Abstract
Indigenous microbial diversity has potential for rapid decomposition of residue through enzyme activities that is alternative, effective, and environment friendly strategy to accelerate degradation of lignocellulose in agricultural residues and make composting process economically viable. Keeping this view, the main objective of the present study was isolation and characterization of lignocellulosic degrading microbial diversity from long-term residue management practice experiments and to develop potential microbial consortium for rapid degradation of lignocellulosic biomass. In this study, twenty-five bacteria, nine fungi, and four actinomycetes isolates were obtained from the soil samples of different residue management fields from Ludhiana, Punjab, India. All isolates were qualitatively and quantitatively screened for enzyme activities, i.e., cellulase, xylanase, laccase, and lignin peroxidase. On the basis of quantitative estimation of enzyme activities, 3 fungal (S1F1, S2F4, and S6F9), 2 actinomycetes (S1A1 and S6A4), and 2 bacterial strains (S6B16 and S6B17) were further selected for in vitro bio-compatibility assay. Selected bio-compatible microbial strains were identified as Streptomyces flavomacrosporus (S6A4), Aspergillus terreus (S2F4), and Bacillus altitudinis (S6B16) through 16S rRNA and 18S rRNA sequencing. Furthermore, single and developed microbial consortium (S6B16 + S6A4 + S2F4) were screened for quantitative estimation of cellulase, xylanase, laccase, and lignin peroxidase enzymes with 23 biochemically different cereal, legume, and oil seed crop residues for optimization of enzyme activities at different time intervals. Results revealed that Vigna radiata followed by Cajanus cajan and Arachis hypogaea straw residue powder @ 1% in culture broth are a promising carbon source for B. altitudinis, S. flavomacrosporus, and A. terreus to produce higher ligno-cellulolytic microbial degrading enzymes due to variable range of carbon (C):nitrogen (N) ratio and higher ligno-cellulolytic content in the studied crop residues. Thus, the application of indigenous microbial consortium with efficient lignocellulose hydrolysis enzyme machinery might be an attractive alternative for ex situ crop residue management practices under sustainable manners.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Crop residues, as essential renewable resources and by-products of crop production, contain abundant essential macro- and micro-nutrients for plant growth promotion (Zhang et al. 2017). In India, about 550 million tons (Mt) of crop residues are produced annually and about 90% of production are concentrated in the Indo-Gangetic Plains (IGP) (Devi et al. 2017). Majority of agricultural residues were discarded or burned, wasting resources and generating significantly environmental risks (Shen and Chen 2009; Sharma et al. 2021). As a result, the appropriate exploitation of agricultural residues is critical for a long-term agricultural production system. In the long term, leaving residues in the field is a beneficial agricultural strategy for increasing organic matter, soil nutrient availability, alleviating soil degradation and essential nutrient depletion, reducing agrochemical inputs, increasing crop production, and reducing pollution (Singh et al. 2018; Wang et al. 2018; Sharma et al. 2020).
Soil is one of the most precious natural resources of earth and to maintain its health is the moral responsibility of mankind. Microorganisms are well known for maintaining the stability, sustainability, and productivity of soil ecosystems through their contributions to the plant-microbial interactions system (Kuijken et al. 2015; Zhang et al. 2018). The agricultural residue inputs can serve as a direct source of energy and a habitat for soil microbial activities (Zhang et al. 2017). Moreover, crop residue input can also alter soil physio-chemical parameters such as nutrient and moisture availability, which might indirectly affect microbial proliferation and physiological activities (Prendergast-Miller et al. 2014; Banerjee et al. 2016). Different crop residues decompose as a result of complicated microbial metabolic processes influenced by a variety of environmental conditions (Kriauciuniene et al. 2012). Native soil organic matter (SOM), organic inputs, and their microbial interactions can impact the functional diversity of the microbial community and other early decomposing rhizospheric microorganisms (Georgieva et al. 2005). Even bio-chemicals such as soluble hemicellulose, cellulose, lignin fraction, C:N ratio, and recalcitrant C fractions in the different crop residues are generally considered sufficient to induce difference in the nature of decomposer organisms (Cheshire et al. 1999). Plant materials are composed of standard components, but their ratios vary, affecting the rates of degradation of agricultural residues (Martin 2002; Hadas et al. 2004). Decomposition is influenced by the morphological characteristics of residue, which interact with the biochemical components to regulate the rate of decomposition (Wolf and Snyder 2003). Many researchers have confirmed the role of various physico-chemical properties to predict the initial residue N content, lignin, cellulose, hemicellulose, polyphenol, and soluble C concentrations, which are useful indicators of different crop residue quality. When the N requirements for the metabolic activities of the soil decomposers are not provided by the residue, N availability may limit the kinetics of decomposition of crop residues, especially those with a high C:N ratio such as cereals (Oglesby and Fownes 1992; Recous et al. 1995; Sharma et al. 2021).
In view of reducing the harmful effects of in situ burning of crop residues, as well as the convenience of farmers, an economical and environment friendly residue management approach should be adopted for effective utilization of different crop residues. Inoculation of ligno-cellulolytic microorganisms is an effective technology to accelerate the degradation potential of lignocellulose in agricultural waste. Shukla et al. (2014) demonstrated that using fungal inoculation to turn crop residues into good quality compost provides a potential ex situ solution to its disposal issues. Using a wide range of agricultural wastes (i.e., paddy straw, soybean trash, pearl millet, maize, and mustard), the hypercellulolytic fungal consortium (such as Aspergillus nidulans, Trichoderma viride, Phanerochaete chrysosporium, and Aspergillus awamori) enhanced compost production on the basis of their ligno-cellulytic enzyme production potential (Lee et al. 2011; Song et al. 2010; Choudhary et al. 2016). Among microbial agents of decomposition, fungi, bacteria, and actinomycetes are the most important group in the rhizosphere and play an essential role in the biodegradation of the lignocellulosic organic waste. Several fungal and bacterial species such as Rhizopus oryzae, A. oryzae, A. fumigates, A. terreus, P. fusisporous, Micromonospora, Bacillus sp., Trichonympha, and Clostridium and their consortium have been reported in literature (Goyal and Sindhu 2011; Borah et al. 2016). Kausar et al. (2010) reported ligno-cellulolytic actinobacteria, Micromonospora carbonacea with potential to degrade cellulose, hemicelluloses, and lignin significantly over the control. As a result, current research has concentrated on microbial consortia degrading plant biomass, with the assumption that the expected diversity of microbial secreted enzymes will result in effective breakdown rates (Schwarz 2001). As a result, microbial consortia may be better able to cope with physiological changes. Understanding, designing, and testing efficient bio-degradative agents under a sustainable agriculture system requires examining bio-compatible microbial consortia grown on lignocellulosic diverse plant biomass (Zuroff and Curtis 2012). As a result, we hypothesized that in the Indo-Gangetic Plains (IGPs) zone of Punjab, India, the majority of farmers practice in situ burning of agricultural residues, which degrades soil health and environmental quality. Indigenous microbial communities have the ability to degrade lignocellulose and can be exploited to decompose crop residues quickly in situ or ex situ. Following these divergent lines of reasoning, the main objective of this study were (i) to isolate indigenous microbial diversity from different locations of Ludhiana, Punjab, and with 16S/18S rRNA gene sequencing; (ii) to produce potential microbial consortia on the afore mentioned with synthetic substrate; and (iii) to test biochemically diverse residues as substrates for specific enzyme activities with developed microbial consortia up to 20th days of incubation to drive the establishment of best substrate for rapid microbial lignocelluloses degrading consortia.
