Abstract
Swine manure treatment plants are important reservoirs of plasmid-harboring antibiotic resistance genes (ARGs) and physicochemical contaminants, but the changes in the abundances of plasmids and ARGs, and their interactions with the physicochemical properties of manure, are still unclear. Thus, in the present study, plasmidome and metagenome analyses were conducted for samples collected at different stages in the swine manure treatment process. The results indicated that anaerobic digestion and aerobic digestion were the most efficient stages for reducing the abundances of ARGs in swine manure. However, the plasmids associated with ARGs were not effectively removed in these stages. Through the whole treatment process, the IncL/M, IncQ1, IncHI2A, IncA/C, and IncN plasmid groups had strong correlations (r > 0.8, P < 0.01) with most ARG types, thereby indicating that these plasmids play important roles in the persistence of ARGs in this environment. Furthermore, the pH, total nitrogen, total phosphorus, and four heavy metals (Cu, Zn, As, and Fe) significantly affected the abundances of seven ARG subtypes (tetB(P), ant(6)-Ia, tet44, aph(3′′)-Ib, mefB, tet(L), and tet(39)). In particular, florfenicol had the most positive correlations with ARGs. Our results indicated that nutrients, heavy metals, and antibiotics all contributed to the presence and persistence of plasmid-harboring ARGs. This study provides insights into the fate of plasmids and ARGs, and related factors during the swine manure treatment process, thereby facilitating the development of a new treatment technique for removing ARGs and reducing the public health risk associated with livestock production.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In China, more than 600 million swine are produced each year and about 618 billion kg of swine manure (Wang et al. 2006). Many veterinary antimicrobials are applied in swine husbandry for the treatment and prevention of disease (Selvam et al. 2012; Zhu et al. 2013a). As a consequence, swine manure is a major source of residual antimicrobials. Furthermore, studies increasingly indicate that swine feedlot manure comprises a huge reservoir of antibiotic resistance genes (ARGs) (Heuer et al. 2011; Van den Meersche et al. 2019; Wen et al. 2019), most of which are carried and spread by plasmids (Bennett 2008; Che et al. 2019; Loftie-Eaton et al. 2016). These ARG-harboring plasmids with a high capacity for transmission in the environment are a risk to global public health (Carlet 2015; Cheng et al. 2013; Dyson et al. 2019; He et al. 2016; Kime et al. 2019).
However, the current large-scale swine feedlot manure treatment methods are often based on solid–liquid separation, anaerobic digestion, and precipitation processes (Liang-wei 2001; San-cheng et al. 2010), and they are mainly applied to avoid polluting the environment with high concentrations of organic matter and to convert manure into fertilizer and biogas. The effective elimination of plasmids and ARGs has not been achieved during the manure treatment process. Moreover, changes in the chemical and biological contents (including nitrogen, phosphorus, potassium, copper, zinc, arsenic, iron, doxycycline (DO), oxytetracycline (OTC), enrofloxacin (ENR), and florfenicol (FFC)) of sewage and waste may have different effects on the fate of plasmids and ARGs. Some previous studies investigated the effects of various physicochemical properties on ARGs (Baker-Austin et al. 2006; Zhang et al. 2017, 2020; Zhou et al. 2017), but most focused on the impact of a single factor on a specific type of ARG. Thus, the effects of multiple physicochemical properties on plasmids and ARGs in complex communities are not well understood.
Therefore, in the present study, we conducted simultaneous plasmidome and metagenome analyses, which are characterized as high throughput and cultivation independent (Szczepanowski et al. 2008), in order to obtain information about the extrachromosomal elements in the whole community, irrespective of the encoded traits (Dib et al. 2015). In particular, we aimed (i) to monitor the variations in the plasmid and antimicrobial resistome in each swine manure treatment stage; (ii) to assess the performance of the current swine manure treatment techniques at eliminating ARGs in the different treatment stages; and (iii) to determine the changes in the physicochemical properties during the treatment process, and the correlations between these factors and the abundances of ARGs. Our results provide insights into the occurrence of plasmids and ARGs, and various physicochemical properties in the swine manure treatment stages, as well as their correlations. These findings may facilitate improvements to the waste treatment techniques to enhance reductions in the abundances of ARGs and reduce the public health risk associated with livestock production.
