Abstract
This study assessed the bacterial populations in a non-sanitary landfill around Guarani Aquifer recharge zone in Brazil. Samples from two different positions (sites 1 and 2) at three different depths were evaluated, totaling six solid waste samples; two samples from an impacted stream were also collected. 16S rRNA sequencing was performed using the Ion S5TM XL platform; 3113 operational taxonomic units (OTUs) and 52 phyla were identified. Proteobacteria (37%) and Firmicutes (28%) were the most abundant phyla in the landfill, whereas Proteobacteria (~ 50%) and Bacteroidetes (~ 10%) were more profuse in surface water samples. Canonical correlation analysis (CCA) enabled us to clearly separate the samples according to their spatial location (site 1 or 2) or environmental matrix (surface water or solid waste samples), showing that microbiological populations are strongly associated with site-specific conditions and the kind of environmental matrix they come from. Environmental factors that mostly influenced the microbial communities were organic matter, oxidation–reduction potential, moisture, alkalinity, nitrogen (TKN), sodium, potassium, and zinc. Exiguobacterium (phylum Firmicutes) was overwhelmingly dominant at site 1 and was associated with higher concentrations of organic matter and potassium. Differently, site 2 did not present such dominant genera and was more diverse having lower concentrations of organic matter and nutrients. Distinct environments co-exist inside the same waste deposit, including zones which are representative of active and closed landfills and the occurrence of considerable physicochemical and microbiological shifts within short distances. Those shifts indicate that microbial populations are well adapted to the heterogeneity typical of urban solid waste, which is possibly beneficial to contaminant degradation.

Graphical abstract
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Waste deposits are sources of environmental pollution as they can generate highly contaminated leachates that percolate through the subsoil, reaching the aquifers and altering them for decades or centuries after ending disposal activities (Bichet et al. 2016; Aharoni et al. 2017; Fetter 2018). In Brazil, dumpsites and non-sanitary landfills still receive about 40% of the total collected solid waste (Abrelpe 2017); nevertheless, the long-term impacts of those areas are not sufficiently characterized, leading to several areas being abandoned as a final closure action (Zolnikov et al. 2018).
Waste deposits can be regarded as reactors in which several chemical and microbiological degradation processes occur, unleashing different byproducts (Belevi and Baccini 1989). Hence, such areas should be properly characterized aiming at their appropriate management and optimization, as well as at predicting the pollutant’s behavior and planning effective remediation actions (Sawamura et al. 2010; Quintaes et al. 2014).
Different environmental variables—such as residue physical composition, rainfall regime, percolation of pluvial water, evapotranspiration, waste compaction and age, presence of contaminants, and availability of nutrients—influence landfill ecosystems, interfering in the richness and dynamism of microbial communities, and consequently in pollutant degradation processes (Sawamura et al. 2010; Quintaes et al. 2014). Several studies have been conducted in waste deposits from different cultural and socioeconomical backgrounds, as well as from distinct landfilling and climatic conditions, showing considerable variations in the observed physicochemical parameters and microbial communities (Kochling et al. 2015; Song et al. 2015; Stamps et al. 2016; Wang et al. 2017; Xu et al. 2017; Rajasekar et al. 2018; Zainun and Simarani 2018; Liu et al. 2019).
Since microbial communities are significantly heterogeneous and depend on environmental aspects, their presence in landfills still needs to be characterized so as to better understand the factors influencing degradation processes, and consequently enabling their management to reduce environmental impacts. This is especially relevant for developing countries, where improper landfills and dumpsites are the most common practice for disposal of urban solid waste (Mangimbulude 2013; Kumar 2016; Lavagnolo 2018; Zolnikov et al. 2018; Idowu et al. 2019) and where microbiological studies could enhance the understanding of natural attenuation processes and permit the development of alternative and lower-cost technologies for waste treatment and disposal.
It is important to highlight that identifying microbial populations in landfills and impacted environments has significantly benefitted from the use of molecular biology techniques as they have enabled the discovery of several organisms and the inference of the roles played by them in such environments (Wang et al. 2017; Zainun and Simarani 2018; Liu et al. 2019). These findings contribute to the development of biotechnological applications, which promote the mitigation of environmental impacts in landfills—including the reduction of greenhouse gas emissions—and enhance wastewater and leachate treatments (Santos et al. 2009; Jayanthi et al. 2016; Pan et al. 2019).