Material and methods
Sampling site
Soil samples (0–15 cm) were randomly collected from different long-term experimental fields at the soil research farm, Punjab Agricultural University, Ludhiana, Punjab (30° 56′ N and 75° 52′ E) with almost similar physiographic conditions, representative of the study region from the fields under crop residue management practices in rice–wheat cropping system. The region has a sub-tropical climate, wet summers with hot, and dry winters with 876 mm mean annual precipitation and temperature range 23.7–39.2 °C during summer (Singh et al. 2010). The basic soil conditions of Typic Ustochrept sandy loam soil at the experimental farm of Punjab Agricultural University, Ludhiana, Punjab, are provided in Supplementary Table 1. After wheat harvesting in May–June 2018, soil samples were collected with a metallic soil core sampler (inner diameter 7.3 cm). Each soil sample was passed through 2-mm sieve for analysis of soil chemical properties, while another portion was retained without sieving and stored immediately at 4 °C for isolation of microbial diversity.
Soil analysis
Soil samples were analyzed for pH1:2 (1:2; soil:water suspension) using a glass electrode, electrical conductivity (E.C.1:2) (1:2; soil:water supernatant) with a conductivity meter (Jackson 1973), and particle size distribution by international pipette method (Piper 1966). Olsen’s-P in soil samples was determined by extracting the soils with 0.5 M sodium bicarbonate (Olsen et al. 1954). Available-K in soil samples was extracted with 1 N (ammonium acetate (Mervin and Peech 1950) followed by flame photometric determination.
Biochemical analysis of different crop residues
Cell wall components of different crop residues were determined using standard protocol described by AOAC (1995) under crude fiber extraction unit (FOSS, FT-122 FibertecTM). The oven-dried different crop residues were ground to pass 40 meshes for analyzing diverse cell wall fractions. The components of cell wall from different crop residues such as cellulose, hemicellulose, and lignin were measured via standard protocol described by Van Soest et al. (1991). C and N were estimated by Kel Plus unit and C to N ratio was calculated as per the procedure described by Choudhary et al. (2016).
Isolation and morpho-biochemical characterization of microbial diversity
One gram of each composite soil sample was suspended in 9 ml of autoclaved distilled water as blank through serial dilution method. Suspension of serial diluted samples (10−1 to 10−6 times), 100 µl of 10−5 aliquot was spread on carboxyl methyl cellulase (CMC), xylan agar, and lignin agar media plates for isolation of cellulase, xylanase, and laccase producing microbial diversity (Puentes-Tellez and Salles 2018). The isolates forming clear zone around the colonies on the specific medium were selected as cellulase, xylanase, and laccase producers. The fungal, bacterial, and actinomycetes isolates were maintained on slant of rose bengal, nutrient agar, and Kenknight medium, respectively, at 4 °C. The diversity of microbial isolates were microscopically analyzed for morphological characteristics, i.e., colony shape, cell motility, and Gram’s reaction under the OLMPUS-CX21 bio-inocular microscope (Cowan 1974; Vincent 1970). All the isolates were biochemically characterized by different tests (catalase, oxidase, methyl red, Voges Proskauer’s, nitrate reduction, and starch hydrolysis).
Quantitative enzyme activities
The carboxyl methyl cellulase (CMCase) enzyme activity was estimated with standard protocol described by Mandel and Sternberg (1976). After 10th days of incubation culture CMC broth, 0.5 ml of culture supernatant, filter paper strip (1 × 6 cm) was added to 500 µL of 1% CMC prepared in 0.1 M sodium citrate buffer (pH 4.8 ± 0.2) in the culture tube and incubated in water bath at 50 °C for 1 h. After the incubation, reaction of enzyme activity was terminated by adding 3.0 ml of di-nitrosalicylic acid (DNS) and subsequently incubated reaction tubes at 100 °C for 20 min in a water bath. One milliliter of sodium potassium tartrate solution (Rochelle salt solution) was added to stabilize the color and after cooling diluted with 2 ml of distilled water. The absorbance/optical density (OD) was noticed against the blank of 0.1 M sodium citrate buffer at 575 nm. The corresponding enzyme activity was calculated from standard curve of glucose and expressed as µg/ml.
The enzyme activities including xylanase (Sandhu and Kalra 1982), laccase (Turner and Green 1974), and lignin peroxidase (LiP) (Singh et al. 1988; Tien and Kirk 1983) were estimated according to standard protocol described. The crude enzyme extract was taken from different culture inoculated in the specific broth (xylan and lignin broth) supplemented with 1% xylan for xylanase and 4 mM guaiacol for laccase as well as lignin peroxidase enzyme activities after 10 days of incubation at 28 °C. All the enzyme activities were defined and calculated according to method of Xu and Yang (2010) and Seo et al. (2013).
Molecular identification and phylogenetic analysis of potential indigenous microbial isolates
Selected bio-compatible potential microbial (bacteria, actinomycetes, and fungus) isolates selected on the basis of higher enzyme activities were subjected to genus level identification by using 16S rRNA and 18S rRNA gene sequencing. Microbial DNA was extracted with standard CTAB protocol described by Huang et al. (2013). Quality and quantity of genomic DNA were measured by gel electrophoresis in 0.8% agarose gel and visualized under gel documentation systems. The 16S rRNA gene fragment (1500 bp) was PCR-amplified from selected bacteria and actinomycetes using the following universal primers: 8F (5′AGAGTTTGATCCGTGGCTC-3′) and 1492R (5′ACGGCTACCTTGTTACGACTT-3′) (Giongo et al. 2010). PCR reaction was performed (Applied Biosystems, Thermo-Fisher Scientific) in a 20.0-µl final volume containing 2 µl (30–60 ng) of template DNA, 2.5 µl of 10X Taq polymerase buffer (100 mM of Tris- HCl and 15 mM MgCl2), 1.5 µl (100 pM/µl) of each universal primer, 200 µM of dNTP’s, 0.5 µl Taq DNA polymerase (Promega, USA), and 10.0 µl of nuclease free autoclavable double distilled water. An initial denaturation stage at 94 °C for 1.30 min was followed by 35 amplification cycles of denaturation at 94 °C for 1 min, annealing at 52 °C for 40 s, primer extension at 72 °C for 1 m, and a final extension at 72 °C for 7 min. To assess the quantity and quality of amplified PCR product at 1.5%, agarose gel stained with ethidium bromide dye were used.
Amplification of the 18S rRNA gene fragment was performed in a 20-µl final volume containing 2 µl (30–60 ng) of fungal DNA, 0.125 µl of each (ITS-1 & ITS-4) primers (100 pM µL−1), 2.5 µl of 10X Taq polymerase buffer (100 mM of Tris -HCl and 15 mM MgCl2), 200 µM of dNTP’s, 0.5 µl Taq DNA polymerase (Promega, USA), and 10.0 µl of nuclease free autoclavable double distilled water. Amplifying PCR product (500–600 kb fragment) of 18S rRNA gene, the following primers were used: ITS-1 (5′-TCCGTAGGTGAACCTGCGC-3′) and ITS-4 (5′TCCTCCGCTTATTGATATGC-3′) (Gardes and Bruns 1993). The PCR profile employed was as follows: primary denaturation for 1.30 min at 94 °C, 35 cycles of denaturation at 94 °C for 1 min, annealing at 52 °C for 40 s, extension at 72 °C for 1.30 min, and an additional reaction for 10 min at 72 °C. The PCR products were detected on 1.5% agarose gel to confirm its amplification, quantity, and size of 18 S rRNA genes. All the amplified PCR products from bacteria, actinomycetes, and fungal isolates were sent to Medauxin Biotech Laboratory, Bangalore, India, for sequencing. The 16S rRNA (from bacteria and actinomycetes) and 18 SrRNA sequences (from fungal DNA) were compared with different 16S rRNA and 18S rRNA gene sequences available in NCBI GenBank by using the BLASTN program (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and aligned with similar sequences by using Clustal Omega program. The phylogenetic tree was generated using the Neighbor-Joining method with the MEGA 6.1 tool with Kimura-2 parameters and 1000 bootstrap replicates.