Methods
Sampling
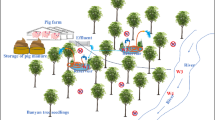
Sampling was conducted in a swine manure treatment plant located in Jianyang, Sichuan Province, China, where the swine industry is well developed. The plant processed approximately 20 tons of manure each day. The main sewage storage facilities had a capacity of 4322 m3, and the composting sites covered about 800 m2. The solid and liquid components of fresh manure were separated in septic tanks with a volume of 2 m3. The dehydrated manure was transported to the composting site (CO) for stacking and the liquid was transported to regulating tanks with a volume of 20 m3, integrated anaerobic digestion tanks (anaerobic digestion tanks, AN) with a volume of 800 m3, aeration tanks (aerobic digestion, AE) with a volume of 960 m3, oxidation tanks (planted with aquatic plants) with a volume of 21,400 m3, sand filter tanks (SF) with a volume of 800 m3, and storage pools with a volume of 350 m3. Triplicate samples were collected (Guo et al. 2017; Wang et al. 2021) at each stage of the manure treatment process comprising fresh manure (FM) from a manure septic tank, dewatered manure from CO, and effluent from AN, AE, primary oxidation pond (PO), secondary oxidation pond (SO), and SF (Fig. 1). We collected 100-g and 1-L samples of compost and sewage, respectively, where samples from the same stage were mixed, and then divided into two parts for total DNA extraction and plasmid DNA extraction. All samples were stored at 4 °C until DNA extraction.
Schematic illustration of the sampling site and swine manure treatment stages. The samples comprised fresh manure (FM) from the manure septic tank, dewatered manure from the composting site (CO), and effluent from the anaerobic fermentation tank (AN), aeration tank (AE), primary oxidation pond (PO), secondary oxidation pond (SO), and sand filter tank (SF)
Total DNA extraction
Total genomic DNA was extracted from compost samples using a DNeasy® PowerSoil® Pro Kit (Qiagen, Germany). DNA was extracted from the liquid samples using a Water DNA Kit (Omega, USA) according to the manufacturer’s protocol. Three DNA samples extracted from the same stage were mixed. Next, the quantity and quality of the DNA mixture were determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). DNA samples were stored at – 20 °C for subsequent analyses.
Plasmid DNA extraction
Two compost samples weighing 10 g were used for plasmid DNA extraction in 40 mL of 0.01 M phosphate buffer in 50-mL centrifuge tubes. The samples were mixed thoroughly and centrifuged at 1000 × g for 5 min. The supernatant was removed and placed in a 50-mL centrifuge tube. The precipitate was mixed with 40 mL of 0.01 M phosphate buffer and the previous step was repeated two times, where the supernatant was retained after centrifugation at 10,000 × g for 5 min and the precipitate was collected. Phosphate buffer was used to wash the precipitate until it was clear. Finally, the precipitate was used for plasmid DNA extraction.
For the liquid samples, the precipitate was collected following centrifugation at 10,000 × g for 3 min and then re-suspended in pre-chilled 50-mL poly (β-amino) ester buffer. Large particles and eukaryotic cells were removed after centrifugation at 200 × g for 6 min. The supernatant was then centrifuged at 10,000 × g for 6 min, and the precipitated microbial biomass was collected.
The precipitates obtained as described above were used for plasmid DNA extraction with a Large-Construct Kit (QIAGEN, Germany) (which is a genomic DNA-free kit) according to the manufacturer’s protocol, where three replicates were mixed to produce one sample. The quality and concentration of the DNA samples were assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA), before storing the DNA extracts at – 20 °C.
High-throughput sequencing
Total DNA and plasmid DNA samples were sequenced by the Illumina Hiseq X Ten platform (Illumina, USA) with the 150-bp paired-end strategy. Low-quality reads and adapter sequences were removed using Trimmomatic (v0.36) (Bolger et al. 2014) with the default parameters “trimmomatic-0.36.jar PE -phred64 LEADING:10 TRAILING:10 MINLEN:50 TOPHRED33.” We assembled the clean reads with SPAdes (v2.03) (Antipov et al. 2019) with “–meta –plasmid” as parameters, and systematically assessed the quality of the results using QUAST (v5.0.2). Open reading frames (ORFs) in the assembled contigs from each sample were predicted using Prokka (v1.13.3). CD-HIT (v2.7) was used to remove redundant metagenomic data and plasmidomic data with the parameters: “-c 0.95 -G 0 -aS 0.9 -g 1 -d 0.”
Annotation of microbial communities and plasmids
MetaPhlAn (v2.7.8) (Segata et al. 2012) was used for taxonomic profiling. Complete sequences of plasmids up to July 2019 were downloaded from the NCBI website. The “makeblastdb” command (BLAST + 2.9) was used to format these sequences and to construct a plasmid database. A python script was written to add metadata for these plasmids, including accession, organism source, and plasmid name. All contigs in the plasmidome were clustered with identity ≥ 95% using CD-HIT (v2.7), and the longest sequence in each group of clusters was output as the representative sequence. The representative sequences were aligned against the constructed plasmid database using BLASTN with: identity ≥ 95%, coverage ≥ 80%, and E-value ≤ 1e − 5. Sequences that hit a same known plasmid was defined as a “plasmid type” and designated with the accession number of the matching known plasmid. Plasmid replicon information was annotated using Plasmidfinder (v2.0.1) (Carattoli et al. 2014) with identity ≥ 95% and coverage ≥ 60%. Plasmid mobility was also annotated using Plascad (Che et al. 2021).