The present study aimed at evaluating the microbial populations of an abandoned non-sanitary landfill (dumpsite) in a Guarani Aquifer outcrop zone in Brazil (Wendland et al. 2007), under subtropical conditions. Since the closure of this deposit, several studies have been conducted, mainly focused on the physicochemical characterization of the surrounding water resources (Bossolan 1993; Menezes 1995; Gadotti 1997; Álvares 2000; PMSC 2011; Morita and Wendland 2019a; Morita et al. 2020a; Morita et al. 2020b), showing that the impacts are restricted to areas closer to the landfill. On the other hand, the characterization of the buried solid waste content, conducted by Shinzato (2014), indicated that there are still zones within the deposit with highly concentrated leachate. Thus, it is supposed that the degradation and attenuation processes lead to a decrease in the concentrations of organic matter, dissolved ions, and metals, making it possible to reach acceptable levels within small distances from the deposit (Morita and Wendland 2019a).
Adopting molecular biology techniques to identify microbial populations in such a deposit is highly recommended, leading to results which could be considered as references in Brazil—a country where landfills are still abundant and constantly abandoned (Cetrulo et al. 2018), and where it is necessary to link the lack of financial resources to technological alternatives to protect the environment. It is important to emphasize that relatively few published studies used high-throughput sequencing to evaluate microbial populations of landfills in Brazil (Kochling et al. 2015; Moreira 2019).
Methodology
Study area
The study area is a non-sanitary landfill situated in the southeast of Brazil, São Carlos city (22.1° S, 47.8° W; see Fig. 1), in a Guarani Aquifer outcrop zone, fundamentally constituted by sandstone and sandy soils. The local climatic conditions are classified as Cfa by Koppen and Geiger (humid subtropical climate), with annual rainfall of about 1440 mm and an average temperature of 19.7 °C.
Solid waste disposal occurred from 1980 to 1996, when domestic, industrial, construction and demolition, and health service wastes were disposed of inside an old gully, totaling about 440,000 m3 of residues. No structure for collecting and treating leachate or gas was adopted, and the location of the water table inside the deposit (at around 6-m deep) may have favored groundwater contamination. This led to the site being included in the list of contaminated areas in the state of São Paulo (CETESB 2017).
Sample collection
Solid waste samples from two different sites separated from each other by about 30 m (sites 1 and 2, see Fig. 1) and three different depths (30 cm, 1 m, and 2 m; see Fig. 1) were collected, using a manual driller. Samples were named 1_1, 1_2, 1_3, 2_1, 2_2, and 2_3, in which the first number refers to the spatial position shown in Fig. 1 and the second number refers to the location in depth (1—30 cm, 2—1 m, 3—2 m). The first samples (1_1 and 2_1) were collected at about 30-cm deep in order to exclude the soil cover and consider only solid waste samples for analysis.
The sites for the sample collection were selected based on previous knowledge of the waste and soil layer deposition in the region (Shinzato 2014). Therefore, the sample collection until 2-m deep was based on pockets of accumulated leachate below this depth, which would make collection even more difficult. It is important to highlight that the collection of solid waste samples in landfills is considerably problematic, as non-degraded materials—especially plastic—become attached to the drills and prevent drilling from evolving.
Despite the mentioned difficulties, the sample collection until 2-m deep was made in order to comprehend the interface between soil cover and solid waste (samples 1_1 and 2_1); the transitional environment, with a gradual increase of moisture (1_2 and 2_2); and, finally, the environment with an accumulation of liquids (1_3 and 2_3).
Before collecting each sample, appropriate care was taken in order to clean the drill and avoid contamination. About 500 g of solid waste from each sampling point was collected by using the quartering method, kept in sterilized plastic containers, and placed in an icebox. Samples were then kept at − 20 °C until microbiological and physicochemical analyses were performed.
Additionally, surface water samples from a small stream (at around 15-cm deep), at locations upstream and downstream to the deposit (named Up and Dw, respectively; see Fig. 1), were collected. Surface water samples had their pH, electrical conductivity (EC), and oxidation–reduction potential (ORP) measured in situ with a multiparameter water quality sonde (YSI 6920) and were kept in sterilized PVC bottles inside an icebox. After collection, the suspended solids were left to sediment and were separated for microbiological analysis (at − 20 °C,) whereas the liquid portion was kept for physicochemical analysis (at 4 °C).