Bio-compatibility assay of the potential isolates
Selected indigenous potential microbial isolates, B. altitudinis (S6B16), and S. flavomacrosporus (S6A4) containing crop residue degrading enzymes were screened for bio-compatibility with A. terreus (S2F4) on modified succinate agar (MSA) medium (Subramanian et al. 2015). Furthermore, bio-compatibility was measured between B. altitudinis (S6B16), S. flavomacrosporus (S6A4), and A. terreus (S2F4) for synergistic growth (initial inoculum concentration 107 cfu/ml and used inoculums as 1:1:1 ratio) at 600 nm wave length after 3rd, 6th, and 9th days of incubation in modified succinate broth (MSB) as single or combined microbial inoculants at 28 ± 2 °C in triplicate (Mishra et al. 2009).
Impact on enzyme activities of different biochemical crop residues with indigenous potential microbial consortia
Twenty-three representative plant residues (straw and roots) from legumes, cereals, and oil seed in Punjab (India) were collected and dried at 65 °C for 2–3 days (Table 1). Furthermore, each crop residue grinded and sieve with 2 mm. The potential cultures as single as well a consortium was inoculated into the specific broth medium supplemented with 1% cereals, legumes, and oil seed crop residue as cellulosic, hemi-cellulase, and lignin substrate and incubated at 28 ± 2 °C for 20th days. The crude enzyme extract was collected at 10th day interval up to 20th days. All the enzyme (cellulase, xylanase, laccase, and lignin peroxidase) activities were estimated as per standard protocol described above (Mandel and Sternberg 1976). The corresponding enzyme activity was calculated with standard curve and expressed as µg/ml.
Statistical analysis
The data were statistically analyzed using analysis of variance (ANOVA) technique using SPSS software for windows 21.0 (SPSS Inc., Chicago, USA). The significant difference between mean of single and combined inoculation treatments was analyzed with high-range statistical domain (HSD) using Tukey’s tests. The treatment means were separated by the least significant difference (LSD test at p < 0.05).
Results
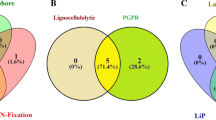
The parameters of chemical composition of straw and roots of 23 representative crop residues are summarized in Table 1. The residue C and N contents ranged from 10.5 to 76.8 g kg−1 and 0.47 to 2.52 g kg−1, respectively, resulting in C:N ratio ranging from 5.06 to 78.9. The cellulose, hemi-cellulose, and lignin content varied from 4.32 to 39.3, 2.49 to 30.4, and 2.03 to 11.0 g kg−1. A total of 5 actinomycetes, 11 fungus, and 72 bacteria were isolated from soil samples of long-term experimental fields. Furthermore, all the microbial diversity was screened qualitatively on the specific media such as CMC agar, xylan agar, and lignin agar media. Out of 5, 11, and 72 (actinomycetes, fungus, and bacteria, respectively), 80% (4) actinomycetes, 82% (9) fungus, and 34.7% (25) bacteria were positive for lignocellulytic enzyme activities such as cellulase, xylanase, and laccase producers (Supplementary Table 2). On the basis of Bergey’s Manual, out of 38 microbial isolates, 73.7% and 26.3%, were Gram positive and Gram negative, respectively. The biochemical characterization of microbial diversity revealed that 78.9%, 71.1%, 68.4%, 65.8%, 15.8%, and 13.2% were positive for nitrate reduction, oxidase, starch hydrolysis, catalase, methyl red, and Voges Proskauer’s test, respectively.
All the microbial isolates were positive for cellulase, xylanase, laccase, and lignin peroxidase enzyme activities in the heat map with clustering and also depicted overall results in the Fig. 1A, B and C. After qualitative screening, furthermore, all the microbial isolates were screened quantitatively for enzyme activities at 10 days of inoculation. Maximum cellulase enzyme activity was recorded for fungal isolates, with S2F4 (72.9 µg/ml) followed by S6F9 (53.9 µg/ml) and S1F1 (45.9 µg/ml). Among bacterial isolates, highest cellulase activity was recorded with S7B23 (21.2 µg/ml) followed by S6B17 (17.0 µg/ml) and S6B16 (16.1 µg/ml) and minimum enzyme activity were observed with S6B21 (7.1 µg/ml) followed by S3B4 (8.1 µg/ml). Similarly, for actinomycetes, significantly high cellulase activity was recorded with S6A4 (13.4 µg/ml) followed by S1A1 (8.3 µg/ml).
For xylanase enzyme activities, fungal isolate S2F4 showed maximum xylanase activity (425.8 µg/ml) followed by S6F9 (401.8 µg/ml) and S3F5 (394.9 µg/ml). From the bacterial diversity, highest xylanase enzyme activity was noticed with S1B1 (447.7 µg/ml) followed by S6B15 (427.3 µg/ml) and S6B22 (322.6 µg/ml) and minimum was detected with S6B20 (94.9 µg/ml) followed by S5B11 (119.8 µg/ml). Similarly, for actinomycetes, significantly high xylanase enzyme activity was recorded S1A1 (279.2 µg/ml) followed by S3A2 (262.4 µg/ml) at 10th day of incubation. At the 10th day of incubation, maximum laccase and lignin peroxidase enzyme activities were noticed with fungal strain S2F4 (16.6 µg/ml and 13.2 µg/ml, respectively) followed by strain S6F9 (15.7 µg/ml and 12.9 µg/ml, respectively). Among bacterial isolates, significantly high laccase and lignin peroxidase was detected in bacterial strain S6B16 (13.0 µg/ml and 11.5 µg/ml, respectively) followed by strain S6B17 (12.8 µg/ml and 11.0 µg/ml, respectively) and minimum was recorded with S2B2 (5.1 µg/ml and 4.4 µg/ml, respectively). On the basis of quantitative estimation of different enzyme activities, 3 fungal strains (S1F1, S2F4, and S6F9), 2 actinomycetes (S1A1 and S6A4), and 2 bacterial isolates (S6B16 and S6B17) were selected for in vitro bio-compatibility assay.
Bio-compatibility interaction assay
On the basis of quantitative estimation of different enzyme activities, 3 fungal strains (S1F1, S2F4, and S6F9), 2 actinomycetes (S1A1 and S6A4), and 2 bacterial isolates (S6B16 and S6B17) were selected for in vitro bio-compatibility interaction on modified succinate agar (MSA) medium (Table 2). Selected strains S. flavomacrosporus (S6A4) and A. terreus (S2F4) with B. altitudinis (S6B16) did not show harmful impact on growth towards each other on specific medium in disc plate, while the remaining selected microbial isolates (S1F1, S6F9, S1A1, and S6B17) showed inhibitory effect against each other on MSA medium plates which were non-compatible and discarded from further studies. Furthermore, mutual proto-cooperation was detected between S. flavomacrosporus (S6A4), A. terreus (S2F4), and B. altitudinis (S6B16) in terms of optical density (OD at 600 nm). Maximum optical density was detected with S2F4 + S6A4 + S6B16 (1.76) in comparison to single inoculants of S2F4 (1.01), S6A4 (0.40), and S6B16 (1.10) on 9th day of inoculation (Fig. 2).