Detection and quantification of ARGs
ARGs were annotated with the Comprehensive Antibiotic Resistance Database (CARD) (v3.8.1; (Alcock et al. 2020) and ResFinder database (v3.8; (Zankari et al. 2012) using BLASTN with E-value ≤ 1e − 5, sequence identity ≥ 90%, and alignment length ≥ 75 nt, and the optimal alignment was selected. Salmon (v1.2.0; (Patro et al. 2017)) was used to calculate the lengths of the resistance genes in each sample and to calculate their abundances based on the number of reads in the alignment. The formula used to calculate the abundance of genes (Xiong et al. 2018) is as follows:
- N mappedreads :
-
represents the number of reads relative to the target gene ORF.
- L reads :
-
represents the length of reads.
- L targetgene :
-
represents the ORF length for the target gene.
- S :
-
represents the data set size in gigabytes.
Salmon was used to obtain abundance information directly for plasmids (Wagner et al. 2012).
Determination of physicochemical properties
The physicochemical properties of the manure were measured during the treatment process by Qingdao Sci-tech Innovation Quality Testing Co., Ltd. The pH value and total organic carbon (TOC) concentration were measured with a pH meter (PHS-3C, INESA, China) and TOC analyzer (TOC-V CPH, Thermo, USA), respectively. The concentrations of total nitrogen (TN), total phosphorus (TP), total potassium (TK), and heavy metals (copper (Cu), zinc (Zn), arsenic (As), and iron (Fe)) were measured by inductively coupled plasma mass spectrometry (Optima, 2000 DV, Perkin Elmer, USA) (Yuan et al. 2004). The concentration of four antimicrobials comprising DO, OTC, ENR, and FFC were determined according to National Standard SN/T 4747.3–2017 using ultra-performance liquid chromatography-tandem mass spectrometry (Batt et al. 2008).
Data analysis and visualization
The means, standard deviations, and significant differences in high-throughput data and physicochemical properties were analyzed using R (v3.5.0) with the base packages (v3.5.0) and aov function. Pheatmap (v1.0.12) in R was used to draw heatmaps representing the abundances and diversity of microorganisms, plasmids, and ARGs. Venn diagrams were prepared to assess the numbers of shared and unique plasmid types in samples from different stages with the Venn package (v1.9) in R. Other diagrams were generally prepared with ggplot2 (v3.3.2). Network analysis was conducted to investigate the associations among ARGs and plasmids. Hmisc (v4.0–2) in R was used to calculate the Spearman’s correlation coefficients between plasmids and ARG subtypes (strong correlation value at |r|> 0.8 and significant difference at P < 0.05). Gephi (v0.9.2) software was used for network visualization. The correlations between plasmids and physicochemical properties were investigated using GraphPad Prism 6 by analyzing the Pearson’s correlation coefficients. Among the coefficients obtained, r > 0 indicated a positive correlation, r < 0 indicated a negative correlation (P < 0.05 denotes a significant correlation), 0.5 ≤ ∣r∣ ≤ 0.8 indicated a moderate correlation, and ∣r∣ > 0.8 indicated a high correlation. The correlation heatmap was also drawn using GraphPad Prism 6.
Results
Diversity of plasmids in PMTP
According to the results, 30.17% (565 of 1873), 16.92% (581 of 3433), 7.48% (97 of 1297), 5.89% (217 of 3682), 14.18% (114 of 804), 10.89% (205 of 1882), and 6.35% (389 of 6263) of the representative sequences matched known plasmids in CO, AN, AE, PO, SO, and SF, respectively. After clustering these contigs based on the known target plasmids, 440, 375, 233, 117, 59, 97, and 192 plasmid types were found in FM, CO, AN, AE, PO, SO, and SF, respectively (Fig. 2). During the overall treatment process, only 10 plasmid types were present from FM to SF, and most of their hosts belonged to the phyla Proteobacteria and Bacteroidetes (Table S6).
