Abstract
Textile effluent is one of the most hazardous industrial pollutant sources. It is generated in huge volumes and contains a wide array of toxicants. Reactive azo dyes, which are xenobiotic compounds, are predominantly utilized by textile industries for dyeing cotton, viscose, wool, and silk. The conventional physicochemical treatments used by industrial effluent treatment plants are ineffective in dye degradation. The present study thus attempted to find a potential treatment for reactive azo dyes. A novel bacterial consortium VITPBC6 was constructed with the most potent and compatible reactive orange 16 (RO-16) decolorizing isolates of tannery and textile effluents, and the isolates were identified as Bacillus flexus VITSP6, Bacillus paraflexus VITSPB7, Bacillus megaterium VITSPB9, Bacillus firmus VITEPB1, B. flexus VITEPB2, and Bacillus aryabhattai VITEPB3. The physicochemical factors of RO-16 decolorization were optimized by response surface methodology. Consortium VITPBC6 was able to tolerate a high concentration of RO-16 up to 800 mg L−1. A cocktail of enzymes including azoreductase, tyrosinase, laccase, lignin peroxidase, and manganese peroxidase was involved in RO-16 degradation by VITPBC6. Consortium VITPBC6 degraded RO-16 following zero-order reaction. The enzymes of consortium VITPBC6 had a Vmax of 352 mg L−1 day−1 for RO-16 degradation; however, the Km value was high. VITPBC6 biodegraded RO-16 resulting in the formation of small aromatic compounds. Lastly, different toxicity assays conducted with untreated RO-16 and its corresponding biodegraded metabolite revealed that the toxicity of biodegraded metabolites was significantly lower than the untreated dye.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Environmental pollution is a persistent global phenomenon, which consequently affects biodiversity, ecosystems, and human health. Globally, the textile industry is the second largest polluter of water and contributes 20% in the production of the world’s wastewater (Yaseen and Scholz 2019). The major hindrance in treating textile effluent is caused by industrial dyes. According to global estimations, 280,000 tons of textile dyes are released along with the effluents annually (Madamwar et al. 2019). Reactive azo dyes in particular are popularly utilized in textile industries as they are water soluble, anionic, cost-effective, available in bright shades of wide range, simple in application, and excellent in terms of light-fastness and wash-fastness. Reactive azo dyes are compounds that form covalent linkage with the textile fibers and contain the azo group as their chromophores. Azo dyes constitute a major fraction (95%) of reactive dyes (Pal 2017). These can be chemically characterized as dyes containing the functional azo group “–N = N–” joining two symmetrical and/or asymmetrical similar or non-azo alkyl or aryl radical. These dyes form the second largest class among all other dyes (Benkhaya et al. 2020). However, a major concern with the use of reactive dyes is their poor fixation property. The dyeing process leads to the loss of approximately 20–50% of unfixed reactive dyes along with effluent (Ghaly et al. 2014). The reactive azo dyes are stable and xenobiotic compounds. These remain unaffected during conventional effluent treatment processes. Therefore, the discharge of dye containing effluent directly into environment can cause serious damage to nearby water-bodies, aquatic organisms, soil, animals, and human beings. Dyes discharged in water bodies obstruct sunlight penetration, disturb physicochemical factors (e.g., dissolved oxygen (DO), biological oxygen demand (BOD), chemical oxygen demand (COD), total dissolved solids (TDS), pH), hamper neurosensory and metabolic system in fishes thereby affecting their mortality rate, and destabilize the food chain by suffocating flora and fauna (Ayed et al. 2017; Hassan and Carr 2018). On the other hand, soil contaminated with dye affects its fertility, disrupts photosynthesis in plants, hinders seed germination and growth rate of plants, and lowers plant respiration (Rehman et al. 2018; Ikram et al. 2022a). These persistent hazardous compounds affect biomagnification (Erkurt 2010). Dyes are known to be allergenic, mutagenic, and carcinogenic to human beings. Frequent cases of contact dermatitis, allergic conjunctivitis, rhinitis, asthma, and other allergic responses have been reported after occupational exposure. Colon and bladder cancer occurs commonly upon dye exposure (Ikram et al. 2022b).
Bioremediation by microorganisms is a potential and promising approach for dye removal. Microbial organisms, including bacteria, algae, filamentous fungi, yeasts, and actinomycetes, have demonstrated absorption, transformation, reduction, degradation, or mineralization of hazardous compounds (Saratale et al. 2011, 2013; Ajaz et al. 2020). Bacterial azo dye degradation and mineralization has been the topic of research for a very long time. Bacteria are of particular interest due to their high rate of dye degradation. Some effective azo dye decolorizing bacterial strains include Bacillus sp., Pseudomonas stutzeri, Aeromonas hydrophila, Enterococcus sp., Providencia spp., and Pseudomonas rettgeri (Bhatia et al. 2017; Joshi et al. 2020; Saha and Rao 2020; Srinivasan and Sadasivam 2021). It was observed that mixed bacterial cultures or consortia were able to degrade azo dyes to a greater extent than that of the pure cultures. Microbial communities are metabolically diverse, which reflects in their ability to utilize various dye molecules as carbon sources (Stolz 2001; Shade et al. 2012). The synergistic relationship between the different microbial species results in these species attacking dye molecule at different positions. The degradation product thus generated can further be used by remaining microbial species. In contrast, pure bacterial cultures are less versatile in terms of their biodegradation potential, as they can degrade specific dye molecules (Jadhav et al. 2010; Holkar et al. 2016). Guo et al. (2020a) formulated consortium CG-1 consisting of Zobellella, Rheinheimera, and Marinobacterium that efficiently degraded a variety of dye substrates (metanil yellow, direct blue B, acid black ATT, and acid violet 7). In another study, azo dye reactive red 170 was effectively degraded by a consortium of Bacillus subtilis, Brevibacillus borsstelensis, and Bacillus firmus (Barathi et al. 2022).
In the current study, a bacterial consortium that was adapted to industrial effluents was constructed for reactive orange 16 (RO-16) degradation. The physicochemical factors affecting bacterial dye decolorization were optimized to maximize dye decolorization. The mechanism of RO-16 degradation was studied in detail. The enzymes responsible for dye degradation were identified, and reaction kinetics were studied. Furthermore, the dye degraded metabolites were characterized to uncover the probable dye degradation pathway. Finally, toxicity assays were performed to assess any toxicity present in the dye degraded metabolites.
Materials and methods
Chemicals used
Pure reactive dye RO-16 was procured from a textile industry in Mysuru, Karnataka. Concentrated dye stock solution was prepared at 20 g L−1 in milli-q water. Stock solutions were sterilized through 0.22-µm cellulose acetate (CA) syringe filters and stored in sterile polyethylene bottles. Dilutions were constituted from the stock solution for further studies. Maximum absorbance of each of the dyes was noted by scanning the region 200–700 nm in a UV–Vis spectrophotometer (Shimadzu UV-1800).
Microbiological media were purchased from the HiMedia Laboratories Pvt. Ltd., India. Different inorganic and organic chemicals were bought from Sisco Research Laboratories Pvt. Ltd., India; Thomas Baker (chemicals) Pvt. Ltd, India; and Fischer Scientific India Pvt. Ltd. All the chemicals were analytical grade and pure.
Industrial sample collection
An industrial effluent sample was collected from a tannery industry at Ambur, Tamilnadu (12.7904° N and 78.7166° E). Samples were collected in polyethylene bottles using the grab sampling technique. The samples were then stored in iced coolers, and transported to laboratory for further processing (Simpson 2017).
Industrial sample characterization
The collected effluent samples were filtered through Whatman filter paper no. 42 (diameter 90 mm) to eliminate large particulate matters. The resulting filtrate was then used to determine different physicochemical parameters. The quantity of color was measured based on the colorimetric, ADMI method and the specific color of the samples was identified by the spectroscopic method as described by APHA (1992). Temperature and pH were measured with a thermometer (Thomas Scientific) and pH meter (Hanna instruments) respectively. Conductivity meter (Hanna instruments, HI98312 DIST) operated at 25 ℃ was used to note the electrical conductivity (EC). A DO meter and sensor (Hanna Instruments, Portable Dissolved Oxygen Meter — HI9146) were used for detecting DO. BOD, COD, total solids (TS), TDS, and total suspended solids (TSS) were measured as described by APHA (1992) and USEPA (1978). The cationic constituents (Ca2+, Mg2+, Na+, Cu+, Zn2+, Fe3+, total Cr content, Cd2+, and Ni2+) of the sample were quantified using a flame atomic absorption spectrophotometer (AAS) (Agilent, model 55B AA). Anionic constituents Cl− and SO42− were determined using the Mohr method and turbidimetric method respectively (Rice et al. 2017).
Isolation and screening and quantification of the decolorization potential of dye decolorizing bacteria from industrial samples
Indigenous bacteria from the leather industry sample were isolated following enrichment method, as was reported in our previous study (Saha and Rao 2020). Bacterial colonies with distinct morphology were selected and preserved as pure cultures. The isolated bacteria were then primarily screened in test tubes containing Luria Bertani (LB) broth amended with 50 mg L−1 of RO-16. After inoculation (1%), the test tubes were kept in an incubator at 37 °C (TECHNICO Digital Bacteriological Incubator TLPPL 104) until visible decolorization occurred (6 days).
Decolorization assay was carried out with potent isolates in decolorizing media (DM), as described in our previous study (Saha and Rao 2020). Decolorization was quantified in percentage using the following equation:
Compatibility assay of all the dye decolorizing potent isolates
Compatibility of three potent isolates that were identified in our previous study (Saha and Rao 2020) and three potent isolates identified in this study was used for the purpose of building up of a consortium. This assay was carried out in Mueller Hinton (MH) agar plates. Bacterial isolates were inoculated in nutrient broth (NB) and incubated at 37 ℃ for 24 h to obtain seed cultures. Two bacterial isolates were swabbed and streaked against each other in each plate. The plates were then incubated at 37 ℃ for 48 h. Compatibility between two isolates was confirmed by the absence of any inhibition zone.
Bacterial consortium build-up for efficient RO-16 dye decolorization
A bacterial consortium consisting of six potent bacterial isolates was constituted. Bacterial seed cultures of the individual strains were prepared by inoculating them in LB broth at 37 ℃ for 12 h. The prepared seed cultures were then inoculated in equal proportion (1:1:1:1:1:1) in 50 mL LB broth containing dye supplementation (Mohanty and Kumar 2021).
Identification of the dye decolorizing bacterial isolates
Bacterial isolates that were selected for consortium build-up were identified. The genus of the bacterial isolates was first identified tentatively by characterizing their morphological and biochemical features, according to Bergey’s Manual of Determinative Bacteriology (Holt et al. 1993).