Molecular identification of potential microbial isolates
Selected bio-compatible microbial strains (i.e., S2F4, S6A4, and S6B16) were subjected to molecular identification at genera level by using partial 16S rRNA and 18S rRNA genes sequencing through nucleotide blast using blastin software tool. The partial sequences of microbial strains S2F4, S6A4, and S6B16 showed 92.4, 92.4%, and 85.2% homology with Aspergillus sp., Streptomyces sp., and Bacillus sp., respectively, and submission to NCBI, MD, USA, is under process. Based on consensus sequence homology of 16S rRNA (bacteria and actinomycetes) and 18S rRNA (fungus) gene sequence available in the NCBI Gen Bank database, strain S2F4, S6A4, and S6B16 were identified as A. terreus, S. flavomacrosporus, and B. altitudinis, respectively. The 16S rRNA and 18S rRNA gene sequences of S2F4, S6A4, and S6B16 were aligned with other relevant sequences of microbial species in the GenBank database. Phylogenetic tree of selected potential microbial stains generated through MEGA 6.1 software revealed their homology with microbial strains of the respective species and submission under processing at NCBI, MD, USA (Supplementary Figs. 1 and 2).
Impact of indigenous single as well as microbial consortia for enzyme activities with biochemically different crop residues
Different cereals, leguminous, and oil seed crop residues were supplemented in specific medium to determine the specific enzymatic activities with potential compatible single as well as microbial consortium at 10th days interval up to the 20th day of incubation at 28 ± 2 °C. The present study observed that synthetic substrate, i.e., CMC, xylan, and guaiacol supplemented at the rate of (@) 1% in specific media, showed significantly (p < 0.05) high cellulase, xylanase, laccase, and lignin peroxidase enzyme activities with microbial consortium S6B16 + S6A4 + S2F4 (82.0, 457.4, 192.9, and 173.7 µg/ml, respectively) at the 20th day of incubation (Tables 3, 4, 5, and 6).
The cellulase activity of microbial consortium was significantly (p < 0.05) high as compared to single microbial inoculants with biochemically different crop residues (Table 3). In leguminous crop residues, at the 20th day of incubation, significantly high cellulase activity was recorded in CMC medium supplemented with 1% of V. radiata straw with consortium S6B16 + S6A4 + S2F4 (76.9 µg/ml) followed by 1% A. hypogea and C. cajan straw residue (71.0 and 65.0 µg/ml, respectively) as compared to single inoculant treatments (Supplementary Fig. 3). Among cereals and oil seed crop residues, significant (p < 0.05) interaction was recorded with microbial treatments and different crop residues. Significantly high cellulase enzyme activity was noticed in CMC medium containing 1% S. indicum straw with microbial consortium of S6B16 + S6A4 + S2F4 (50.1 µg/ml) followed by S. indicum root and O. sativa straw (48.7 and 41.4 µg/ml, respectively) as compared to single inoculants at the 20th day of incubation (Fig. 3a, b). Minimum cellulase activity was noticed in C. cajan root residue (34.3 µg/ml) followed by Zea mays straw (28.5 µg/ml) with microbial consortium (S6B16 + S6A4 + S2F4) at 20th days of incubation. Higher cellulase enzyme activity was noticed in the different leguminous crop residue straw as compared to leguminous roots residue.
Effect of crop residues on cellulase enzyme activity with potential microbial inoculants at different time intervals at (A) 10th day and (B) 20th day. T1 Oryza sativa straw; T2 Oryza sativa root; T3 Zea mays straw; T4 Zea mays root; T5 Gossypium hirsutum straw; T6 Gossypium hirsutum root; T7 Sesamum indicum straw; T8 Sesamum indicum root; T9 Triticum aestivum straw
At the 20th day of incubation, xylanase activity in consortium bio-inoculant (S6B16 + S6A4 + S2F4) ranged from 178.4 to 323.5 µg/ml in specific medium supplemented with 14 biochemically different leguminous crop straw and root residue depicted in Table 4. Significantly, high xylanase activity was noticed in media containing 1% V. radiata straw powder with microbial consortium S6B16 + S6A4 + S2F4 (323.5 µg/ml) followed by C. cajan straw (285.5 µg/ml) and S. bispinosa root (264.0 µg/ml) and minimum xylanase enzyme activity was observed with C. juncea root (178.4 µg/ml) followed by C. juncea straw (182.8 µg/ml) over the single inoculants. In the present study, xylanase activity was enhanced by 8.4% with microbial consortium (S6B16 + S6A4 + S2F4) in media containing 1% V. radiata straw powder over the single bacterial (S6B16) inoculant at the 20th day of incubation. Similarly, among cereal and oil seed crop residues, significantly higher xylanase activity was recorded with microbial consortium (S6B16 + S6A4 + S2F4) supplemented with 1% S. indicum root (286.8 µg/ml) followed by S. indicum straw (279.5 µg/ml) and T. aestivum straw (270.5 µg/ml) over the single inoculation treatment at the 20th day of incubation. In cereal and oil seed crops, xylanase enzyme activity was increased by 10.4% with 1% S. indicum straw powder followed by 4.0% in S. indicum root powder with microbial consortium (S6B16 + S6A4 + S2F4) over the single bacterial inoculant (S6B16) at the 20th day of incubation. Minimum xylanase activity (192.0 µg/ml) was noticed with O. sativa and Z. mays @ 1% root powder inoculated with consortium bio-inoculants (S6B16 + S6A4 + S2F4) at the 20th day of incubation (Fig. 4a, b). The interaction effect between microbial inoculation and biochemically different crop residues were found to be significant for laccase and lignin peroxidase activities (Tables 5 and 6). The laccase and lignin peroxidase activities (192.9 and 173.7 µg/ml, respectively) were significantly higher @ 1% guaiacol with microbial consortium (S6B16 + S6A4 + S2F4) treatment over the single inoculation treatment. At the 20th day, significant enhancement of laccase and lignin peroxidase was noticed with microbial consortium (S6B16 + S6A4 + S2F4) @ 1% of V. radiata straw (177.4 and 181.2 µg/ml, respectively) followed by 1% C. cajan straw (162.4 and 151.5 µg/ml, respectively) and A. hypogaea straw (159.4 and 145.9 µg/ml, respectively). Minimum laccase activity was noticed with consortium microbial inoculant in V. mungo root (90.2 µg/ml) and lignin peroxidase with C. juncea root (88.0 µg/ml) at the 20th day of incubation. Whereas in single inoculants, actinomycetes (S6A4) showed minimum laccase and lignin peroxidase enzyme activities with biochemically different crop residue as compared to single bacterial (S6B16) and fungal (S2F4) inoculants.
Effect of crop residues on xylanase enzyme activity with potential microbial inoculants at different time intervals at (A) 10th day and (B) 20th day. T1 Oryza sativa straw; T2 Oryza sativa root; T3 Zea mays straw; T4 Zea mays root; T5 Gossypium hirsutum straw; T6 Gossypium hirsutum root; T7 Sesamum indicum straw; T8 Sesamum indicum root; T9 Triticum aestivum straw
Similarly, among cereal and oil seed crop residues, highest laccase and lignin peroxidase enzyme activities were observed at 10th days of intervals up to 20th days of incubation. Laccase and lignin peroxidase enzyme activities are expressed in the Figs. (5a, b, and 6a, b). At the 20th day of incubation, significantly high laccase and lignin peroxidase enzyme activities were recorded with microbial consortium (S6B16 + S6A4 + S2F4) with 1% of Sesamum indicum root (171.4 and 127.8 µg/ml, respectively) followed by S. indicum straw (161.7 and 122.9 µg/ml, respectively) and Triticum aestivum straw (159.4 and 121.8 µg/ml, respectively). Furthermore, minimum laccase and lignin peroxidase enzyme activities were noticed with microbial consortium with 1% Oryza sativa root powder (121.8 and 88.0 µg/ml, respectively) than single bacterial inoculants. In the present study, compatible consortium bio-inoculant (S6B16 + S6A4 + S2F4) secreted more enzymes, i.e., cellulase, xylanase, laccase, and lignin peroxidase, as compared to single bio-inoculants. Overall mean basis, similar trend was noticed at 10th and 20th days of incubation and maximum enzyme activities were observed by consortium bio-inoculant (S6B16 + S6A4 + S2F4) followed by single inoculants as fungus (S2F4), bacteria (S6B16), and actinomycete (S6A4), respectively, with specific broth supplemented with twenty-three biochemically different legumes, cereals, and oil seed crop residues.