Different plasmid types detected during swine manure treatment stages. Red, orange, yellow, light green, grass green, blue, and purple denote plasmid types in the fresh manure, compost, anaerobic tank, aeration tank, primary oxidation tank, secondary oxidation tank, and sand filter tank, respectively. The overlapping areas indicate plasmid types shared in the corresponding stages. The numbers indicate the number of specific plasmid types
From FM to PO, the diversity of plasmids tended to decrease and the lowest point was reached in PO (n = 59). By contrast, from PO to SF, the diversity of plasmid types increased, with 192 in the last stage (SF). In addition, the number of unique plasmid types tended to increase from FM to SF, with 188, 130, 51, 16, two, 15, and 84 different plasmid types in FM, CO, AN, AE, PO, SO, and SF, respectively (Fig. 2).
Analysis at the family level also showed that the diversity of the plasmid hosts changed greatly. In particular, Campylobacteraceae, Enterobacteriaceae, and Moraxellaceae were dominant in FM, CO, and AN, respectively, whereas Burkholderiaceae and Rhodocyclaceae were dominant in AE and PO. The plasmid host communities were more diverse in SO and SF (Fig. S3).
Changes in abundances of ARGs during swine manure treatment process
In total, 185 ARG subtypes were detected in the plasmidome and metagenome, and most conferred resistance to aminoglycosides, tetracyclines, diaminopyrimidines, fluoroquinolones, β-lactams, macrolides, phenicols, sulfonamides, lincosamides, MLS group antibiotics, and multidrug resistance antibiotics (resistance genes belonging to 3 or more CARD drug classes at the same time). Among these subtypes, 126 ARG subtypes were shared by the metagenome and plasmidome, and 30 subtypes were only found in the plasmidome (Fig. S1a). Furthermore, 18 subtypes in the plasmidome were shared in all stages, i.e., cfrC, floR, aph(3′)-IIIa, ant(6)-Ia, aph(3′′)-Ib, aac(6′)-IIa, aadA, aadA2, tetM, tetX, tet(A), tet(C), tet(G), tetO, ermF, msrE, qnrD1, and sul2.
The abundances of ARGs in the plasmidome and metagenome differed in FM, CO, AN, AE, PO, SO, and SF (Fig. 3a). The total abundances of ARGs in the plasmidome decreased by the most in AN (40.0%) and AE (86.0%), and reached the lowest level in PO (2242.4 × /Gb), before increasing in the last two stages in SO (69.8%) and SF (77.4%). The diversity of ARGs in the metagenome was 115, but it decreased to 49 in AN, and remained at about 49 in the later stages. The total abundance of ARGs decreased from FM (6623.94 × /Gb) until the last stage in SF (662.26 × /Gb).
Changes in abundances of ARGs in different swine manure treatment stages. a Relative abundances of main ARGs detected in the plasmidome and metagenome. b Changes in the relative abundances of the top 70 most abundant ARG subtypes during swine manure treatment. The samples were from the fresh manure (FM), compost (CO), anaerobic tank (AN), aeration tank (AE), primary oxidation tank (PO), secondary oxidation tank (SO), and sand filter tank (SF)
We also found that the abundance of sulfonamide resistance genes increased greatly in AE and subsequent stages according to the plasmidome, especially sul1. In particular, sul1 was not detected in FM but its abundance increased throughout the treatment process and it was one of the most abundant ARG subtypes (1901.33 × /Gb) in SF (Fig. 3a, b; Table S5).
Moreover, cfrC was the most abundant ARG subtype in both the plasmidome and metagenome, but its abundance (18,594.20 × /Gb) in the plasmidome was more than 16 times that in the metagenome (1125.86 × /Gb) (Fig. 3b).
Association among plasmids and ARGs in the treatment process
We compared the changes in plasmids and ARGs during different stages in order to understand their associations. Interestingly, we found similar trends in the diversity of plasmids and the abundances of ARGs in the plasmidome, where they tended to decrease initially and then increase, with the lowest levels in PO. In particular, at the inflection point in PO, we found that the number of plasmids was lowest (n = 59) and the diversity (n = 37) and abundance of ARGs were also at their lowest levels (2242.4 × /Gb). In the final treatment stage in SF, the number of plasmid types increased greatly (n = 84), and the abundance of ARGs also increased (Fig. 3, Fig. S4).
We conducted correlation analysis between plasmids and ARGs, and obtained a co-occurrence network diagram to understand their associations. The network representing the pair-wise connections between plasmids and ARG subtypes comprised 57 nodes and 150 edges. The modularity value was 0.263 for this network, thereby indicating that plasmids and ARGs were generally associated (module value greater than 0.4 including the module structure), and the number of communities was 12 (Fig. S5). The IncL/M, IncN, IncI2, IncA/C, IncQ1, and IncHI2A plasmid groups were located at the center of the network and connected with most ARG types (Fig. 4).