Furthermore, molecular biology techniques (16S rRNA sequencing) were applied to identify the bacterial isolates until the species level. Bacterial gDNA was isolated (phenol/chloroform method described by Wilson (2001)) and amplified by providing the primers 27 F and 1492 S in a polymerase chain reaction (PCR) (Bio-Rad T100 Thermal Cycler). The 16S rDNA amplicons were subjected to purification followed by sequencing (Thermo-Fisher Sanger Sequencing kit in Applied Biosystems 3730 × l Genetic Analyzer). A consensus sequence was obtained from the forward and reverse sequences using an aligner software (BioEdit). The basic local alignment search tool (BLAST) provided by the National Centre for Biotechnology Information (NCBI) database was used for a comparative analysis of the obtained consensus sequence with other submitted sequences of the database. The sequences with the highest similarity to the consensus sequence were aligned along with the consensus sequence using MUSCLE, a phylogenetic tree (neighbor-joining tree) was constructed, and bootstrapping (1000 replicates) was performed in MEGA 7. The p-distance method was applied to calculate the evolutionary distances. The analysis involved a total of 20 nucleotide sequences. The obtained sequence was deposited in NCBI and denoted with accession numbers for future reference.
Selection of suitable media, salt, carbon, and nitrogen sources for RO-16 decolorization by consortium
Different media, such as LB broth, minimal salt medium (MSM) (g L−1 composition: K2HPO4 4, KH2PO4 4, (NH4)2SO4 2, MgSO4.7H2O 5, CaCl2 0.01, and FeSO4.7H2O 0.01), and DM, were studied. Salt sources, such as sodium chloride (NaCl) and sodium sulfate (Na2SO4), were used. Carbon sources studied were glucose, xylose, fructose, arabinose, lactose, sucrose, and starch. Nitrogen sources included were beef extract, malt extract, peptone, ammonium chloride (NH4Cl), ammonium sulfate (NH)2SO4, and potassium nitrate (KNO3). Overnight bacterial cultures (3% v/v) were inoculated in media and kept in an incubator at 37 ℃ for 4 days. Media, salt, carbon, and nitrogen sources that promoted efficient decolorization potential were selected for response surface methodology (RSM).
Statistical design for optimization of RO-16 decolorization by consortium — RSM (D-optimal)
The D-optimal design was used for optimizing the decolorization of RO-16 by the consortium in RSM (Rauf et al. 2008). Seven independent variables, viz., temperature, pH, initial dye concentration, arabinose concentration (carbon source), peptone concentration (nitrogen source), Na2SO4 concentration (salt source), and inoculum volume were chosen to design the experiment. Table 1 demonstrates the different levels of the independent variables used for this study. The D-optimal design generated 46 runs of the experiment. The experimental design was obtained in Design-Expert 7.0.0, Stat-Ease, Inc., Minneapolis, USA. After conducting the experiments, a response surface experiment was done to create a prediction model for deduction of the curvature, interaction among the independent variables, and identification of the local optimum independent variables which causes maximum dye decolorization. The experimental model to estimate response surface is given by the second-order polynomial equation as follows:
where “Y” is the decolorization percentage, “n” is the number of factors, “β0” is the constant coefficient (intercept term), “βi” is the linear coefficient of “xi,” “βij” is the interactive coefficient between “xi” and “xj,” “βii” is the quadratic coefficient, and “xi” and “xj” are the input factors.
Maximum dye tolerance assay of RO-16
Initial RO-16 concentration was altered within the range of 100 to 800 mg L−1 to study the dye tolerance ability of the consortium. The media was supplemented with different dye concentrations and decolorization was monitored under optimized conditions. The flasks were incubated for 4 days under static condition. Decolorization percentage was then determined.
Preparation of cell-free extract for enzyme assays
Cell-free extracts of the individual strains and consortium were prepared and used for examining the presence of different enzymes. Individual bacterial isolates and the consortium were inoculated in 40 mL LB broth supplemented with RO-16 (50 ppm) and incubated until complete decolorization occurred. Bacterial broth was then harvested at 12,000 × g for 15 min (REMI C-24 Plus). The resulting supernatant was passed through a 0.22-µm CA filter to obtain cell-free extracellular enzymes. On the other hand, cell pellet was washed thoroughly in 50 mM phosphate buffer (pH 7) and then stored at − 20 ℃ for future use. For obtaining intracellular enzymes, the frozen pellet was dissolved in equal volume of chilled phosphate buffer and subjected to ultrasonication (SONICS, Vibracell). Instrument was set at an amplitude of 40% and 10 strokes (30 s) were applied at an interval of 2 min. The ultrasonicated solution was centrifuged at 12,000 × g for 20 min to remove cell debris. Supernatant containing cytosolic proteins was passed through a 0.22-µm CA filter to obtain cell-free intracellular enzymes (Telke et al. 2009; Leelakriangsak and Borisut 2012).
Enzyme assays
Extracellular and intracellular enzyme extracts were used to check for the presence of different dye degrading enzymes.
Azoreductase was detected in a reaction using RO-16 dye as substrate. Enzyme activity was assayed in the addition of nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FAD) as explained by Bafana et al. (2008). Decolorization of the reaction mixture was monitored spectrophotometrically at 492 nm. Tyrosinase was identified with the help of substrate catechol as described by Parshetti et al. (2010). Absorbance was monitored spectrophotometrically at 410 nm.
Laccase, lignin peroxidase, and manganese peroxidase were primarily screened in plate assay, followed by quantitative estimation. Bacterial isolates were primarily screened for laccase by streaking on petriplates containing nutrient agar (NA) supplemented with 0.5 mM guaiacol and incubated at 37 ℃ for 96 h (Sheikhi et al. 2012). Primary screening of bacterial isolates for lignin and manganese peroxidase was performed by streaking the isolates on azure B agar plates (composed of NA supplemented with 0.01% azure B and 20% glucose) and incubated at 37 ℃ for 96 h (Archibald 1992). ABTS (2,2′-azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid)) substrate was used to quantitate laccase enzyme activity as described by Sheikhi et al. (2012). Absorbance was determined at 420 nm. Lignin peroxidase enzyme activity was quantitated using azure B substrate as described by Paliwal et al. (2015). Absorbance was measured at 651 nm. Manganese peroxidase enzyme activity was quantitated using the substrate manganese sulfate (MnSO4) (Paszczyński et al. 1988). Absorbance was determined at 270 nm.
An enzyme unit was defined as the quantity of enzyme that catalyzed 1 µmol substrate transformation per minute, under stated temperature, pH, ionic strength, and substrate conditions (Labuda et al. 2018). Enzyme activity was calculated by the following equation:
where “EA” is enzyme activity, “A” is absorbance change, “V” is volume of the reaction mixture, “t” is the incubation time, “e” is the extinction coefficient of the substrate, and “v” is the volume of enzyme.
Protein content of the enzyme extract was measured as described in the Lowry’s method (Lowry et al. 1951). Specific enzyme activity was defined as enzyme units per milligram of protein content (Földesi, 2019).
Enzyme kinetic study of RO-16
The enzyme kinetics of RO-16 degradation by the consortium was studied. The consortium was cultured in 20 mL LB broth (under optimized conditions) and incubated for 102 h under static condition. Different initial dye concentrations within the range of 30 to 300 mg L−1 were used for batch decolorization by the bacterial consortium. Decrease in absorbance was monitored at λmax of RO-16 at an interval of 6 h. Graphs for different reaction orders were plotted with final substrate concentration (Ct in mg L−1) and time (t in h) to identify the reaction order with closest fit. A graph for zero-order reaction (k0) was plotted as Ct vs. t, first-order reaction (k1) was plotted as ln Ct vs. t, and second-order (k2) reaction was plotted as 1/Ct vs. t. The following equations define the different reaction orders (Connors 1990; Atkins and de Paula 2006):
where “C0” is the initial substrate concentration (mg L−1).
To determine the Michaelis constant (km) and maximum substrate consumption rate (Vmax), dye decolorization rate was estimated at altered initial concentrations of dyes. Lineweaver–Burk double reciprocal model was then plotted as it was the best fit (Berg et al. 2002). The corresponding equation for the model has been given below. The Km and Vmax values were obtained from the double reciprocal plot.
where “V” is the substrate concentration rate (mg L−1 h−1) and “S” is the substrate concentration (mg L−1).
Analytical study of the dye decolorization metabolites
UV-Vis spectrophotometric analysis
The bacterial consortium was used for decolorization of RO-16 in 40 mL LB broth under optimized conditions. The culture flasks after inoculation were incubated for 4 days. Cellular content was removed from the broth after centrifugation at 15,300 × g for 15 min. Spectral scan (200–800 nm) was performed with the resulting supernatant. Dye biotransformation or biodegradation was indicated by a shift in the λmax of the solution.
Extraction of metabolites after dye decolorization
Bulk decolorization was performed with the consortium in 1.5 L of LB broth under optimized conditions. The culture broth was incubated 4 days for decolorization. Decolorized broths were harvested at 12,000 × g for 15 min. The supernatant containing dye decolorized metabolites was subjected to filtration (0.22 µm CA filter). Liquid–liquid extraction was performed to collect metabolites from the supernatant. Acetonitrile resulted in maximum recovery of decolorized RO-16 metabolites by the consortium. Extracts were collected and water was eliminated using anhydrous Na2SO4. The organic solvent extract was then dried in a rotary evaporator (BUCHI, Rotavapor R-300) (Dawkar et al. 2008). The dye decolorized metabolites were obtained in dried powder form and used for further studies.
Fourier transform infrared spectroscopy (FTIR)
FTIR (Thermo Nicolet AVATAR 330) of the samples were performed using the KBr disc method. Samples were analyzed within the wavelength range of 400–4000 cm−1 (mid-IR region) at scan speed 16. At different wavelengths, the variations in percentage transmittance were noted.
Ultra high-performance liquid chromatography (UHPLC)
Reverse phase UHPLC (Waters, ACQUITY H class) was performed. Mobile phase chosen for conducting the analysis was composed of HPLC grade carbinol and water in proportions of 1:1. The dye and metabolite samples were dissolved (0.1 mg L−1) in a solution having the same composition as the mobile phase. Separation was performed in a C18 reverse phase column equipped with a photo diode array (PDA) detector. The samples were injected at a flow-rate of 0.8 mL min−1, and spectrum was recorded at the wavelength range of 200–700 nm. The sample separation was carried out for 20 min.
Liquid chromatography high-resolution mass spectrometry–quadrupole time-of-flight (LCHRMS–QTOF)
RO-16 dye and its corresponding degraded metabolites were subjected to LCHRMS–QTOF (Agilent technologies, 1290 Infinity UHPLC System, TOF 6500 series). Separation was performed in a HYPERSIL Gold C18 column (100 × 2.1 mm, 3 µm). The samples were dissolved (1 µg µL−1) in deionized water and passed through a 0.22-µm CA filter to remove any undissolved particle. Mobile phase consisted of water. It was flown at 0.2 mL min−1 for 30 min. Electrospray ionization (ESI) was performed in both positive and negative mode under the flow of nitrogen (N2) gas (1 mL min−1), and fragment voltage was 175 V. Mass spectra were thus generated. The probable degradation products were identified based on this analysis.