Effect of crop residues on laccase activity with potential microbial inoculants at different time intervals at (A) 10th day and (B) 20th day. T1 Oryza sativa straw; T2 Oryza sativa root; T3 Zea mays straw; T4 Zea mays root; T5 Gossypium hirsutum straw; T6 Gossypium hirsutum root; T7 Sesamum indicum straw; T8 Sesamum indicum root; T9 Triticum aestivum straw
Effect of crop residues on lignin peroxidase activity with potential microbial inoculants at different time intervals at (A) 10th day and (B) 20th day. T1 Oryza sativa straw; T2 Oryza sativa root; T3 Zea mays straw; T4 Zea mays root; T5 Gossypium hirsutum straw; T6 Gossypium hirsutum root; T7 Sesamum indicum straw; T8 Sesamum indicum root; T9 Triticum aestivum straw
Discussion
Microbial populations as diverse as bacteria, actinomycetes, and fungi have the ability to produce enzymes that breakdown a specific component of various crop residues. Composting additives obtained from plant material include organic substrate and bulking agents (Zhe et al. 2008). As a result, the ability of indigenous potential microorganisms to immobilize organic matter is based on their ability to secrete the enzyme required for degradation of substrate constituents such as cellulose, hemicelluloses, and lignin. The enzyme system required becomes more complex and extensive as the substrate becomes more varied. Micro-organisms play a major role in rapid decomposition process with appropriate substrate utilization. Therefore, main objective of present study was isolation, characterization of indigenous microbial strains for enzyme activities with utilization of biochemically different cereals, legumes, and oil seeds crop residues as a substrate and to develop efficient microbial consortium for rapid lignocellulosic decomposition. A total of 88 isolates (72 bacteria, 11 fungus, and 5 actinomycetes) were obtained from different long-term residue management experiments. Our results revealed that the lignocellulolytic enzyme ability is common in microbial diversity from top soil of crop residue management field and especially in the compost. Guniathilake et al. (2013) isolated lignocellulosic microbial diversity from soil, compost, and leaf litter. Similarly, Sahni and Phutela (2013) also diversity of ligno-cellulosic degrading fungi isolated from soil compost, digest slums, and crop residue using paddy straw agar medium supplemented with antibiotic. Vimal et al. (2016) isolated various cellulase-producing bacteria from industrial and agricultural areas and the microbial isolates were tentatively identified to be Bacillus species based on the cultural and morpho-biochemical analysis. Similarly, Rasi and Mahalingam (2014) characterized lignocellulolytic microbial communities conventionally by morphological and biochemical tests and tentatively assigned as Bacillus, Burkholderia, Micrococcus, Serratia, Staphylococcus, Clostridium, Pseudomonas, Achromobacter, Acitobacter, Klebsiella, Proteus, and Enterobacter.
Present results showed significant interaction between microbial consortium and different crop residues for all the enzyme activities at different time intervals. Present study demonstrated that maximum cellulase enzyme activity was noticed in the different leguminous crop residue straw as compared to leguminous roots residue due to the wider range of C:N ratio observed in straw residue over the root residue that showed rapid decomposition of substrate for utilization of nutrients through microbial metabolic activities as well as secretion of large quantity of cellulase enzymes. Kumar et al. (2014) conducted similar study by screening fungal species isolated from forest plant for cellulase enzyme activity and observed significant amount of cellulase production (1.76 U/ml) by Trichoderma viridii on 5 days of incubation at 28 ± 2 °C and pH 5.0. Similarly, Shaikh et al. (2013) isolated 34 lignocellulytic micro-organisms from different regions, i.e., industrial waste, wood wastage material, and sugarcane crop residue. Out of 34, CDB-24 (8.42 U/mg) and CDB-30 (6.01 U/mg) were found to be as most prominent lignocellulytic enzyme producers and the isolates, CDB-24 and CDB-30, were tentatively assigned as Pseudomonas sp. and Bacillus sp., respectively. Diversity of actinomycetes, i.e., Streptomyces, Thermoactinomyces, and Thermomonospora, are capable of degrading cellulose and interfering with lignin structure, although their ability to solubilize lignin is restricted. According to Singh (2017), Streptomyces sp. has been found to play a role in delignification of rice straw, making it more vulnerable to cellulose degrading enzymes. Similarly, Kumar et al. (2008) identified the most promising fungal cultures as Humicola sp. (Th10), Aspergillus nidulans (Th4), Scylalidium thermophilum (T5), and showed significantly high activity of FPase by Th10, cellobiose by Th4, CMCase, and xylanase. Our observations are coherent with Shukla et al. (2016) who also noticed highest production of CMCase, xylanase, FPase, and cellobiose enzyme activities with paddy straw + 1% urea + bacterial consortia + fungal consortia + commercial fungal consortium as compared to control treatment. Mixed compatible microbial consortium have greater effect on substrate accumulation through resistance to environmental contamination and increased enzyme production as compared to single inoculants (Vazquez et al. 2015; Mishra and Malik 2014; Martinez-Sanz et al. 2014; Datta et al. 2017). Present results are in close agreement with Akhter (2014) who reported that 5 PGPR (ARLS510, S412, S414, S4, and FVC70) and 2 Rhizobium strains (AB4 and AB17) were selected for their compatibility. Pseudomonas strain ARLS510 was found compatible with Rhizobium AB4 and AB17 strain in agar spot plate assay. PGPR S414 and S4 strains were interpreted as incompatible with Rhizobium sp. as they induced a clear inhibition zone around their colony. According to Prasad and Babu (2017), they also revealed Azospirillum brasilense strain TNAU and Pseudomonas fluorescens strain PF1 of groundnut as compatible inoculant by streak plate method. Similarly, Kumawat et al. (2021) also found positive interaction between Rhizobium sp. LSMR-32 and Enterococcus mundtii LSMRS-3 of spring mungbean. Shareef et al. (2015) isolated cellulose-degrading bacteria and fungi from agricultural wastes, with Trichoderma viridii and Aspergillus niger producing cellulase activity ranging from 450.39 to 1861.61 IU ml−1, respectively. Our study reported that V. radiata followed by C. cajan and A. hypogaea straw residue in leguminous crops are better alternative agricultural based substrates for microbial enzyme activities after synthetic substrate, i.e., CMC, xylan, and guaiacol, for in vitro microbial decomposition of lignocellulosic biomass. Similarly, in the cereal and oil seed crops, S. indicum followed by T. aestivum and O. sativa straw residue are best substrate for higher microbial enzyme activities for rapid degradation of agricultural residues. Present results concluded significantly higher amount of specific enzyme (i.e., cellulase, xylanase, laccase, and lignin peroxidase) activities estimated in the leguminous crop residues as compared to cereal and oil seed crop residues. This study also showed rapid decomposition of lignocellulosic biomass of legume crop through higher microbial metabolic activities for enzyme production over the cereals and oil seed crop residues under agricultural residue management practices in rice–wheat cropping system. Kumar et al. (2008) demonstrated that thermophilic fungal consortium of A. nidulans, Scytalidium thermophilum, and Humicola sp. were more efficient in decomposition of soybean trash and paddy straw through lignocellulytic enzyme activities. Our study concluded that maximum xylanase activity was noticed with biochemically different crop residue straw powder followed by root powder with potential microbial consortium at the 20th day of incubation due to the wider range of C:N ratio and rapid decomposition for microbial metabolic activities. Cunha et al. (2017) reported significantly high xylanase activity (11.84 U ml−1) with Aspergillus foetidus in soybean residue. Similarly, Delabona et al. (2013) noticed xylanolytic activity for Aspergillus fumigates, 1055.6 U/g and 558.3 U/g in wheat bran and soybean, respectively, after 5 day of incubation. In the same research, using a residual wheat bran, soybean straw, and wheat bran with sugarcane bagasse, he discovered that Aspergillus niger had specific xylanolytic activity of 1285.0 U/g, 484.2 U/g, and 1050.0 U/g, respectively. Shi et al. (2020) reported novel synergetic lignocelluloses degrading mechanisms between T. lanuginosus and T. fusca regulating by the accessibility and concentration of substrate. Previous research not recorded xylanolytic enzyme activity with biochemically different cereals, leguminous, and oil seeds crop residues through developed indigenous microbial consortium (Farinas et al. 2010; Yang et al. 2015).