Correlation network analysis based on plasmids and the top 40 most abundant ARG subtypes (strong correlation: |r|> 0.8; significant difference: P < 0.05). Larger nodes indicate that more points are connected with greater importance. All types of plasmids are denoted by purple nodes. Other colors represent different types of resistance genes. The red nodes labeled “multidrug” are resistance genes to various types of antibiotics according to the CARD classification
Changes in physicochemical properties during swine manure treatment process
After all of the treatment stages, the TN, TP, and TK concentrations decreased by 47.83%, 64.95%, and 63.77%, respectively. The Cu, Zn, and Fe concentrations decreased to below the detection limit. In the different stages, the pH was always around 7.5 and TOC was about 400 mg/L, and they did not change significantly. The average concentrations of antimicrobials followed the order of OTC (0.2172 mg/L) > DO (0.0759 mg/L) > ENR (0.0008 mg/L) > FFC (0.0006 mg/L). From FM to SF, the concentrations of ENR and FFC decreased to below 0.0005 mg/L (FFC was not detected in PO and subsequent stages), whereas those of DO and OTC were still 0.015 mg/L and 0.029 mg/L in the last SF stage, respectively, thereby indicating that these two antibiotics persisted throughout the whole treatment process (Fig. 5a).
Correlations between ARGs in the plasmidome and physicochemical properties. a Changes in the concentrations of various physicochemical properties. b Correlations between pH, total nitrogen (TN), total phosphorus (TP), total potassium (TK), total organic carbon (TOC), copper (Cu), zinc (Zn), arsenic (As), iron (Fe), doxycycline (DO), oxytetracycline (OTC), enrofloxacin (ENR), florfenicol (FFC), and ARG subtypes. ** indicates P ≤ 0.01, * indicates P ≤ 0.05
Associations among physicochemical properties and ARGs
We analyzed the correlations between the physicochemical properties and ARGs in the plasmidome and metagenome to determine whether changes in the physicochemical properties of the manure affected ARGs. We found that ARGs on plasmids had highly significant positive associations with the changes in the physicochemical properties (Fig. S6, Fig. 5).
Surprisingly, physicochemical properties such as pH, TN, TP, Cu, Zn, and As all had significant positive correlations with “other” types of ARGs in the plasmidome. In particular, pH, TN, TK, and four heavy metals had significant positive correlations (r > 0.8, P < 0.05) with seven ARG subtypes (tetB(P), ant(6)-Ia, tet44, aph(3′′)-Ib, mefB, tet(L), and tet(39)). We found that FFC had the most positive correlations with ARGs. In addition, TOC had negative correlations with tetB(P), tet44, and mefB (Fig. 5b).
Discussion
Effects of different swine manure treatment stages on the elimination of ARGs
Twelve main types of ARGs were found in both data sets, where they mainly comprised aminoglycoside, tetracycline, multidrug, chloramphenicol, and sulfonamide resistance genes. These results are similar to the ARG types detected in previous studies (Zhou et al. 2016; Zhu et al. 2013b), thereby indicating that the prevalent ARGs in this environment are generally consistent.
For example, Yuan et al. found that the abundances of most ARGs decreased after treatment by analyzing the total DNA (Yuan et al. 2019). Moreover, Tao et al. used real-time qPCR to show that the average ARG removal efficiency in the treatment stages ranged between 33.30 and 97.56% (Tao et al. 2014). These findings are consistent with our results, but metagenomic analysis can provide a more comprehensive understanding of the reductions in the overall abundances of ARGs during the manure treatment process. By analyzing the plasmidome, we found that many ARGs were not effectively eliminated according to metagenomics and their abundances increased again after the AN stage. Thus, plasmids probably serve as “safe havens” for resistance genes in the ARG elimination process. In particular, IncN, IncL/M, IncQ1, and other broad-spectrum host plasmids (Rozwandowicz et al. 2018) can carry multiple ARGs and facilitate their widespread transmission. Thus, provided that ARGs are not completely eliminated, they can continue to enter new hosts and confer antimicrobial resistance.
We also found that the efficiency of ARG elimination was highest in the anaerobic fermentation tank and aeration tank. Many previous studies have suggested that changes in the structure of the bacterial community are mainly responsible for the variations in the abundances of ARGs during anaerobic digestion (Tong et al. 2016; Zhang et al. 2016). However, our results also indicated close relationships between the diversity and abundance of plasmids and ARGs. Moreover, the reductions in the abundances of ARGs appeared to have important relationships with the reductions in the concentrations of TN, TK, TK, Cu, Zn, and Fe.