Toxicity assessment study
The toxicity profile of untreated RO-16 and the corresponding degradation products by the consortium were analyzed.
Phytotoxicity assay
Phytotoxicity assay was performed by assessing the seed germination of Vigna radiata (common name: moong) and Cicer arietinum (common name: chickpea) and root growth from the bulbs of Allium cepa (common name: onion).
Seeds for germination assay were pre-treated with 0.1% HgCl2 for 10 min to ensure surface sterilization, followed by a thorough wash with treated water. The seeds received three treatment types (untreated dye, dye degraded metabolite, and distilled water (control)). The treatment sets were subdivided based on their concentration, i.e., 500 ppm and 1500 ppm. The experiment was conducted in sterilized petriplates lined with filter paper. Each petriplate contained ten seeds randomly chosen. The seeds received a treatment of 10 mL on the first day, followed by 5 mL every day for a period of 7 days. Moong seeds were incubated at room temperature (35 ℃) and chickpea seeds were incubated at 15 ℃ (Rao and Prasad 2014; Salian et al. 2018). The seeds were observed every day. At the end of the treatment period, seedling characteristics were noted and relative seed germination (RSG) and germination index (GI) were calculated as follows:
where “Ms” is the number of seeds germinated under test treatment, “Ds” is the number of seeds germinated under control treatment, “Rs” is the mean root length under test treatment, and “Rc” is the mean root length in the control treatment.
Onion bulbs, after a thorough wash with treated water, were grown in hydroponic condition. These were grown in 250-mL beakers filled with 150 mL of treatment solution. Like the germination assay, bulbs were divided into three sets of treatment including dye solutions, degraded metabolite solution, and distilled water (control). Treatment concentrations used were 500 ppm and 1500 ppm. The bulbs were kept at a temperature of 23 ℃ for a period of 7 days (Jadhav et al. 2010; Haq et al. 2016). Root growth was monitored every day to note down the toxic effect of the treatments. The number of roots that sprouted from each bulb was counted, and root length was recorded as the mean length of the longest five roots from each bulb.
Cytogenotoxicity assay
Onion bulb roots grown during phytotoxicity were further analyzed for cytotoxic and genotoxic effects. Roots of about 2–2.5 cm were cut from the tip and observed under a microscope using the meristematic root tip squash technique with acetocarmine (Prasad et al. 2013). Roots were divided into four groups, including dye-treated roots, degraded dye metabolite–treated roots, distilled water-treated roots (negative control), and H2O2-treated roots (positive control). Meristematic cells were observed under a light microscope (Magnus microscopes, model MLX). Mitotic index was recoded per 1000 cells scored and aberration index was also noted.
where “MI” is mitotic index, “d” is total dividing cells, “c” is total analyzed cells, “AI” is aberration index, and “a” is total chromosomal aberrations identified.
Microbial toxicity assay
The bacterial growth curves of Escherichia coli, Pseudomonas aeruginosa, and Bacillus cereus were used as an indicator of toxicity in this assay. The bacteria were divided into three groups, viz. dye-treated bacteria, degraded dye metabolite–treated bacteria, and control group without any treatment. The bacteria (2%, v/v) were inoculated in 40 mL LB. Treatments (dye and degraded dye metabolite) were supplemented in NB at 200 ppm concentration. E. coli and P. aeruginosa were incubated at 37 ℃ and B. cereus was incubated at 30 ℃ for 8 h. Bacterial growth was observed by measuring the O.D of the culture broth in a colorimeter (Systronics digital colorimeter 112) at an interval of 1 h. The O.D of the RO-16 dye-treated culture broths were measured at 492 nm, and degraded dye metabolite–treated and control broths at 600 nm (Asses et al. 2018).
Biotoxicity assay
Artemia salina (common name: brine shrimp) was chosen as the test organism for lethality assay (Prasad and Rao 2013; Bekhit et al. 2020). Commercially bought Artemia cysts (4 g) were hatched in 1 L of sea water in a glass beaker. The glass beaker containing cysts was incubated at 25 ℃ under a continuous illumination condition for 84 h. Eggs were hatched, and the nauplii were grown until instar II and III stages. The brine shrimps (~ 500) were then shifted in 250 mL glass beakers containing 150 mL sea water. Brine shrimps were subjected to four kinds of treatment, viz., sea water amended with dye, sea water amended with degraded dye metabolite, sea water amended with potassium dichromate (K2Cr2O7) (positive control), and sea water without any amendment (negative control). The treatments were again examined with two different concentrations, i.e., 0.125% and 0.5%. Brine shrimps under different treatments were incubated under the same conditions as was used for hatching, for a period of 24 h. After incubation, the shrimps were observed under a light microscope for any physiological change. The number of survivors was counted in each treatment as follows:
where “S” is survival percentage, “s” is number of survivors, and “t” is total number of shrimps.
Statistical analysis
All the experiments were carried out in triplicate. The data in this research has been expressed as mean ± standard deviation. The corresponding data generated were analyzed by one-way ANOVA, two-way ANOVA, and coefficient of determination (R2) as per requirement. The significance of the results was established based on probability (P-value). Test results with probability value (P-value) ≤ 0.05 were considered significant.
Results and discussion
Effluent sample characterization
The physicochemical characterization of the leather effluent sample has been listed down in Table 2. The sample was observed as gray colored and consisted of 189 ADMI color units. A strong odor was present, which might have arisen from the presence of volatile organic compounds. The pH was somewhat alkaline but suitable for aqueous organisms’ survival. The sample temperature was almost equivalent to normal room temperature. BOD was elevated, thus making the sample appropriate for release solely in sewers. A high BOD reduced the DO content. DO is essential for maintaining fish life and other aqueous organisms, such as caddisfly larvae and mayfly nymphs. COD was also found at an elevated level which made the effluent unsuitable for disposal. A high COD is a direct indication of the presence of excessive quantities of organic and inorganic chemicals. EC was very high and above the limit set by surface water regulations (1989). EC, a function of TDS, is directly correlated to the presence of ionic content in the effluent sample. The TDS content was above average but within the acceptable limit, whereas the TSS content was excessive and more than the acceptable limit for irrigation purpose or discharge into inland surface water. The hazardous implications of a high TSS content include reduced sunlight penetration and damage to fishes and may form septic and unaesthetic deposits in lakes and river beds. High TDS and TSS content resulted in an increased TS content. Among the dissolved metals, the contents of Ca2+, Mg2+, Cu+, Zn2+, SO42−, and Cd2+ were low, whereas the contents of Na+, Fe3+, Cl−, total Cr content, and Ni2+ were very high. The presence of increased salinity in effluent is detrimental, when used for agricultural purposes. Overexposure to Fe content has not been associated to any harmful effect in humans; however, laboratory studies have reported harmful effects of Fe3+ on aquatic life. Cr is an essential dietary requirement; however, overexposure can be carcinogenic. Ni2+ is considered carcinogenic to humans, as well as being hazardous to aquatic life forms, including plants and fishes (EPA 2001; Central Pollution Control Board 2021).
Isolation, screening, and quantification of the decolorization potential of dye decolorizing bacteria
The leather industry sample yielded 8 bacterial isolates, designated as VITEPB1–VITEPB8. The visible decolorization of RO-16 was achieved by three isolates VITEPB1–VITEPB3 after 6 days under static condition. These bacterial isolates decolorized > 85% of RO-16 after 6 days under static condition. VITEPB2 was the highest dye decolorizing isolate (87.16%), followed by VITEPB1 (85.55%) and VITEPB3 (85.02%). Effluent and sludge samples from the leather industry, textile industry, paper mill, and wastewater treatment facility have been previously used for the isolation of dye degrading organisms (Thakur et al. 2012; Roy et al. 2018; Al-Ansari et al. 2022). The stressed conditions and myriad of chemicals present in industrial samples acclimatize the microorganisms that survive under those conditions. As a result, microorganisms develop the ability to counteract different hazardous chemicals for their survival. The presence of dyes in the abovementioned industrial effluents and sludges thus suggests the probability of isolating dye degrading organisms from the industrial samples.
Compatibility assay of the potent dye decolorizing bacteria
An essential criterion for the building up of a microbial consortium is that the microbial strains should not exhibit antagonistic activity against each other. Compatibility assay was conducted with the six most efficient dye-decolorizing bacterial isolates VITEPB1–VITEPB3, VITSP6, VITSP7, and VITSP9. Bacterial isolates VITSP6, VITSP7, and VITSP9 were potent dye decolorizing bacteria obtained from textile industry effluent in our previous study (Saha and Rao 2020). No inhibition zones were observed in the MH agar assay plates at 37 ℃ after 48 h. Thus, the isolates were compatible with each other and were used to build up a consortium designated as VITPBC6. Compatibility assay in MH agar has been used in previous studies to determine the compatibility of microorganisms (Soundararajan et al. 2012; Shinkafi et al. 2016; Krishnamoorthy et al. 2018). On the other hand, Haque et al. (2021a, b) performed bacterial compatibility assay in yeast extract peptone agar plates following the protocol as described by Furuya et al. (1997). However, it is noteworthy to mention that several dye degradation studies involving microbial consortium were conducted without a compatibility assay (Lade et al. 2012; Patel et al. 2012; Khan et al. 2014; Eslami et al. 2019; Krithika et al. 2021).
Identification of the individual bacterial isolates of the consortium
The potent bacterial isolates used to build the consortium, i.e., VITEPB1–VITEPB3, VITSP7, and VITSP9 were identified based on morphological, biochemical, and molecular biology techniques (16S rRNA sequencing). According to the results of microscopic and biochemical tests of the isolates (Online Resource 1), all the isolates VITEPB1–VITEPB3, VITSP7, and VITSP9 belonged to genus Bacillus.
The highest sequence similarity of the contig sequence of isolates VITEPB1, VITEPB2, VITEPB3, VITSPB7, and VITSPB9 with other existing sequences in the NCBI database was 99.12%, 99.07%, 99.65%, 98.58%, and 98.23% respectively. The isolates were thereby identified as Bacillus firmus VITEPB1 (accession number: MZ934656), B. flexus VITEPB2 (accession number: MZ951161), Bacillus aryabhattai VITEPB3 (accession number: MZ951124), Bacillus paraflexus VITSPB7 (accession number: MZ817967), and Bacillus megaterium VITSPB9 (accession number: MZ934417).