It is important to highlight that we used the suspended solids for DNA extraction (and not the surface water samples) in order to permit the use of the same methodology adopted for solid waste samples, as well as the same kit (FastDNA™ SPIN Kit for Soil), and permit more representative comparisons. We consider that, even though there is a partitioning of contaminants in the environment, increases of concentrations in one matrix (sediment or water) lead to increases in the other, due to chemical balance. Since the objective was to relate higher and lower concentrations of contaminants with microbial communities, it is believed that analyzing water and suspended solids from the same sample can give results which are representative of the analyzed environment and ongoing microbiological and physicochemical processes, permitting the comparison of the collected samples.
Physicochemical analysis
For physicochemical analysis, the solid waste samples were solubilized following the Brazilian norm NBR 10.006. According to it, about 400 g from each sample was mixed and large particles (glass, stones, plastic bags) were manually removed. After that, the samples were weighed and dried at 42 °C for 24 h. The dry samples were also weighed, making it possible to calculate the humidity. About 250 g of the dried samples was weighed and placed in beakers, and 1000 mL of distilled water was added to each one. The solution was mixed for 5 min and covered with PVC plastic film for 7 days at 25 °C; after this period, pH and ORP were immediately measured, and samples were filtered in a 0.45-μm filter prior to physicochemical analysis.
Concerning the surface water samples (Up and Dw), filtration in a 0.45-μm filter was also conducted after collection, and physicochemical analysis was performed.
The following physicochemical parameters were analyzed in all samples: alkalinity, nitrite (N-NO2−), nitrate (N-NO3−), ammoniacal nitrogen (N-NH4−), total Kjeldahl nitrogen (TKN), sulfate (SO42−), phosphate (PO43−), fluoride (F−), chloride (Cl−), chemical oxygen demand (COD), total organic carbon (TOC), aluminum (Al), antimony (Sb), barium (Ba), cadmium (Cd), calcium (Ca), lead (Pb), cobalt (Co), copper (Cu), chromium (Cr), strontium (Sr), iron (Fe), magnesium (Mg), manganese (Mn), nickel (Ni), potassium (K), silver (Ag), selenium (Se), sodium (Na), and zinc (Zn). All the analyses were performed according to the Standard Methods for the Examination of Water and Wastewater (APHA 2012).
DNA extraction and sequencing using the platform Ion S5 XL, SE600
The genomic DNAs of solid waste and sediment samples (suspended solids from Up and Dw) were extracted using the FastDNA™ SPIN Kit for Soil DNA Extraction (MP Biomedicals), using 0.5-g subsamples. It is important to highlight that this method uses solid samples, so that the analysis of surface water samples was made after sedimentation of the suspended solids, as previously mentioned. DNA purification was also performed, using the Geneclean ® Turbo Kit (MP Biomedicals). The DNA concentration was measured in a NanoDrop 2000 spectrophotometer (Nanodrop Wilmington, DE, USA), and its integrity was assessed by performing an agarose (0.8%) gel electrophoresis.
After DNA extraction, the samples had their 16S rRNA genes of distinct regions (V3–V4 466 bp) amplified using the domain bacteria (341F CCTAYGGGRBGCASCAG; 806R GGACTACNNGGGTATCTAAT) with the barcode. Library preparation and sequencing libraries were generated using the Ion Plus Fragment Library Kit 48 rxns (Thermo Scientific) following the manufacturer’s recommendations. The library quality was assessed using the Qubit@ 2.0 Fluorometer (Thermo Scientific). Finally, the library was sequenced on an Ion S5TM XL platform, and 400-bp/600-bp single-end reads were generated.
Quality filtering on the raw reads was performed under specific filtering conditions to obtain the high-quality clean reads according to the Cutadapt quality control process (Martin 2011). The reads were compared with the reference database (Quast et al. 2013) using the UCHIME algorithm (Edgar et al. 2011) in order to detect and remove chimera sequences (Haas et al. 2011) and obtain the clean reads.
Sequence analysis was performed by using the Uparse software (Edgar 2013), and sequences with ≥ 97% similarity were assigned to the same operational taxonomic units (OTUs). Alpha- and beta-diversity analyses were performed using the QIIME (Version1.7.0) and R (Version 2.15.3) software. The 16S rRNA sequencing and bioinformatics were performed by GenOne Biotechnologies, situated in Rio de Janeiro, Brazil.