Correlations between physicochemical properties and ARGs
Previous studies have shown that the presence of high nutrient contents is conducive to horizontal gene transmission (Guo et al. 2015). Similarly, we found that the concentration of certain nutrients influenced the abundances of ARGs. In particular, TN and TP were significantly positively correlated with the ARGs comprising tetB(P), aph(3′′)-lb, and mefB in plasmid DNA (r ≥ 0.8, P ≤ 0.01), possibly because they provided sufficient nutrients for the growth of antibiotic-resistant bacteria. In addition, under nutrient-deficient conditions, the energy required for plasmid conjugation is limited. Thus, negative correlations were found between TOC and some ARGs. Previous research into the effects of carbon resource on bacterial growth (Yang et al. 2019) has shown that different types of carbon resources and their interactions with various factors can affect bacterial growth and functional gene expression, thereby explaining the negative correlations between TOC and resistance genes.
Moreover, it has been demonstrated that co-selection relationships exist between metal resistance genes and ARGs (Dickinson et al. 2019), which may lead to the co-selection of heavy metals and some ARGs. We also detected co-selection where four heavy metals (Cu, Zn, As, and Fe) had significant positive correlations (r ≥ 0.8, P ≤ 0.01) with tetB(P), aph(3′′)-lb, and mefB.
Previous studies have often found correlations between antimicrobials and ARGs (Zhao et al. 2017). However, in the present study, we found no correlations between DO, OTC, and ENR and their corresponding resistance genes. By contrast, FFC had highly significant positive correlations with a large number of ARGs in plasmid DNA, including chloramphenicol resistance genes (cfrC, cmx, and catQ) and other types of antimicrobials (aminoglycosides, macrolides, tetracyclines, and link amides) (Fig. 5b). We found that FFC affected FFC-related resistance genes, but it also selected for other types of ARGs. The co-transmission of ARGs may explain why antimicrobials affect many different types of resistance genes. Thus, significant correlations were found between multiple resistance genes (cfrC, tetM, ant(6)-la, tet44, aph(3′′)-lb, tetA(P), ant(6)-la, OXA-347, Isa(E), Inu(B), Inc(C), tet32, and mel) and fluoroquinolone residues in the swine manure treatment process.
Conclusion
This is the first comprehensive survey of ARGs, especially ARGs in plasmid DNA, and the effects of several physicochemical properties in the swine manure treatment process. We found that the current swine manure treatment technique can only partially eliminate ARGs, and the presence of plasmids may be important for ensuring their persistence. Thus, there is an urgent need to improve manure treatment techniques in order to address the continuing challenge of ARG removal. Nutrients, heavy metals, and antibiotics together contribute to the long-term persistence of plasmid-harboring ARGs. Further studies are required to explore the specific mechanisms that allow various physicochemical properties to influence the transmission of plasmids and ARGs.
Data availability
All of the sequences in the plasmid database analyzed in the present study are available via the NCBI RefSeq repository (https://ftp.ncbi.nlm.nih.gov/refseq/release/plasmid/) (2020–11-16). The nucleotide sequences in the plasmidome and metagenome were deposited in the GSA database, and they are publicly available under accession number PRJCA004346.
Code availability
The code used in this study to generate the plasmid database is available at Github (https://gist.github.com/Alice-shui/88d39ffe3509172e32826772b5b88aa6).
References
Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen AV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG (2020) CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48(D1):D517-d525
Antipov D, Raiko M, Lapidus A, Pevzner PA (2019) Plasmid detection and assembly in genomic and metagenomic data sets. Genome Res 29(6):961–968
Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV (2006) Co-selection of antibiotic and metal resistance. Trends Microbiol 14(4):176–182
Batt AL, Kostich MS, Lazorchak JM (2008) Analysis of ecologically relevant pharmaceuticals in wastewater and surface water using selective solid-phase extraction and UPLC− MS/MS. Anal Chem. 80(13):5021–5030
Bennett P (2008) Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol 153(S1):S347–S357