Isolate VITSPB6 was identified in our previous study (Saha and Rao 2020) as B. flexus strain VITSP6 (accession number: MG407663).
The phylogenetic tree depicts the evolutionary relationship of the sequences of the potent isolates with each other and their corresponding closely related sequences (Fig. 1).
Evolutionary relationship of taxa. The evolutionary history was deduced with the help of neighbor-joining approach. The optimal tree has a branch length sum of 0.12818173 as shown above. Replicate tree percentages that had clusters of associated taxa in the bootstrap test have been indicated near the branches. The evolutionary distances are expressed as the number of base differences per site (P-distance method)
Selection of media, salt, carbon, and nitrogen sources for maximum RO-16 decolorization by the consortium
The consortium designated as VITPBC6 showed efficient decolorization in LB broth (87.85%), with Na2SO4 (86.94%), arabinose (87.97%), and peptone (85.08%) supplementation.
RSM optimization (D-optimal) for maximum RO-16 decolorization by the consortium
RSM is a sequence of processes with the main objective to carry out experiments rapidly and efficiently that leads to the determination of optimum operational conditions (Mohana et al. 2008). Table 3 contains the data obtained from 46 runs of the D-optimal design containing the seven independent variables, including temperature, pH, initial RO-16 concentration, arabinose concentration, peptone concentration, Na2SO4 concentration, and inoculum volume. Decolorization percentage was the response corresponding to the combined effect of the seven variables. Decolorization varied significantly from 0 to 100%, under the test conditions. No decolorization was observed in runs 10, 11, 12, 19, 32, and 33. On the other hand, complete decolorization was observed in run 20. The actual decolorization percentage was approximately the same as that of the predicted values. The predicted decolorization percentage has been obtained, by applying multiple regression analysis, from the second-order polynomial equation given below in terms of coded factors as:
The actual and predicted decolorization percentage data indicate that the empirical models actually fit the obtained data with an R2 value of 0.9978 and adjusted R2 value of 0.9901. The adjusted R2 value suggests that 99.01% variability in decolorization percentage can be explained with the help of its relation with the input variables included in the model. The predicted R2 value 0.9305 was also in reasonable agreement with the adjusted R2 value 0.9901. The adequate precision ratio of 30.193 indicates sufficient signal for navigating the design space.
The fitting results of the quadratic response surface model in terms of ANOVA have been shown in Table 4. F-value of the model was measured at 130.07, and p-value was less than 0.0001. Thus, it is evident that the model is significant. Significant model terms were indicated by p-values less than 0.05. In this case, the model terms A, B, C, D, E, F, G, AC, AD, AE, AF, AG, BC, BD, BE, BG, CD, CE, DE, DF, EF, EG, FG, B2, D2, E2, F2, and G2 were significant. Lack of fit F-value of the model was 0.99 that is insignificant relative to the pure error. This implies that the quadratic model is statistically significant for predicting the decolorization percentage.
The 3D surface plots depicting interactive effects of the different pairs of the independent variables on decolorization percentage by the consortium, while maintaining the remaining variables at or near the central level, have been represented in Fig. 2. According to the constructed model, the optimum values for achieving the maximum decolorization were 40 ℃ temperature, pH 7.38, 218.75 mg L−1 dye, 0.61% arabinose, 0.55% peptone, 2.5% Na2SO4, and 7.5% inoculum.
3D surface response plots of reactive orange 16 decolorization by bacterial consortium VITPBC6 as an effect of the interaction of the independent variables. a pH and dye concentration. b Carbon source and salinity. c Nitrogen source and pH. d Inoculum and pH. e Salinity and carbon source. f Inoculum and salinity
Maximum RO-16 tolerance assay by consortium VITPBC6
Dye concentration plays an important role in inhibiting bacterial decolorization because of dye toxic effect, inadequate cell to dye ratio, and blockage of enzyme active sites by dye molecules (Saratale et al. 2011). In this study, dye concentration effect was thus studied on the RO-16 decolorization of consortium VITPBC6 (Fig. 3). Consortium VITPBC6 could maintain efficient decolorization (> 90%) even at the highest concentration (800 mg L−1). Compared to the recent studies on bacterial consortium, consortium VITPBC6 possessed better decolorization property even at very high dye concentration. Eskandari et al. (2019) constituted two bacterial consortia PsGo (Pseudoarthrobacter and Gordonia) and StSp (Stenotrophomonas and Sphingomonas) capable of decolorizing only 36% and 25% RB-5 at the highest concentration of 100 mg L−1 respectively. Guo et al. (2020b) studied metanil yellow G (MY-G) decolorization by the mixed bacterial consortium ZW1 (mainly composed of Halomonas (49.8%), Marinobacter (30.7%), and Clostridiisalibacter (19.2%)). The highest concentration of MY-G (500 mg L−1) exhibited an inhibitory effect on ZW-1, which led to a very low decolorization (8.2%). Guo et al. (2021) further continued his study with MY-G using a different halo-thermophilic bacterial consortium HT-1 composed of Bacillus, norank_f_Bacillaceae and Piscibacillus. The highest MY-G concentrations (300 mg L−1 and 400 mg L−1) inhibited bacterial growth resulting in no dye decolorization.
Enzyme assays
Azo dye degradation is mediated by bacteria with the help of several oxidoreductive enzymes, such as azoreductase, DCIP reductase, amino pyrine N-demythlase, tyrosinase, veratryl alcohol oxidase, laccase, lignin peroxidase, and manganese peroxidase (Dave et al. 2015). Enzymatic assays were thus conducted to shed an insight into the biodegradation mechanism of RO-16. The assays were conducted with the individual bacterial strains and consortium VITPBC6 to compare the specific enzyme activities (Tables 5 and 6). Tyrosinase had high specific activity in all the strains, and strain VITEPB2 had the highest specific activity. Tyrosinase had the second highest specific activity in consortium VITPBC6. Manganese peroxidase had the highest specific activity in strain VITEPB1, followed by strains VITEPB2, VITSPB9, VITEPB3, and VITSPB7. Manganese peroxidase activity was not detected in strain VITSP6. The highest specific activity of lignin peroxidase was noted in strain VITEPB1, followed by strains VITEPB3, VITSPB9, and VITSPB7l, whereas it was not produced by VITSP6 and VITEPB2. However, both the manganese and lignin peroxidase activity in VITPBC6 was higher than any of the individual isolates. Lower amount of azoreductase compared to the other enzymes was produced by all the individual strains and VITPBC6. Laccase had the lowest specific activity and was produced by strains VITEPB1–VITEPB3 and consortium VITPBC6. The induction and activity of enzymes produced by a consortium is not the sum of the induction and activity of enzymes produced by the individual strains constituting the consortium. Consortium GG-BL (composed of Galactomyces geotrichum and Brevibacillus laterosporus) expressed veratryl alcohol oxidase activity induction by 304% more than that of the individual strain (B. laterosporus) producing it, while the induction of riboflavin reductase, tyrosinase, NADH–DCIP, and azoreductase activities was less in the consortium than in the individual strains producing these enzymes (Waghmode et al. 2011; Kurade et al. 2012).
Enzyme kinetic study of RO-16 decolorization by the bacterial consortium
Determination of reaction order
The effect of different initial concentrations on RO-16 decolorization by consortium VITPBC6 is depicted in Table 7. The table presents Ct obtained at different time intervals for the specified C0 values. During RO-16 batch decolorization, there was a slow linear decrease in Ct during the initial phase; however, the Ct values dropped rapidly during the latter phase. The rapid drop in Ct for the C0 values of 50, 100, 150, 200, 250, and 300 occurred at 66 h, 72 h, 84 h, 72 h, and 96 h respectively. Figures 4a–c represent the different reaction order kinetic models for RO-16 decolorization. A zero-order kinetic model plotted as Ct vs. t followed a more linear relationship compared to that of the other kinetic models. Furthermore, the reaction rate constants of different kinetic models at varied initial dye concentrations and the corresponding R2 values have been estimated and presented in Table 8. The table shows that the R2 values of the zero-order kinetic model ranged from 0.8753 to 0.9522 and were closer to unity than that of the other kinetic models. Therefore, it can be concluded that VITPBC6 decolorized RO-16 following zero-order reaction kinetics. Very few studies have reported zero-order reaction kinetics for bacterial azo dye degradation. Sarioglu et al. (2007) reported an anaerobic mixed culture that degraded basic red 46 following first-order reaction kinetic at low dye concentration (50–250 mg L−1), and then shifted to zero-order kinetic at high dye concentrations (500–1000 mg L−1). Tan et al. (2013) reported a microbial community composed of dominant microbial strains belonging to phyla Actinobacteria, Bacteroidetes, Proteobacteria, and Firmicutes decolorized acid brilliant scarlet GR following a zero-order reaction. In another study, halo-alkaliphilic bacterium Nesterenkonia lacusekhoensis EMLA degraded reactive red 35 following zero-order kinetic during the initial 4 h, and then followed a first-order reaction thereafter (Prabhakar et al. 2021). On the contrary, majority of kinetic studies on bacterial azo dye degradation have reported a first-order reaction. In two subsequent previous reports, Karunya et al. (2014) and Nachiyar et al. (2016) constituted consortium CN-1 composed of Citrobacter freundii (2 strains), Moraxella osloensis, Pseudomonas aeruginosa, and P. aeruginosa BL22 and consortium CN-1A composed of M. osloensis, P. aeruginosa, Citrobacter freundii, Bacillus sp., and Bacillus thuringiensis that decolorized mordant black 17 and acid black 24 respectively following first-order reaction kinetics. Das and Mishra (2019) reported a bacterial consortium MA12C composed of Bacillus pumilus, Zobellella taiwanensis, and Enterococcus durans that decolorized remazol navy blue by following first-order kinetics. Similarly, another indigenous bacterial consortium decolorized direct blue 2B following first-order kinetics under increasing temperatures and initial dye concentrations (Cao et al. 2019). The zero-order reaction kinetics obtained for RO-16 decolorization in our study essentially means that the rate of dye decolorization is independent of the initial dye concentrations. It is notable that the dye concentrations might have been very high compared to that of the enzymes produced by the consortium, which might have resulted in a zero-order dye decolorization in both cases.