Sharma et al. (2015) reported that thermophilic bacterial consortium isolated from mature compost of sugarcane showed efficient degradation of rice straw as compared to microbial consortium isolated from rice straw compost. Similarly, Odier and Roch (1983) demonstrated that degradation of poplar wood was stimulated by microbial inoculation with reduced glucose concentration at mesophilic condition. Wang et al. (2011) isolated 14 thermophilic bacteria from soil which can degrade 99%, 81.3%, and 76.9% of filter paper, rice, and cotton straw, respectively, under static conditions, suggesting the potential of indigenous microbial bio-inoculants to degrade lignocelluloses at higher temperature conditions. Wongwilaiwalin et al. (2013) noticed that the crop biomass degrading capability is based on the functional and structural stability of a microbial consortium. After 40 days of incubation, Xu et al. (2009) found 20% weight loss of maize stover in a culture of the white rot fungus Irpex lacteus. Baldrian and Gabriel (2003) also reported a 30% weight loss of wheat biomass during Pleurotus ostreatus growth after 20 days of incubation. Very less information is available on the degradation mechanisms of lignocellulose by microbial diversity. Nonetheless, it is obvious that lignin can be degraded by a variety of microbial genera and species because they erode the secondary cell wall and reduce the concentration of acid insoluble components (Klason lignin) from agricultural waste. Elevated oxygen levels, which accelerate the rate of lignin biodegradation by producing hydrogen peroxides as an extracellular oxidant and inducing ligninolytic activity, are another mechanism linked to lignocellulosic biomass breakdown (Kirk and Cullen 1998; Sánchez 2009). On the basis of above studies, we concluded that mixed microbial consortia, i.e., consisting of bacteria, actinomycetes, and fungal partners, have potentially higher biodegradable performance than single potential bio-inoculants.
Our indigenous potential microbial diversity S. flavomacrosporus strain S6A4 (actinomycete), B. altitudinis strain S6B16 (bacteria), and A. terreus strain S2F4 (fungus) showed significant extracellular (cellulolytic/xylanolytic/ligninolytic) enzyme activities, suggesting that these microbial consortia have metabolic roles in plant polymer degradation. Our in vitro experiment firstly developed and identified lignocellulosic degrading microbial consortium using twenty-three bio-chemically different leguminous, cereals, and oil seed crop residue as a substrate for specific enzyme activities. Lignocellulose degrading microbial consortium were selected and tested with high capability to decompose cellulose, hemicelluloses, and lignin under in vitro conditions with different crop residue up to 20 days at time intervals of 10 days. Superior results for specific enzyme activities was noticed with developed consortium microbial inoculants supplemented with 1% V. radiata followed by C. cajan and A. hypogaea straw residues and could be suitable in accelerating rice wheat straw decomposition without causing any harmful effect on the environment. Furthermore, understanding the biotic and abiotic interaction in the indigenous developed consortium will pave the way for the establishment of an efficient multispecies based process for rapid lignocelluloses degradation under in vivo conditions with different inoculums concentration.
Conclusions
Our study demonstrated that V. radiata followed by C. cajan and A. hypogaea straw residues are a promising carbon source for S6B16 (B. altitudinis), S6A4 (S. flavomacrosporus), and S2F4 (A. terreus) to produce higher lignocellulosic microbial degrading enzymes due to wide range of C:N ratio and higher lignocellulosic content present in the crop residue. The combined use of B. altitudinis, S. flavomacrosporus, and A. terreus as indigenous developed consortium with V. radiata substrate enzymes resulted in a significant synergistic enhancement in enzymatic activity at 20 days of incubation under in vitro conditions. However, the different specific enzyme activities with V. radiata substrate were generally lower than commercially available synthetic substrate such as CMC, xylan, and guaiacol. The use of developed indigenous microbial consortium (S6B16 + S6A4 + S2F4) in low-cost bioremediation projects might be attractive given their highly efficient lignocellulose hydrolysis enzyme machinery with biochemically different agricultural waste residue.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Akhter S (2014) Interaction between Rhizobium, antagonistic bacteria and fungal pathogens in faba bean. Dissertation (Thesis), Swedish University of Agricultural Sciences
AOAC (1995) Official methods of analysis, 16th edn. Association of official analytical chemists, Washington DC
Baldrian P, Gabriel J (2003) Lignocellulose degradation by Pleurotus ostreatus in the presence of cadmium. FEMS Microbiol Lett 220:235–240
Banerjee S, Kirkby CA, Schmutter D, Bissett A, Kirkegaard JA, Richardson AE (2016) Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol Biochem 97:188–198
Borah N, Barua R, Nath D, Hazarika K, Phukon A, Goswami K, Barua DC (2016) Low energy rice stubble management through in situ decomposition. Procedia Environ Sci 35:771–780
Cheshire MV, Bedrock CN, Williams BL, Chapman SJ, Solntseva I, Thomsen I (1999) The immobilization of nitrogen by straw decomposition in soil. Euro J Soil Sci 50:329–341
Choudhary M, Sharma PC, Jat HS, Nehra V, McDonald AJ, Garg N (2016) Crop residue degradation by fungi isolated from conservation agriculture fields under rice-wheat system of North-West India. Int J Recycl Org Waste Agric 5:349–360
Cowan ST (1974) Cowan and steel’s manual for the identification of medical bacteria, 2nd edn. Cambridge, pp 67–83
Cunha L, Martarello R, Souza PM, Freitas MM, Barros KVG, Filho EXF, de-Mello MH, Magalhães PO (2017) Optimization of xylanase production from Aspergillus foetidus in soybean residue. Enzy Res 6597017: 7.https://doi.org/10.1155/2018/6597017
Datta R, Kelkar A, Baraniya D, Molaei A, Moulick A, Meena R, Formanek P (2017) Enzymatic degradation of lignin in soil. Sustain 9:1163. https://doi.org/10.3390/su9071163
Delabona PDS, Pirota RDPB, Codima CA, Tremacoldi CR, Rodrigues A, Farinas CS (2013) Effect of initial moisture content on two Amazon rainforest Aspergillus strains cultivated on agro-industrial residues: biomass-degrading enzymes production and characterization. Ind Crops Products 42:236–242
Devi S, Gupta C, Jat SL, Parmar MS (2017) Crop residue recycling for economic and environmental sustainability: the case of India. Open Agric 2:486–494
Farinas CS, Loyo MM, Baraldo A, Tardioli PW, Neto VB, Couri S (2010) Finding stable cellulase and xylanase: evaluation of the synergistic effect of pH and temperature. New Biotechnol 27:810–815