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Carattoli A, Zankari E, Garcìa-Fernandez A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H (2014) PlasmidFinder and pMLST: in silico detection and typing of plasmids. Antimicrob Agents Chemother
Carlet J (2015) The world alliance against antibiotic resistance: consensus for a declaration. Clin Infect Dis 60(12):1837–1841
Che Y, Xia Y, Liu L, Li A-D, Yang Y, Zhang T (2019) Mobile antibiotic resistome in wastewater treatment plants revealed by Nanopore metagenomic sequencing. Microbiome 7(1):44
Che Y, Yang Y, Xu X, Břinda K, Polz MF, Hanage WP, Zhang T (2021) Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc Nat Acad Sci 118(6):e2008731118
Cheng W, Chen H, Su C, Yan S (2013) Abundance and persistence of antibiotic resistance genes in livestock farms: a comprehensive investigation in eastern China. Environ Int 61:1–7
Dib JR, Wagenknecht M, Farías ME, Meinhardt F (2015) Strategies and approaches in plasmidome studies—uncovering plasmid diversity disregarding of linear elements? Front Microbiol 6(463)
Dickinson AW, Power A, Hansen MG, Brandt KK, Piliposian G, Appleby P, O’Neill PA, Jones RT, Sierocinski P, Koskella B, Vos M (2019) Heavy metal pollution and co-selection for antibiotic resistance: a microbial palaeontology approach. Environ Int 132:105117
Dyson ZA, Klemm EJ, Palmer S, Dougan G (2019) Antibiotic resistance and typhoid. Clin Infect Dis 68(Suppl 2):S165-s170
Guo MT, Yuan QB, Yang J (2015) Distinguishing effects of ultraviolet exposure and chlorination on the horizontal transfer of antibiotic resistance genes in municipal wastewater. Environ Sci Technol 49(9):5771–5778
Guo H, Nasir M, Lv J, Dai Y, Gao J (2017) Understanding the variation of microbial community in heavy metals contaminated soil using high throughput sequencing. Ecotoxicol Environ Saf 144:300–306
He L-Y, Ying G-G, Liu Y-S, Su H-C, Chen J, Liu S-S, Zhao J-L (2016) Discharge of swine wastes risks water quality and food safety: Antibiotics and antibiotic resistance genes from swine sources to the receiving environments. Environ Int 92–93:210–219
Heuer H, Schmitt H, Smalla K (2011) Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol 14(3):236–243
Kime L, Randall CP, Banda FI, Coll F, Wright J, Richardson J, Empel J, Parkhill J, O'Neill AJ (2019) Transient silencing of antibiotic resistance by mutation represents a significant potential source of unanticipated therapeutic failure. mBio 10(5)
Liang-wei DJCB. 2001. Models of pig waste treatment. 1
Loftie-Eaton W, Yano H, Burleigh S, Simmons RS, Hughes JM, Rogers LM, Hunter SS, Settles ML, Forney LJ, Ponciano JM, Top EM (2016) Evolutionary paths that expand plasmid host-range: implications for spread of antibiotic resistance. Mol Biol Evol 33(4):885–897
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14(4):417–419
Rozwandowicz M, Brouwer M, Fischer J, Wagenaar J, Gonzalez-Zorn B, Guerra B, Mevius D, Hordijk J (2018) Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73(5):1121–1137
San-cheng Y, Xue-bin L, Zhi-ping HJSA and Sciences V (2010) Observation of different patterns for pig waste treatment and utilization in Sichuan. 6
Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C (2012) Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9(8):811–814
Selvam A, Xu D, Zhao Z, Wong JW (2012) Fate of tetracycline, sulfonamide and fluoroquinolone resistance genes and the changes in bacterial diversity during composting of swine manure. Bioresour Technol 126:383–390
Szczepanowski R, Bekel T, Goesmann A, Krause L, Krömeke H, Kaiser O, Eichler W, Pühler A, Schlüter A (2008) Insight into the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to antimicrobial drugs analysed by the 454-pyrosequencing technology. J Biotechnol 136(1–2):54–64
Tao CW, Hsu BM, Ji WT, Hsu TK, Kao PM, Hsu CP, Shen SM, Shen TY, Wan TJ, Huang YL (2014) Evaluation of five antibiotic resistance genes in wastewater treatment systems of swine farms by real-time PCR. Sci Total Environ 496:116–121
Tong J, Liu J, Zheng X, Zhang J, Ni X, Chen M, Wei Y (2016) Fate of antibiotic resistance bacteria and genes during enhanced anaerobic digestion of sewage sludge by microwave pretreatment. Biores Technol 217:37–43
Van den Meersche T, Rasschaert G, Haesebrouck F, Van Coillie E, Herman L, Van Weyenberg S, Daeseleire E, Heyndrickx M (2019) Presence and fate of antibiotic residues, antibiotic resistance genes and zoonotic bacteria during biological swine manure treatment. Ecotoxicol Environ Saf 175:29–38
Wagner GP, Kin K, Lynch VJ (2012) Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Bio 131(4):281–285