Determination of Vmax and Km
The decolorization rate was calculated for different initial RO-16 concentrations and has been tabulated in Table 9. Furthermore, the data obtained was applied to the reciprocal form of the Michaelis–Menten equation that is the Lineweaver–Burk equation. The applicability of this equation was verified by plotting a graph with 1/V vs. 1/S (Fig. 5). The experimental data clearly showed a good fitting to the Lineweaver–Burk model equation, and the R2 value was recorded as 0.9733. The graph has an interception of 1/Vmax and a slope of Km/Vmax that was used to derive the Vmax and Km values of the enzymes involved in RO-16 decolorization. The Vmax and Km values obtained were 352 mg L−1 day−1 and 1453.426 mg L−1 respectively. Comparatively, very less kinetic studies have been conducted with bacterial consortium. The Vmax values found in this study were comparable to that reported for Remazol black B decolorization by a bacterial consortium composed of the predominant strains of P. aeruginosa, Rhodobacter sphaeroides, P. mirabilis, Bacillus circulance, NAD 1, and NAD 6 (Dafale et al. 2008). The obtained values were much higher than that reported for direct black 38 decolorization by a bacterial consortium composed of Pseudomonas stutzeri and Cardiobacterium hominis, whereas lower than that reported for reactive green 19 decolorization by a bacterial consortium composed of Zobellella taiwanensis and Bacillus pumilus (Bafana et al. 2007; Das and Mishra 2017). The high Km value indicates the low affinity of the enzymes towards the substrate dye molecule, and it may stem from the fact that the reactions were conducted at high initial dye concentrations. Previous enzyme kinetic studies conducted with lower initial dye concentration exhibited low Km values (Cai et al. 2015; El-Kader et al. 2019), whereas those conducted with higher initial dye concentration showed high Km values (Cossolin et al. 2019; Saha et al. 2022).
Analytical study of RO-16 decolorized metabolites
UV-Vis spectrophotometric analysis
The spectra of RO-16 dye and its corresponding decolorized metabolites obtained with consortium VITPBC6 have been represented in Fig. 6. RO-16 dye showed maximum absorbance at 492 nm, whereas the maximum absorbance peak shifted for the degraded RO-16 metabolites. The degraded metabolites exhibited maximum absorbance at 250 nm and 290 nm respectively. In addition, after dye decolorization, the intensity of the peaks was significantly decreased. The examination of UV–Vis’s spectra is the most preliminary step to identify the nature of dye decolorization that can be either biosorption or degradation. As pointed out by Chen et al. (2008), the absence of the major visible light absorption peak (noted with dyes) or the presence of a new peak indicates the occurrence of dye degradation. Thus, it can be presumed that the decolorization of RO-16 by consortium VITPBC6 occurred due to cleavage or transformation of the dye chromophore, i.e., biodegradation of the dye molecule leading to its decolorization.
FTIR analysis
The FTIR spectrum of RO-16 has been illustrated in Fig. 7a. The hydroxyl group (“–OH”) appeared at 3369.64 cm−1, and secondary amine (“–NH”) bend was observed at 1577.77 cm−1. The peaks at 3070.68 cm−1 and 1492.9 cm−1 correspond to aromatic “–CH” and ring stretch respectively. The carbonyl group (“–C = O”) appeared at 1286.52 cm−1, while the sulfonate group (–SO3) was seen at 1132.21 cm−1. The chromophore “–N = N–” peaked at 1658.78 cm−1. The RO-16 spectrum fingerprint region exhibited peaks at 833.25 cm−1, 754.17 cm−1, and 617.22 cm−1 representing “C–H” out of plane deformation vibration, “N–H” out of plane deformation vibration, and “–OH” out of plane bending vibration.
The FTIR spectrum of degraded metabolites of RO-16 by consortium VITPBC6 has been represented in Fig. 7b. A change in the spectrum was noticed when compared to that of the RO-16 spectrum. “–OH” group peak was shifted to 3309.85 cm−1. Asymmetrical “–CH3” stretching vibration was observed at 2962.66 cm−1 and 2881.65 cm−1. A peak at 1635.64 cm−1 corresponds to the carboxylic acid group (“–COOH”), and 1398.39 cm−1 represents “–CO” stretching vibration. Asymmetric “–SO3” stretching vibration peaked at 1093.64 cm−1. The peak of the “–N = N–” group was absent. The significant changes in the spectrum of the degraded metabolite, when paralleled with that of the RO-16 spectrum, presumably suggest that the changes occurred due to transformation or degradation of the dye molecule.
UHPLC analysis
The UHPLC elution profile of RO-16 dye has been depicted in Fig. 8a. The chromatogram reveals a single sharp peak, and the dye molecule eluted at 1.756 min. The single peak in this case indicates the purity of the dye molecule (Saha and Rao 2020). On the other hand, the chromatogram of the degraded metabolites exhibited multiple peaks (Fig. 8b). The degraded metabolites eluted at 1.117 min, 2.227 min, 1.305 min, and 5.565 min. The disappearance of the major peak that was observed in RO-16 chromatogram and the appearance of multiple other peaks in the degraded metabolite chromatogram clearly indicated that the dye molecule transformed or formed different compounds after biodegradation by VITPBC6.
LCHRMS–QTOF analysis
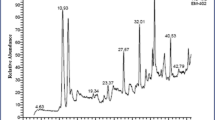
LCHRMS QToF analysis was performed to identify the unknown compounds that were formed after RO-16 degradation. The chromatogram and mass spectrum of degraded RO-16 metabolites by consortium VITPBC6 have been represented in Fig. 9a–d. The dye molecule in this case underwent an initial reduction of the “–N = N–” bond that led to the formation of two compounds with m/z 318.0286 (M + 2 peak) and 280.9955 (M peak) respectively. The compounds further underwent reactions, such as aromatic ring opening, reductive cleavage, desulfurization, and activation of C–O and C–N bonds resulting in the formation of compounds with m/z values of 284.0299 (M peak), 246.2 (M peak), 200.0381 (M peak), 168.0245 (M − 1 peak), and 156.0245 (M + 1 peak) respectively. The probable pathway of the degradation has been illustrated in Fig. 9e. The dye molecule was therefore degraded by the consortium VITPBC6 into different simpler aromatic compounds than the complex parent dye compound.
Toxicity assessment study
Seed germination assay
Data obtained with the seeds of P. mungo have been presented in Table 10. Seeds treated with RO-16 dye exhibited lower RSG, GI, and root and shoot length compared to that of the degraded RO-16 metabolite by consortium VITPBC6. Degraded RO-16 metabolite–treated seeds had an RSG and shoot length comparable to that of the control seeds. However, the root length was less than that of the control seeds. Statistical analysis suggested that the effects of different treatments on the plant seeds were significantly different from each other (P-value 1.62741E-06 < 0.05, Fstatistics > Fcritical). However, concentration variation did not have a significant effect on the plant seeds (P-value 0.178958 > 0.05, Fstatistics < Fcritical). Table 10 also represents the data obtained with seeds of C. arietinum. Seeds treated with RO-16 showed lower RSG, GI, radicle, and plumule length compared to that of the corresponding degraded dye metabolite. Therefore, it can be presumed that the degraded dye metabolites significantly reduced the toxicity of the dye molecules but were not completely non-toxic. Data obtained with different treatments on C arietinum seeds were statistically significant with P-value 0.00281 < 0.05 and Fstatistical > Fcritical. However, similar to that observed in P mungo seeds, concentration variation did not produce any significant effect on C. arietinum seeds (P-value 0.365 > 0.05, Fstatistical < Fcritical). Plant bioassays have been widely used to assess different environmental contaminants. Previous studies have been reported with P. mungo seeds used for assessing the toxicity of dye degradation, such as remazol red, reactive blue 172, methyl red, and RB-5 degradation, wherein the degraded dye metabolites showed a non-toxic effect on the seeds (Saratale et al. 2013; Lade et al. 2015; Waghmode et al. 2019; Al-Tohamy et al. 2020). Similarly, earlier studies performed with C. arietinum seeds for evaluating the toxicity of dye degradation, such as methyl orange, tartrazine, amaranth, procion red-H3B, and dye mixture (azo, anthroquinone, and triphenylmethane) degradation showed that the degraded metabolites significantly reduced the toxicity of the dyes (Dixit and Garg 2018; Bera and Tank 2021; Kumar et al. 2022).
Root growth assay
The number of roots and root length of A. cepa bulbs under different treatments has been recorded in Table 11. The highest number of roots was observed in control treatment followed by degraded RO-16–treated bulbs. The lowest number of roots and root growth was noted in RO-16-treated bulbs. When root length was considered, RO-16 degraded metabolite–treated roots showed comparable results to that of the control bulbs. The lowest root length was recorded in RO-16-treated bulbs. The different treatments produced significant changes on root length (P-value 0.014027057 > 0.05, Fstatistical > Fcritical); however, there was no significant change in the number of roots (P-value 0.076000658 > 0.05, Fstatistical < Fcritical respectively). The number of roots and root lengths were not affected significantly by concentration variation in both dye and degraded dye metabolite–treated bulbs (P-values 0.99264686 > 0.05 and 0.964533655 > 0.05 respectively; Fstatistical < Fcritical in both cases). Similar studies have been reported that analyzed the toxicity of degraded dye metabolites on A. cepa root growth. The toxicity of degradation of dyes, such as Congo red 21, reactive blue 198, and textile effluent was evaluated with A. cepa root growth, and it was found that root growth inhibition was significant after dye treatment, while the inhibition was negligible after degraded dye treatment (Jadhav et al. 2010; Poonkuzhali et al. 2011; Chakravarthi et al. 2020).
Cytogenotoxcity assay
The cytogenotoxic effects of all the test compounds have been tabulated in Table 12. The highest MI was observed in roots treated with degraded RO-16 metabolites and negative control. Positive control roots exhibited the lowest MI, followed by the roots treated with RO-16. The increased MI of degraded dye–treated roots than dye-treated roots imply healthy cell proliferation in the former case. AI of the dividing cells was also calculated. Positive control root cells exhibited the highest AI followed by roots treated with RO-16. Negative control root cells had no aberrantions, while cells treated with degraded RO-16 showed low and comparable aberrant index. Thus, it can be said that toxicity was considerably lowered upon dye degradation, which resulted in less aberrant cells in degraded dye–treated roots. The different treatments had a significant effect on mitotic onion root-tip cells (P-values: 0.019008871 < 0.05 (MI) and 0.005905559 < 0.05 (AI); Fstatistical > Fcritical in both cases). The values of MI decreased and AI increased with increasing concentration and vice versa; however, the effect was not statistically significant (P-value 0.30487 > 0.05 (MI) and 0.264719 > 0.05 (AI); Fstatistical < Fcritical in both cases). The common chromosomal abnormalities observed in the dye-treated aberrant cells included chromosomal bridge, disturbed metaphase, nuclear bud, and apoptotic cells. The observations of this assay were concomitant with previous reports (Prasad et al. 2013; Haq and Raj 2018; Gurav et al. 2021).