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes- application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Georgieva S, Christensen S, Stevnbak K (2005) Nematode succession and microfauna–microorganism interactions during root residue decomposition. Soil Biol Biochem 37:1763–1774
Giongo A, Beneduzi A, Ambrosini A, Vargas LK (2010) Isolation and characterization of two plant growth promoting bacteria from the rhizoplane of a legume (Lupinus albescens) in sandy soil. R Bras Ci Solo 34:361–369
Goyal S, Sindhu S (2011) Composting of rice straw using different inocula and analysis of compost quality. J Microbiol 1:126–138
Guniathilake KMD, Rathayaker R, Kulasaoniya SA, Karunaratna DN (2013) Evaluation of cellulose degrading efficiency of some fungi and bacteria and their films. J Nat Sci Foun Srilanka 41:155–163
Hadas A, Kautsky L, Goek M, Kara EE (2004) Rates of decomposition of plant residues and available nitrogen in soil, related to residue composition through simulation of carbon and nitrogen turnover. Soil Biol Biochem 36:255–266
Huang QX, Wang XC, Kong H, Guo YL, Guo AP (2013) An efficient DNA isolation method for tropical plants. Afric J Biotechnol 12:2727–2732
Jackson ML (1973) Estimation of phosphorus content in soil chemical analysis. Printer Hall, New Delhi
Kausar H, Sariah M, Saud HM, Alam Z, Ismail MR (2010) Development of compatible lignocellulolytic fungal consortium for rapid composting of rice straw. Inter Biodeterio Biodegrad 64:594–600
Kirk TK, Cullen D (1998) Enzymology and molecular genetics of wood degradation by white rot fungi. In: Young RA, Akhtar M (eds) Environmentally friendly technologies for the pulp and paper industry. John Wiley & Sons, New York, pp 273–308
Kriauciuniene Z, Velicka R, Raudonius S (2012) The influence of crop residues type on their decomposition rate in the soil: a litterbag study. Zemdirbyste 99:227–236
Kuijken RCP, Snel JFH, Heddes MM, Bouwmeester HJ, Marcelis LFM (2015) The importance of a sterile rhizosphere when phenotyping for root exudation. Plant Soil 387:131–142
Kumar A, Gaind S, Nain L (2008) Evaluation of thermophilic fungal consortium for paddy straw composting. Biodegrad 19:395–402
Kumar U, Tapwal A, Kalkal P, Varghese S, Chandra S (2014) Isolation and screening of cellulase producing fungi from forest waste. Int J Pharma Biol Arch 5:56–59
Kumawat KC, Sharma P, Nagpal S, Gupta RK, Sirari A, Nair RM, Bindumadhava H, Singh S (2021) Dual microbial inoculation, a Game changer? – bacterial bio-stimulants with multifunctional growth promoting traits to mitigate salinity stress in Spring Mungbean. Front Microbiol. https://doi.org/10.3389/fmicb.2020.600576
Lee S, Jang Y, Lee YM, Lee J, Lee H, Kim GH, Kim JJ (2011) Rice straw decomposing fungi and their cellulolytic and xylanolytic enzymes. J Microbiol Biotechnol 21:1322–1329
Mandel M, Sternburg D (1976) Recent advances in cellular technology. J Ferment Technol 54:267–286
Martin H (2002) Review: lignin conversion by manganese peroxidase (MnP). Enzyme Microb Technol 30:454–466
Martinez-Sanz M, Villano M, Oliveira C, Albuquerque MGE, Majone M, Reis M, Lopez-Rubio A, Lagaron JM (2014) Characterization of poly-hydroxy alkanoates synthesized from microbial mixed cultures and of their nano bio composites with bacterial cellulose nano whiskers. New Biotech 31:364–376
Mervin HD, Peech M (1950) Exchangeability of soils potassium in the sand, silt and clay fractions as influenced by the nature of the complementary exchangeable cations. Soil Sci Soc Am Proc 15:125–128
Mishra A, Malik A (2014) Novel fungal consortium for bioremediation of metals and dyes from mixed waste stream. Biores Technol 171:217–226
Mishra PK, Mishra S, Selvakumar G, Bisht JK, Kundu S, Gupta HS (2009) Co-inoculation of Bacillus thuringeinsis-KR1 with Rhizobium leguminosarum enhances plant growth and nodulation of pea (Pisum sativum L.) and lentil (Lens culinaris L.). World J Microbiol Biotechnol 25:753–761
Odier E, Roch P (1983) Factors controlling bio-degradation of lignin in T wood by various white rot fungi. In: Higuchi T, Chang HM, Kirk TK (eds) Recent Advances in Lignin Biodegradation Research. UNI. Publishers Co., Ltd., Tokyo, pp 183–193
Oglesby KA, Fownes JH (1992) Effects of chemical composition on nitrogen mineralization from green manures of seven tropical leguminous trees. Plant Soil 143:127–132
Olsen SR, Cole CV, Watanabe FS, Dean LA (1954) Estimation of available phosphorus in soils by extraction with NaHCO3, USDA Cir. 939. U.S. Washington
Piper CS (1966) Soil and Plant Analysis. Hans Publisher, Bombay
Prasad AA, Babu S (2017) Compatibility of Azospirillum brasilense and Pseudomonas fluorescens in growth promotion of groundnut (Arachis hypogea L.). Ann Braz Acad Sci 89(2):1027–1040
Prendergast-Miller MT, Duvalla M, Sohia SP (2014) Biochar- root interactions are mediated by biochar nutrient content and impacts on soil nutrient availability. Eur J Soil Sci 65:173–185
Puentes-Tellez PE, Salles JF (2018) Construction of effective minimal active microbial consortia for lignocellulose degradation. Microb Ecol 76:419–429
Rasi RPM, Mahalingam (2014) Screening and partial characterization of cellulose degrading bacteria from decayed sawdust. Int J Sci Res 3:328-331
Recous S, Robin D, Darwis D, Mary B (1995) Soil inorganic N availability: effect on maize residue decomposition. Soil Biol Biochem 27:1529–1538
Sahni N, Phutela UG (2013) Role of Coriolus versicolor MTCC 138 in lignosilica complex removal of paddy straw and its implication on biogas production. J Microbiol Res 7:2689–2693
Sánchez C (2009) Lignocellulosic residues: bio-degradation and bioconversion by fungi. Biotechnol Adv 27:185–194
Sandhu DK, Kalra MK (1982) Production of cellulose, xylanase and pectinase by Trichoderma longibrachiatum on different substrates. Transactions British Myco-Logical Soci 79:409–413
Schwarz WH (2001) The cellolosome and cellulose degradation by anaerobic bacteria. App Microbial Biotech 56:634–649
Seo JK, Park TS, Kwon IH, Piao MY, Lee CH, Ha JK (2013) Characterization of cellulolytic and xylanolytic enzymes of Bacillus licheniformis JK7 isolated from the rumen of a native korean goat. Asian-Australas J Anim Sci 26:50–58
Shaikh NM, Patel AA, Mehta SA, Patel ND (2013) Isolation and screening of cellulolytic bacteria inhabiting different environmental and optimization of cellulase production. Uni J Environ Res Technol 3:39–49
Shareef J, Gopinath SM, Satheesh M, Christopher SX (2015) Isolation and identification of cellulose degrading microbes. Int J Innov Res Sci Engineer Technol 4:6788–6793
Sharma A, Shruti, Kumar M, Malik D (2015) Lignocellulose degradation of rice straw by thermophilic microbial consortium isolated from mature compost of sugarcane industry. Int J Appl Pure Sci Agric 1(8):42–48