Wang FH, Ma W, Dou Z, Ma L, Liu XL, Xu JX, Zhang FS (2006) The estimation of the production amount of animal manure and its environmental effect in China [in Chinese]. Zhongguo Huanjing Kexue/China Environ Sci 26:614–617
Wang J, Gu J, Wang X, Song Z, Dai X, Guo H, Yu J, Zhao W, Lei L (2021) Enhanced removal of antibiotic resistance genes and mobile genetic elements during swine manure composting inoculated with mature compost. J Hazard Mater 411:125135
Wen X, Mi J, Wang Y, Ma B, Zou Y, Liao X, Liang JB, Wu Y (2019) Occurrence and contamination profiles of antibiotic resistance genes from swine manure to receiving environments in Guangdong Province southern China. Ecotoxicol Environ Saf 173:96–102
Xiong W, Wang Y, Sun Y, Ma L, Zeng Q, Jiang X, Li A, Zeng Z, Zhang T (2018) Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome 6(1):34
Yang C, Dong Y, Friman VP, Jousset A, Wei Z, Xu Y, Shen Q (2019) Carbon resource richness shapes bacterial competitive interactions by alleviating growth-antibiosis trade-off. Funct Ecol 33(5):868–875
Yuan CG, Shi JB, He B, Liu JF, Liang LN, Jiang GB (2004) Speciation of heavy metals in marine sediments from the East China Sea by ICP-MS with sequential extraction. Environ Int 30(6):769–783
Yuan QB, Zhai YF, Mao BY, Schwarz C, Hu N (2019) Fates of antibiotic resistance genes in a distributed swine wastewater treatment plant. Water Environ Res: Res Publ Water Environ Fed 91(12):1565–1575
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67(11):2640–2644
Zhang J, Chen M, Sui Q, Wang R, Tong J, Wei Y (2016) Fate of antibiotic resistance genes and its drivers during anaerobic co-digestion of food waste and sewage sludge based on microwave pretreatment. Biores Technol 217:28–36
Zhang Y-J, Hu H-W, Gou M, Wang J-T, Chen D, He J-Z (2017) Temporal succession of soil antibiotic resistance genes following application of swine, cattle and poultry manures spiked with or without antibiotics. Environ Pollut 231:1621–1632
Zhang Y, Yang Z, Xiang Y, Xu R, Zheng Y, Lu Y, Jia M, Sun S, Cao J, Xiong W (2020) Evolutions of antibiotic resistance genes (ARGs), class 1 integron-integrase (intI1) and potential hosts of ARGs during sludge anaerobic digestion with the iron nanoparticles addition. Sci Total Environ 724:138248
Zhao X, Wang J, Zhu L, Ge W, Wang J (2017) Environmental analysis of typical antibiotic-resistant bacteria and ARGs in farmland soil chronically fertilized with chicken manure. Sci Total Environ 593–594:10–17
Zhou B, Wang C, Zhao Q, Wang Y, Huo M, Wang J, Wang S (2016) Prevalence and dissemination of antibiotic resistance genes and coselection of heavy metals in Chinese dairy farms. J Hazard Mater 320:10–17
Zhou Z-C, Zheng J, Wei Y-Y, Chen T, Dahlgren RA, Shang X, Chen H (2017) Antibiotic resistance genes in an urban river as impacted by bacterial community and physicochemical parameters. Environ Sci Pollut Res 24(30):23753–23762
Zhu Y-G, Johnson TA, Su J-Q, Qiao M, Guo G-X, Stedtfeld RD, Hashsham SA, Tiedje JM (2013) Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci 110(9):3435–3440
Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, Hashsham SA, Tiedje JM (2013) Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci U S A 110(9):3435–3440
Funding
This study was supported by the General Program of National Natural Science Foundation of China (grant number [U21A20257]), Key R&D Program of Sichuan province (grant numbers [22ZDZX0011], [2020ZYD003], [2020YFN0147], and [2021YFH0192]), and Project of Science and Technology Bureau of Banan District Chongqing (grant number [BN2020-115]).
Author information
Authors and Affiliations
Contributions
Conceptualization: Junrui Shui, Hongmei Tuo, Anyun Zhang; Methodology: Junrui Shui, Hongmei Tuo, Anyun Zhang; formal analysis and investigation: Hongmei Tuo, Junrui Shui; writing—original draft: Junrui Shui, Hongmei Tuo, Jinxin Liu, Anyun Zhang; writing—review & editing: Junrui Shui, Anyun Zhang, Jinxin Liu, Xialan Zhang, Jingyi Feng, Yuxuan Feng, Wen Su, Cong Lin, Haoyu Zhang, Zunfang Tu; funding acquisition: Anyun Zhang, Hongning Wang.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Diane Purchase
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shui, J., Tuo, H., Liu, J. et al. Insights into the fates of plasmids and antimicrobial resistance genes during swine manure treatment and related factors based on plasmidome and metagenome analyses. Environ Sci Pollut Res 29, 69037–69047 (2022). https://doi.org/10.1007/s11356-022-20574-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-022-20574-7