Microbial toxicity assay
The growth curves of E. coli, P. aeruginosa, and B. subtilis treated with RO-16 and degraded RO-16, as well as control without any treatment, have been illustrated in Fig. 10a–c. It is evident from the curves that bacterial growth was inhibited upon treatment with dye solution. After 8 h of incubation, bacterial density was very low with dye treatment resulting in a flat curve. On the contrary, bacteria treated with degraded dye metabolites promoted healthy bacterial growth resulting in a gradually increasing curve similar to that of the control. The bacterial growth curves under the different treatment solution were significantly different from control (P-values: 1.76419E-07 < 0.05 (E. coli), 1.07334E-07 < 0.05 (P. aeruginosa), and 1.88244E-07 < 0.05 (B. subtilis); Fstatistical > Fcritical in all cases). The bacterial growth densities of E. coli, P. aeruginosa, and B. subtilis at the end of the incubation period were 0.345, 0.325, and 0.36 respectively in control media; and 0.335, 0.28, and 0.325 respectively in degraded RO-16 treatment. Thus, it can be presumed that both the degraded metabolites did not induce any toxic effect on the test bacteria. Previous studies have also used microbial growth as an indicator of toxicity after dye degradation. Microorganisms, such as E. coli, Sinorhizobium meliloti, Pseudomonas cedrina ESR12, B. cereus ESD3, P. aeruginosa, and S. aureus, were successfully used for microbial toxicity assay (Asses et al. 2018; Al-Tohamy et al. 2020; Haque et al. 2021a; Li et al. 2021).
Biotoxicity assay
The survival percentages of RO-16 and degraded RO-16–treated shrimps with respect to positive and negative control have been illustrated in Fig. 11. Survival percentage was the highest in negative control (100%) followed by degraded RO-16 (91.22%). The lowest survival percentage was observed in positive control (0.133%) followed by RO-16 (58.97%). The treatments were significantly different from each other (P-value 0.000785 < 0.05, Fstatistical > Fcritical); however, concentration did not induce any significant effect on survival (P-value 0.18523 > 0.05, Fstatistical < Fcritical). Furthermore, dye-treated shrimps showed visible dye accumulation in their midgut. Positive control and degraded metabolite–treated shrimps on the other hand showed no visible dye accumulation in their midgut. Therefore, it can be concluded from the results that degraded RO-16 metabolites had considerably reduced toxicity level than that of the corresponding dye molecules. A salina has been proved as a good indicator for detecting the toxicity of degraded dye metabolites in previous studies (Huang et al. 2015; Bilal et al. 2017; de Almeida et al. 2021).
Conclusion
In this study, contaminated industrial samples gave rise to a highly efficient dye degrading consortium VITPBC6 composed of B. firmus strain VITEPB1, B. flexus strain VITEPB2, B. aryabhattai strain VITEPB3, B. flexus strain VITSP6, B. paraflexus strain VITSPB7, and B. megaterium strain VITSPB9. Optimization of the crucial physicochemical factors affecting dye decolorization by VITPBC6 led to efficient functioning by the consortium. Consortium VITPBC6 was able to tolerate the model dye RO-16 at a very high concentration thereby indicating the fact that high dye concentration in industrial effluents will not be able to lower the decolorization potential of VITPBC6. Various oxidoreductase enzymes aided in the dye degradation by the consortium. Furthermore, the kinetic studies on reaction order and enzyme–substrate interaction shed a deep insight for process control. VITPBC6 successfully degraded RO-16 into small aromatic compounds. The dye degraded metabolites produced by the consortium were also examined for their toxicity. It was found that VITPBC6 reduced dye toxicity significantly after biodegradation. Thus, bioremediation of azo dyes with VITPBC6 will not cause any secondary pollution. Therefore, the consortium VITPBC6 studied here is a potential candidate for textile industry effluent treatment. Future prospect of this research includes scale-up studies in a suitable bioreactor for a more practical approach to real-time textile industry effluent treatment.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Ajaz M, Shakeel S, Rehman A (2020) Microbial use for azo dye degradation—a strategy for dye bioremediation. Int Microbiol 23:149–159. https://doi.org/10.1007/s10123-019-00103-2
Al-Ansari MM, Li Z, Masood A, Rajaselvam J (2022) Decolourization of azo dye using a batch bioreactor by an indigenous bacterium Enterobacter aerogenes ES014 from the waste water dye effluent and toxicity analysis. Environ Res 205:112189. https://doi.org/10.1016/j.envres.2021.112189
Al-Tohamy R, Sun J, Fareed MF, Kenawy ER, Ali SS (2020) Ecofriendly biodegradation of Reactive Black 5 by newly isolated Sterigmatomyces halophilus SSA1575, valued for textile azo dye wastewater processing and detoxification. Sci Rep 10:12370. https://doi.org/10.1038/s41598-020-69304-4
APHA (1992) Standard methods for the examination of water and wastewater. DC (United States), Washington
Archibald FS (1992) A new assay for lignin-type peroxidases employing the dye azure B. Appl Environ Microbiol 58:3110–3116. https://doi.org/10.1128/2Faem.58.9.3110-3116.1992
Asses N, Ayed L, Hkiri N, Hamdi M (2018) Congo red decolorization and detoxification by Aspergillus niger: removal mechanisms and dye degradation pathway. BioMed Res Int 2018:3049686. https://doi.org/10.1155/2018/3049686
Atkins P, de Paula J (2006) Atkins’ physical chemistry. Oxford University Press, Great Britain, UK
Ayed L, Bekir K, Achour S, Cheref A, Bakhrouf A (2017) Exploring bioaugmentation strategies for azo dye CI Reactive Violet 5 decolourization using bacterial mixture: dye response surface methodology. Water Environ J 31(1):80–89. https://doi.org/10.1111/wej.12216
Bafana A, Devi SS, Krishnamurthi K, Chakrabarti T (2007) Kinetics of decolourisation and biotransformation of direct black 38 by C. hominis and P. stutzeri. Appl Microbiol Biotechnol 74:1145–1152. https://doi.org/10.1007/s00253-006-0751-5
Bafana A, Chakrabarti T, Devi SS (2008) Azoreductase and dye detoxification activities of Bacillus velezensis strain AB. Appl Microbiol Biotechnol 77:1139–1144. https://doi.org/10.1007/s00253-007-1212-5
Barathi S, Aruljothi KN, Karthik C, Padikasan IA, Ashokkumar V (2022) Biofilm mediated decolorization and degradation of reactive red 170 dye by the bacterial consortium isolated from the dyeing industry wastewater sediments. Chemosphere 286:131914. https://doi.org/10.1016/j.chemosphere.2021.131914
Bekhit F, Farag S, Attia AM (2020) Decolorization and degradation of the Azo dye by bacterial cells coated with magnetic iron oxide nanoparticles. Environ Nanotechnol Monit Manag 14:100376. https://doi.org/10.1016/j.enmm.2020.100376
Benkhaya S, M’rabet S, El Harfi A (2020) Classifications, properties, recent synthesis and applications of azo dyes. Heliyon 6:e03271. https://doi.org/10.1016/j.heliyon.2020.e03271
Bera SP, Tank SK (2021) Bioremedial approach of Pseudomonas stutzeri SPM-1 for textile azo dye degradation. Arch Microbiol 203:2669–2680. https://doi.org/10.1007/s00203-021-02258-x
Berg JM, Tymoczko JL, Stryer L (2002) Biochemistry. United States, New York
Bhatia D, Sharma NR, Singh J, Kanwar RS (2017) Biological methods for textile dye removal from wastewater: a review. Crit Rev Environ Sci Technol 47:1836–1876. https://doi.org/10.1080/10643389.2017.1393263
Bilal M, Iqbal HM, Hu H, Wang W, Zhang X (2017) Enhanced bio-catalytic performance and dye degradation potential of chitosan-encapsulated horseradish peroxidase in a packed bed reactor system. Sci Total Environ 575:1352–1360. https://doi.org/10.1016/j.scitotenv.2016.09.215
Cai Z, Zhang W, Ma J, Cai J, Li S, Zhu X, Yang G, Zhao X (2015) Biodegradation of azo dye disperse orange S-RL by a newly isolated strain Acinetobacter sp. SRL8. Water Environ Res 87:516–523. https://doi.org/10.2175/106143014X13975035526068
Cao J, Sanganyado E, Liu W, Zhang W, Liu Y (2019) Decolorization and detoxification of Direct Blue 2B by indigenous bacterial consortium. J Environ Manage 242:229–237. https://doi.org/10.1016/j.jenvman.2019.04.067
Central Pollution Control Board (2021) Pollution control acts, rules & notifications issued thereunder. New Delhi, India
Chakravarthi B, Mathakala V, Palempalli UMD (2020) Bioremediation of textile azo dyes by marine streptomyces. In: Ghosh SK, Saha PD, Di Francesco M (eds) Recent trends in waste water treatment and water resource management, 1st edn. Springer, Singapore, pp 129–142
Chen CH, Chang CF, Ho CH, Tsai TL, Liu SM (2008) Biodegradation of crystal violet by a Shewanella sp. NTOU1. Chemosphere 72:1712–1720. https://doi.org/10.1016/j.chemosphere.2008.04.069
Connors KA (1990) Chemical kinetics: the study of reaction rates in solution. VCH Publisher, Inc., New York, Weinheim
Cossolin AS, Reis HCO, Castro KC, Santos BA, Marques MZ, Parizotto CA, Vasconcelos LG, Morais EB (2019) Decolorization of textile azo dye Reactive Red 239 by the novel strain Shewanella xiamenensis G5–03 isolated from contaminated soil. Rev Ambient e Água 14:e2446. https://doi.org/10.4136/ambi-agua.2446
Dafale N, Wate S, Meshram S, Nandy T (2008) Kinetic study approach of remazol black-B use for the development of two-stage anoxic–oxic reactor for decolorization/biodegradation of azo dyes by activated bacterial consortium. J Hazard Mater 159(2–3):319–328. https://doi.org/10.1016/j.jhazmat.2008.02.058
Das A, Mishra S (2017) Removal of textile dye reactive green-19 using bacterial consortium: process optimization using response surface methodology and kinetics study. J Environ Chem Eng 5:612–627. https://doi.org/10.1016/j.jece.2016.10.005