Sharma S, Singh P, Kumar S (2020) Responses of soil carbon pools, enzymatic activity and crop yields to nitrogen and straw incorporation in a rice-wheat cropping system in north-western India. Front Sustain Food Sys. https://doi.org/10.3389/fsufs.2020.532704
Sharma S, Singh P, Choudhary OP, Neemisha (2021) Nitrogen and rice straw incorporation impact nitrogen use efficiency, soil nitrogen pools and enzyme activity in rice-wheat system in north-western India. Field Crops Res 266:108–131
Shen YY, Chen H (2009) The progress of study on soil improvement research with straw stalk. Chin Agric Sci Bull 25:291–294
Shi Z, Han C, Zhang X, Tian L, Wang L (2020) Novel synergistic mechanism for lignocellulose degradation by a thermophilic filamentous fungus and a thermophilic actinobacterium based on functional proteomics. Front Microbiol 11:539438. https://doi.org/10.3389/fmicb.2020.539438
Shukla L, Senapati A, Tyagi SP, Saxena AK (2014) Economically viable mass production of lignocellulolytic fungal inoculum for rapid degradation of agrowaste. Curr Sci 107:1701–1704
Shukla L, Suman A, Verma P, Yadav AN, Saxena AK (2016) Syntrophic microbial system for ex-situ degradation of paddy straw at low temperature under controlled and natural environment. J Appl Biol Biotechnol 4:30–37
Singh B, Gill RIS, Gill PS (2010) Soil fertility under various tree species and poplar-based agroforestry system. J Res Punjab Agric Univ 47:160–164
Singh G (2017) Assessment of microbial consortium for paddy straw degradation. Punjab Agricultural University, Ludhiana
Singh G, Bhattacharyya R, Das TK, Sharma AR, Ghosh A, Das S, Jha P (2018) Crop rotation and residue management effects on soil enzyme activities, glomalin and aggregate stability under zero tillage in the Indo-Gangetic Plains. Soil till Res 184:291–300
Singh RP, Garcha HS, Khanna PK (1988) Laccase production by Pleurotus spp. Ind J Microbiol 28:38–41
Song F, Tian X, Fan X, He X (2010) Decomposing ability of filamentous fungi on litter is involved in a subtropical mixed forest. Mycologia 102:20–26
Subramanian P, Kim K, Krishnamoorthy R, Sundaram S, Sa T (2015) Endophytic bacteria improve nodule function and plant nitrogen in soybean on co-inoculation with Bradyrhizobium japonicum MN – 110. Plant Growth Reg 76:327–332
Tien M, Kirk TK (1983) Lignin peroxidase of Phanerochaete chrysosporium. Methods Enzymol 161:238–249
Turner JC, Green RS (1974) The effect of a polychlorinated biphenyl (Aroclor® 1254) on liver microsomal enzymes in the male rat. Bull Environ Contam Toxicol 12:669–671
Van Soest PJ, Robertson JB, Lewis BA (1991) Symposium: carbohydrate methodology, metabolism and nutritional implications in dairy cattle. Methods for dietary fiber, neutral detergent fiber and non starch polysaccharides in relation to animal nutrition. J Dairy Sci 74:3583–3597
Vazquez MA, De L, Varga D, Plana R, Soto M (2015) Integrating liquid fraction of pig manure in the composting process for nutrient recovery and water re-use. J Clean Prod 104:80–89
Vimal J, Venu A, Joseph J (2016) Isolation and identification of cellulose degrading bacteria and optimization of cellulase production. Int J Res Bios 5:58–67
Vincent JM (1970) A manual for practical study of root nodule bacteria. Blackwell Scientific Publishers, Oxford
Wang M, Pendall E, Fang CM, Li B, Nie M (2018) A global perspective on agro-ecosystem nitrogen cycles after returning crop residue. Agric Ecosyst Environ 266:49–54
Wang ZH, Dong XF, Zhang GQ, Tong JM, Zhang Q, Xu SZ (2011) Waste vinegar residue as substrate for phytase production. Waste Manag Res 29:1262–1270
Wolf B, Snyder GH (2003) Sustainable soils: The place of organic matter in sustainable soils and their productivity. Food Products Press, New York
Wongwilaiwalin S, Laothanachareon T, Mhuantong W, Tangphatsornruang S, Eurwilaichitr L, Igarashi Y, Champreda V (2013) Comparative metagenomic analysis of microcosm structures and lignocellulolytic enzyme systems of symbiotic biomass-degrading consortia. Appl Microbiol Biotechnol 97:8941–8954
Xu C, Ma F, Zhang X (2009) Lignocellulose degradation and enzyme production by Irpex lacteus CD2 during solid-state fermentation of corn stover. J Biosci Bioengineer 108:372–375
Xu J, Yang Q (2010) Isolation and characterization of rice straw degrading Streptomyces griseorubens C-5. Biodegrad 21:107–116
Yang Q, Gao Y, Huang Y (2015) Identification of three important amino acid residues of xylanase from Aspergillus fumigatus for enzyme activity and formation of xylobiose as the major product. Process Biochem 50:571–581
Zhang H, Xu WX, Li YB, Lyu J, Cao Y, He W (2017) Changes of soil microbial communities during decomposition of straw residues under different land uses. J Arid Land 9:666–677
Zhang L, Jing Y, Xiang Y, Zhang R, Haibo Lu (2018) Responses of soil microbial community structure changes and activities to biochar addition: a meta-analysis. Sci Total Environ 643:926–935
Zhe LO, Dwyer JP, Chang VS, Granda CB, Holtzapple MT (2008) Structural features affecting biomass enzymatic digestibility. Bioresour Technol 99:3817–3828
Zuroff TR, Curtis WR (2012) Developing symbiotic consortia for lignocellulosic biofuel production. Appl Microbiol Biotechnol 93:1423–1435
Acknowledgements
The authors wish to thank the Department of Soil Science, Punjab Agricultural University, Ludhiana, India, for providing facilities for completing research project. We are grateful to the National Agricultural Science Fund (NASF), India (Project no. PID. 1109; NASF/NRM-8018/2019-20) for providing supports for this research work.
Author information
Authors and Affiliations
Contributions
SS took part in executing experiment, analyzed samples, processed and interpreted data, and prepared the manuscript and was a major contributor of the manuscript. KCK took part in executing lab and field experiment, prepared microbial consortia, analyzed plant samples, and writing original draft preparation. SK conducted lab experiment in Punjab region, isolated the exotic microorganisms, and analyzed samples and processed data.
Corresponding author
Ethics declarations
Ethics approval
The authors attest that this paper has not been published elsewhere; the work has not been submitted simultaneously for publication elsewhere and the results presented in this work are true and not manipulated.
Consent to participate
All the individual participants involved in the study have received informed consent.
Consent for publication
The participants have consented to the submission of the study to the journal.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Robert Duran
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharma, S., Kumawat, K.C. & Kaur, S. Potential of indigenous ligno-cellulolytic microbial consortium to accelerate degradation of heterogenous crop residues. Environ Sci Pollut Res 29, 88331–88346 (2022). https://doi.org/10.1007/s11356-022-21809-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-022-21809-3