Das A, Mishra S (2019) Complete biodegradation of azo dye in an integrated microbial fuel cell-aerobic system using novel bacterial consortium. Int J Environ Sci Technol 16:1069–1078. https://doi.org/10.1007/s13762-018-1703-1
Dave SR, Patel TL, Tipre DR (2015) Bacterial degradation of azo dye containing wastes. In: Singh SN (ed) Microbial degradation of synthetic dyes in wastewaters. Springer International Publishing, Switzerland, pp 57–83
Dawkar VV, Jadhav UU, Jadhav SU, Govindwar SP (2008) Biodegradation of disperse textile dye Brown 3REL by newly isolated Bacillus sp. VUS J Appl Microbiol 105:14–24. https://doi.org/10.1111/j.1365-2672.2008.03738.x
de Almeida EJR, Halfeld GG, Reginatto V, de Andrade AR (2021) Simultaneous energy generation, decolorization, and detoxification of the azo dye Procion Red MX-5B in a microbial fuel cell. J Environ Chem Eng 9:106221. https://doi.org/10.1016/j.jece.2021.106221
Dixit S, Garg S (2018) Biodegradation of environmentally hazardous azo dyes and aromatic amines using Klebsiella pneumoniae. J Environ Eng 144:04018035. https://doi.org/10.1061/(ASCE)EE.1943-7870.0001353
El-Kader A, El-Chaghaby GA, Khalafalla GM, Refae RI, Elshishtawy HM (2019) A novel microbial consortium from sheep compost for decolorization and degradation of Congo red. Glob J Environ Sci Manag 5:61–70. https://doi.org/10.22034/gjesm.2019.01.05
EPA (2001) Parameters of water quality: interpretation and standards. Environmental Protection Agency. https://www.yumpu.com/en/document/read/22995497/parameters-of-water-quality-interpretation-and-standards
Erkurt HA (2010) Biodegradation of azo dyes. Springer
Eskandari F, Shahnavaz B, Mashreghi M (2019) Optimization of complete RB-5 azo dye decolorization using novel cold-adapted and mesophilic bacterial consortia. J Environ Manage 241:91–98. https://doi.org/10.1016/j.jenvman.2019.03.125
Eslami H, Shariatifar A, Rafiee E, Shiranian M, Salehi F, Hosseini SS, Eslami G, Ghanbari R, Ebrahimi AA (2019) Decolorization and biodegradation of reactive Red 198 Azo dye by a new Enterococcus faecalis–Klebsiella variicola bacterial consortium isolated from textile wastewater sludge. World J Microbiol Biotechnol 35:38. https://doi.org/10.1007/s11274-019-2608-y
Földesi B (2019) Guide to enzyme unit definitions and assay design. https://www.biomol.com/resources/biomol-blog/guide-to-enzyme-unit-definitions-and-assay-design. Accessed 14 Jan 2022
Furuya N, Yamasaki S, Nishioka M, Shiraishi I, Iiyama K, Matsuyama N (1997) Antimicrobial activities of Pseudomonads against plant pathogenic organisms and efficacy of Pseudomonas aeruginosa ATCC7700 against bacterial wilt of tomato. Ann Phytopathol Soc Jpn 63:417–424. https://doi.org/10.3186/jjphytopath.63.417
Ghaly AE, Ananthashankar R, Alhattab MVVR, Ramakrishnan VV (2014) Production, characterization and treatment of textile effluents: a critical review. J Chem Eng Process Technol 5:1000182. https://doi.org/10.4172/2157-7048.1000182
Guo G, Li X, Tian F et al (2020a) Azo dye decolorization by a halotolerant consortium under microaerophilic conditions. Chemosphere 244:125510. https://doi.org/10.1016/j.chemosphere.2019.125510
Guo G, Hao J, Tian F, Liu C, Ding K, Xu J, Zhou W, Guan Z (2020b) Decolorization and detoxification of azo dye by halo-alkaliphilic bacterial consortium: systematic investigations of performance, pathway and metagenome. Ecotoxicol Environ Saf 204:111073. https://doi.org/10.1016/j.ecoenv.2020.111073
Guo G, Liu C, Hao J, Tian F, Ding K, Zhang C, Yang F, Liu T, Xu J, Guan Z (2021) Development and characterization of a halo-thermophilic bacterial consortium for decolorization of azo dye. Chemosphere 272:129916. https://doi.org/10.1016/j.chemosphere.2021.129916
Gurav R, Bhatia SK, Choi TR, Choi YK, Kim HJ, Song HS, Lee SM, Park SL, Lee HS, Koh J, Jeon JM (2021) Application of macroalgal biomass derived biochar and bioelectrochemical system with Shewanella for the adsorptive removal and biodegradation of toxic azo dye. Chemosphere 264:128539. https://doi.org/10.1016/j.chemosphere.2020.128539
Haq I, Raj A (2018) Biodegradation of Azure-B dye by Serratia liquefaciens and its validation by phytotoxicity, genotoxicity and cytotoxicity studies. Chemosphere 196:58–68. https://doi.org/10.1016/j.chemosphere.2017.12.153
Haq I, Kumari V, Kumar S, Raj A, Lohani M, Bhargava R (2016) Evaluation of the phytotoxic and genotoxic potential of pulp and paper mill effluent using Vigna radiata and Allium cepa. Adv Biol 2016:8065736. https://doi.org/10.1155/2016/8065736
Haque MM, Haque MA, Mosharaf MK, Marcus PK (2021a) Novel bacterial biofilm consortia that degrade and detoxify the carcinogenic diazo dye Congo red. Arch Microbiol 203:643–654. https://doi.org/10.1007/s00203-020-02044-1
Haque MM, Haque MA, Mosharaf MK, Marcus PK (2021b) Decolorization, degradation and detoxification of carcinogenic sulfonated azo dye methyl orange by newly developed biofilm consortia. Saudi J Biol Sci 28:793–804. https://doi.org/10.1016/j.sjbs.2020.11.012
Hassan MM, Carr CM (2018) A critical review on recent advancements of the removal of reactive dyes from dyehouse effluent by ion-exchange adsorbents. Chemosphere 209:201–219. https://doi.org/10.1016/j.chemosphere.2018.06.043
Holkar CR, Jadhav AJ, Pinjari DV, Mahamuni NM, Pandit AB (2016) A critical review on textile wastewater treatments: possible approaches. J Environ Manage 182:351–366. https://doi.org/10.1016/j.jenvman.2016.07.090
Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST (1993) Bergey’s manual of determinative bacteriology. Baltimore, USA
Huang G, Wang W, Liu G (2015) Simultaneous chromate reduction and azo dye decolourization by Lactobacillus paracase CL1107 isolated from deep sea sediment. J Environ Manage 157:297–302. https://doi.org/10.1016/j.jenvman.2015.04.031
Ikram M, Naeem M, Zahoor M, Hanafiah MM, Oyekanmi AA, Islam NU, Ullah M, Mahnashi MH, Ali AA, Jalal NA, Bantun F (2022a) Bacillus subtilis: as an efficient bacterial strain for the reclamation of water loaded with textile azo dye, orange II. Int J Mol Sci 23(18):10637. https://doi.org/10.3390/ijms231810637
Ikram M, Naeem M, Zahoor M, Rahim A, Hanafiah MM, Oyekanmi AA, Shah AB, Mahnashi MH, Al Ali A, Jalal NA, Bantun F (2022b) Biodegradation of azo dye methyl red by Pseudomonas aeruginosa: optimization of process conditions. Int J Environmental Res Public Health 19(16):9962. https://doi.org/10.3390/ijerph19169962
Jadhav JP, Kalyani DC, Telke AA, Phugare SS, Govindwar SP (2010) Evaluation of the efficacy of a bacterial consortium for the removal of color, reduction of heavy metals, and toxicity from textile dye effluent. Bioresour Technol 101:165–173. https://doi.org/10.1016/j.biortech.2009.08.027
Joshi AU, Hinsu AT, Kotadiya RJ, Rank JK, Kothari RK (2020) Decolorization and biodegradation of textile di-azo dye Acid Blue 113 by Pseudomonas stutzeri AK6. 3 Biotech 10:214. https://doi.org/10.1007/s13205-020-02205-5
Karunya A, Nachiyar CV, Ananth PB, Sunkar S, Jabasingh SA (2014) Development of microbial consortium CN-1 for the degradation of Mordant Black 17. J Environ Chem Eng 2:832–840. https://doi.org/10.1016/j.jece.2014.02.012
Khan R, Khan Z, Nikhil B, Jyoti D, Datta M (2014) Azo dye decolorization under microaerophilic conditions by a bacterial mixture isolated from anthropogenic dye-contaminated soil. Bioremediat J 18:147–157. https://doi.org/10.1080/10889868.2014.889076
Krishnamoorthy R, Jose PA, Ranjith M, Anandham R, Suganya K, Prabhakaran J, Thiyageshwari S, Johnson J, Gopal NO, Kumutha K (2018) Decolourisation and degradation of azo dyes by mixed fungal culture consisted of Dichotomomyces cejpii MRCH 1–2 and Phoma tropica MRCH 1–3. J Environ Chem Eng 6:588–595. https://doi.org/10.1016/j.jece.2017.12.035
Krithika T, Kavitha R, Dinesh M, Angayarkanni J (2021) Assessment of ligninolytic bacterial consortium for the degradation of azo dye with electricity generation in a dual-chambered microbial fuel cell. Environ Challenges 4:100093. https://doi.org/10.1016/j.envc.2021.100093
Kumar V, Jamwal A, Kumar V, Singh D (2022) Green bioprocess for degradation of synthetic dyes mixture using consortium of laccase-producing bacteria from Himalayan niches. J Environ Manage 310:114764. https://doi.org/10.1016/j.jenvman.2022.114764
Kurade MB, Waghmode TR, Kagalkar AN, Govindwar SP (2012) Decolorization of textile industry effluent containing disperse dye Scarlet RR by a newly developed bacterial-yeast consortium BL-GG. Chem Eng J 184:33–41. https://doi.org/10.1016/j.cej.2011.12.058
Labuda J, Bowater RP, Fojta M, Gauglitz G, Glatz Z, Hapala I, Havliš J, Kilar F, Kilar A, Malinovská L, Sirén HM (2018) Terminology of bioanalytical methods (IUPAC Recommendations 2018). Pure Appl Chem 90:1121–1198. https://doi.org/10.1515/pac-2016-1120
Lade HS, Waghmode TR, Kadam AA, Govindwar SP (2012) Enhanced biodegradation and detoxification of disperse azo dye Rubine GFL and textile industry effluent by defined fungal-bacterial consortium. Int Biodeterior Biodegradation 72:94–107. https://doi.org/10.1016/j.ibiod.2012.06.001
Lade H, Kadam A, Paul D, Govindwar S (2015) A low-cost wheat bran medium for biodegradation of the benzidine-based carcinogenic dye Trypan Blue using a microbial consortium. Int J Environ Res Public Health 12:3480–3505. https://doi.org/10.3390/ijerph120403480
Leelakriangsak M, Borisut S (2012) Characterization of the decolorizing activity of azo dyes by Bacillus subtilis azoreductase AzoR1. Warasan Songkhla Nakharin 34(5):509–516
Li T, Song HL, Xu H, Yang XL, Chen QL (2021) Biological detoxification and decolorization enhancement of azo dye by introducing natural electron mediators in MFCs. J Hazard Mater 416:125864. https://doi.org/10.1016/j.jhazmat.2021.125864
Lowry OH, Rosenbrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Madamwar D, Tiwari O, Jain K (2019) Mapping of research outcome on remediation of dyes, dye intermediates and textile industrial waste. A Research Compendium. https://dbtindia.gov.in/sites/default/files/Compendium_Master-File_1.pdf. Accessed 2 Mar 2022
Mohana S, Shrivastava S, Divecha J, Madamwar D (2008) Response surface methodology for optimization of medium for decolorization of textile dye Direct Black 22 by a novel bacterial consortium. Bioresour Technol 99:562–569. https://doi.org/10.1016/j.biortech.2006.12.033
Mohanty SS, Kumar A (2021) Enhanced degradation of anthraquinone dyes by microbial monoculture and developed consortium through the production of specific enzymes. Sci Rep 11:1–15. https://doi.org/10.1038/s41598-021-87227-6
Nachiyar CV, Ananth PB, Sunkar S, Karunya A, Jabasingh SA (2016) Decolorization and biodegradation of textile azo dye Acid Black 24 by developed bacterial consortium CN-1A. Indian J Biotechnol 15:412–419
Pal P (2017) Industrial water treatment process technology. Butterworth-Heinemann, UK
Paliwal R, Uniyal S, Rai JPN (2015) Evaluating the potential of immobilized bacterial consortium for black liquor biodegradation. Environ Sci Pollut Res 22:6842–6853. https://doi.org/10.1007/s11356-014-3872-x
Parshetti GK, Telke AA, Kalyani DC, Govindwar SP (2010) Decolorization and detoxification of sulfonated azo dye methyl orange by Kocuria rosea MTCC 1532. J Hazard Mater 176(1–3):503–509. https://doi.org/10.1016/j.jhazmat.2009.11.058
Paszczyński A, Crawford RL, Huynh VB (1988) Manganese peroxidase of Phanerochaete chrysosporium: purification. Methods Enzymol 161:264–270. https://doi.org/10.1016/0076-6879(88)61028-7
Patel Y, Mehta C, Gupte A (2012) Assessment of biological decolorization and degradation of sulfonated di-azo dye Acid Maroon V by isolated bacterial consortium EDPA. Int Biodeterior Biodegradation 75:187–193. https://doi.org/10.1016/j.ibiod.2012.10.004
Poonkuzhali K, Sathishkumar P, Boopathy R, Palvannan T (2011) Aqueous state laccase thermostabilization using carbohydrate polymers: effect on toxicity assessment of azo dye. Carbohydr Polym 85:341–348. https://doi.org/10.1016/j.carbpol.2011.02.031
Prabhakar Y, Gupta A, Kaushik A (2021) Microbial degradation of Reactive Red-35 dye: upgraded progression through Box-Behnken design modeling and cyclic acclimatization. J Water Process Eng 40:101782. https://doi.org/10.1016/j.jwpe.2020.101782
Prasad ASA, Rao KVB (2013) Aerobic biodegradation of Azo dye by Bacillus cohnii MTCC 3616; an obligately alkaliphilic bacterium and toxicity evaluation of metabolites by different bioassay systems. Appl Microbiol Biotechnol 97:7469–7481. https://doi.org/10.1007/s00253-012-4492-3
Prasad ASA, Satyanaryana VSV, Rao KVB (2013) Biotransformation of direct blue 1 by a moderately halophilic bacterium Marinobacter sp. strain HBRA and toxicity assessment of degraded metabolites. J Hazard Mater 262:674–684. https://doi.org/10.1016/j.jhazmat.2013.09.011
Rao KVB, Prasad ASA (2014) Biodecolourisation of azo dye reactive red 22 by Bacillus infantis strain AAA isolated from seawater and toxicity assessment of degraded metabolites. Nat Environ Pollut Technol 13:369–374
Rauf MA, Marzouki N, Körbahti BK (2008) Photolytic decolorization of Rose Bengal by UV/H2O2 and data optimization using response surface method. J Hazard Mater 159:602–609. https://doi.org/10.1016/j.jhazmat.2008.02.098
Rehman K, Shahzad T, Sahar A, Hussain S, Mahmood F, Siddique MH, Siddique MA, Rashid MI (2018) Effect of Reactive Black 5 azo dye on soil processes related to C and N cycling. PeerJ 6:e4802. https://doi.org/10.7717/peerj.4802
Rice EW, Baird RB, Eaton AD (2017) Standard methods for the examination of water and wastewater. Standard methods for the examination of water and wastewater, Washington D.C., US
Roy DC, Biswas SK, Saha AK, Sikdar B, Rahman M, Roy AK, Prodhan ZH, Tang SS (2018) Biodegradation of Crystal Violet dye by bacteria isolated from textile industry effluents. PeerJ 6:e5015. https://doi.org/10.7717/peerj.5015
Saha P, Rao KVB (2020) Biotransformation of Reactive Orange 16 by alkaliphilic bacterium Bacillus flexus VITSP6 and toxicity assessment of biotransformed metabolites. Int J Environ Sci Technol 17:99–114. https://doi.org/10.1007/s13762-019-02256-z
Saha P, Madliya S, Khare A, Subudhi I, Bhaskara Rao KV (2022) Enzymatic biodegradation, kinetic study, and detoxification of Reactive Red-195 by Halomonas meridiana isolated from Marine Sediments of Andaman Sea. Environ Technol, India. https://doi.org/10.1080/09593330.2022.2038276
Salian R, Wani S, Reddy R, Patil M (2018) Effect of brewery wastewater obtained from different phases of treatment plant on seed germination of chickpea (Cicer arietinum), maize (Zea mays), and pigeon pea (Cajanus cajan). Environ Sci Pollut Res 25:9145–9154. https://doi.org/10.1007/s11356-018-1218-9
Saratale RG, Saratale GD, Chang JS, Govindwar SP (2011) Bacterial decolorization and degradation of azo dyes: a review. J Taiwan Inst Chem Eng 42:138–157. https://doi.org/10.1016/j.jtice.2010.06.006
Saratale RG, Gandhi SS, Purankar MV, Govindwar SP, Oh SE, Saratale GD (2013) Decolorization and detoxification of sulfonated azo dye C.I Remazol red and textile effluent by isolated Lysinibacillus sp. RGS J Biosci Bioeng 115:658–667. https://doi.org/10.1016/j.jbiosc.2012.12.009
Sarioglu M, Bali U, Bisgin T (2007) The removal of CI Basic Red 46 in a mixed methanogenic anaerobic culture. Dyes Pigm 74:223–229. https://doi.org/10.1016/j.dyepig.2006.02.001
Shade A, Peter H, Allison SD, Baho DL, Berga M, Bürgmann H, Huber DH, Langenheder S, Lennon JT, Martiny JB, Matulich KL (2012) Fundamentals of microbial community resistance and resilience. Front Microbiol 3:417. https://doi.org/10.3389/fmicb.2012.00417
Sheikhi F, Ardakani MR, Enayatizamir N, Rodriguez-Couto S (2012) The determination of assay for laccase of Bacillus subtilis WPI with two classes of chemical compounds as substrates. Indian J Microbiol 52:701–707. https://doi.org/10.1007/2Fs12088-012-0298-3
Shinkafil MS, Mohammed IU, Hayatu JM, Audu AA (2016) Microbial biotechnology for the decolourization and mineralization of organic components of textile wastewater by single and mixed microbial consortium isolated from effluent treatment plant of African textiles industry Kano, Nigeria. IOSR J Environ Sci Toxicol Food Technol 10:32–39. https://doi.org/10.9790/2402-1004013239
Simpson B (2017) Wastewater sampling, SESDPROC-306-R4. Georgia, Athens
Soundararajan N, Gopi V, Upgade A, Begam N (2012) Bioremediation ability of individual and consortium of non-immobilized and immobilized bacterial strains on industrial azo textile effluent. Ann Biol Res 3:1773–1778
Srinivasan S, Sadasivam SK (2021) Biodegradation of textile azo dyes by textile effluent non-adapted and adapted Aeromonas hydrophila. Environ Res 194:110643. https://doi.org/10.1016/j.envres.2020.110643
Stolz A (2001) Basic and applied aspects in the microbial degradation of azo dyes. Appl Microbiol Biotechnol 56:69–80. https://doi.org/10.1007/s002530100686
Tan L, Ning S, Wang Y, Cao X (2013) Aerobic decolorization of Acid Brilliant Scarlet GR by microbial community and the community dynamics during sequencing batch processes. World J Microbiol Biotechnol 29:1763–1771. https://doi.org/10.1007/s11274-012-1249-1
Telke AA, Kalyani DC, Dawkar VV, Govindwar SP (2009) Influence of organic and inorganic compounds on oxidoreductive decolorization of sulfonated azo dye CI Reactive Orange 16. J Hazard Mater 172:298–309. https://doi.org/10.1016/j.jhazmat.2009.07.008
Thakur MC, Khan A, Doshi H (2012) Isolation and screening of dye degrading micro-organisms from the effluents of dye and textile industries at Surat. Am J Environ Eng 2:152–159. https://doi.org/10.5923/j.ajee.20120206.02.html
USEPA (1978) Chemical oxygen demand (titrimetric, high level for saline waters) by titration. https://www.epa.gov/cwa-methods/approved-cwa-test-methods-inorganic-non-metals. Accessed 8 Dec 2021
Waghmode TR, Kurade MB, Govindwar SP (2011) Time dependent degradation of mixture of structurally different azo and non azo dyes by using Galactomyces geotrichum MTCC 1360. Int J Biodeterior Biodegradation 65:479–486. https://doi.org/10.1016/j.ibiod.2011.01.010
Waghmode TR, Kurade MB, Sapkal RT, Bhosale CH, Jeon BH, Govindwar SP (2019) Sequential photocatalysis and biological treatment for the enhanced degradation of the persistent azo dye methyl red. J Hazard Mater 371:115–122. https://doi.org/10.1016/j.jhazmat.2019.03.004
Wilson K (2001) Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol 56:2–4. https://doi.org/10.1002/0471142727.mb0204s56
Yaseen DA, Scholz M (2019) Textile dye wastewater characteristics and constituents of synthetic effluents: a critical review. Int J Environ Sci Technol 16(2):1193–1226. https://doi.org/10.1007/s13762-018-2130-z
Acknowledgements
The authors wish to thank the VIT management for providing a suitable platform to carry out this research work.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Conceptualization, experimentation, and interpretation were performed by Purbasha Saha, Akella Sivaramakrishna, and K.V. Bhaskara Rao. The first draft of the manuscript was written by Purbasha Saha and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: Diane Purchase
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saha, P., Sivaramakrishna, A. & Rao, K.V.B. Bioremediation of reactive orange 16 by industrial effluent-adapted bacterial consortium VITPBC6: process optimization using response surface methodology (RSM), enzyme kinetics, pathway elucidation, and detoxification. Environ Sci Pollut Res 30, 35450–35477 (2023). https://doi.org/10.1007/s11356-022-24501-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-022-24501-8