These data have been uploaded to the National Center for Biotechnology Information Database (NCBI), under Bioproject number PRJNA566349 and BioSample Accessions SAMN12787304, SAMN12787305, SAMN12787306, SAMN12787307, SAMN12787308, SAMN12787309, SAMN12787310, and SAMN12787311.Footnote 1
Statistical analysis
In order to better evaluate the relations between environmental factors and the microbial communities, canonical correlation analysis (CCA) was performed, using the PAST software (version 3.26).
Results and discussion
Physicochemical characterization of the solid waste and surface water samples
The physicochemical parameters of the solid waste and surface water samples are shown in Fig. 2 and in Table 1SM of the Supplementary Material. The results refer to one analysis of each sampling point; no replication was performed due to the difficulties found during the sample collection.
Physicochemical parameters of solid waste and surface water samples (ORP, oxidation–reduction potential; EC, electrical conductivity; COD, chemical oxygen demand; TKN, total Kjeldahl nitrogen; Cl, chloride; Na, sodium; Mg, magnesium; K, potassium; Ba, barium; Zn, zinc; Ca, calcium; Fe, iron; Pb, lead; Mn, manganese). The location of samples is presented in Fig. 1 and explained in the “Sample collection” section
It can be observed that the evaluated parameters varied considerably with depth (mainly from the first layer to the deeper ones) and depending on the site location. The first layers presented similarities among samples and low concentrations of most parameters. On the other hand, it seems that the characteristics of the second and third depths are influenced by specific physicochemical conditions on the site, which are the samples with greatest concentrations from site 1. On the other hand, site 2 seems to have had conditions similar to the first layers at all depths.
Moisture content also varied according to the depth and spatial location. Samples from site 1 generally had more water content (6–23%) than those from site 2 (4–10%). Additionally, the deeper they were, the more moisture there was in both sites. It is important to highlight that at about 2.2-m deep, there is a soil layer in the deposit (Shinzato 2014), which causes an accumulation of leachate above it (see Fig. 1). This condition especially affected the samples collected at 2-m deep (1_3 and 2_3), leading to the verification of more moisture and higher concentrations of dissolved ions (higher electrical conductivity values).
It is interesting to note that the ORP values in the samples from site 1, and especially in 1_2, were much lower than those in the samples from site 2. However, those values may not be representative of the landfill environment—due to the conditions of the solubilization test—and should be used only for the sake of comparison among different samples.
EC, alkalinity, organic matter (COD and TOC), N-NO3−, N-NH4−, SO42−, PO43−, Cl−, TKN, moisture, and the vast majority of metals were higher in samples from site 1. The existence of higher concentrations of organic matter, ions, and metals is a characteristic of the first phases of degradation in landfills (Andreottola and Cannas 1992; Filho and Miguel 2017). On the other hand, lower concentrations of most parameters were found in site 2, which are characteristic of more degraded and older content. Exceptions were the concentrations of NO2−, higher in site 2; Zn, especially high in 2_2; and Al and Fe; higher in 2_1.
The differences encountered between sites 1 and 2 can be attributed to solid waste heterogeneity, which depends on the characteristics of the buried content at each site and is more evident when waste samples, instead of leachate samples, are analyzed. This is because landfills are highly heterogeneous, and processes occur in microenvironments; the leachate, when properly collected in sanitary landfills, is formed by a mixture of contributions from those different zones, representing the dominant process (Cossu et al. 2019).
Regarding such heterogeneities, it is important to mention that the accumulation of leachate in pockets inside the deposit may also lead to an increase in contaminant concentrations in specific locations (Shinzato 2014; Aharoni et al. 2017; Moretto et al. 2017). Therefore, distinct environments could be distinguished inside the same landfill, with zones containing high contaminant concentrations and others with much lower values.
Nevertheless, all the samples—from sites 1 and 2—presented characteristics which are in agreement with the ranges observed in an uncontrolled landfill in Israel (Aharoni et al. 2017) and in a wide number of landfills in Brazil (Souto and Povinelli 2011), and with the stable methanogenic phase studied by Song et al. (2015). This agreement shows the representativeness of the studied case.
It is important to mention that the results of the solubilization process and their comparison with the limits presented by the Brazilian norm NBR 10.006 showed that the buried waste is still non-inert, after about 20 years of ending disposal activities, especially considering Pb, Mn, Fe, Zn, and Al (Morita and Wendland 2019b).
Regarding surface water samples, a clear influence of the waste deposit on water resources was observed, significantly changing the values of pH, ORP, EC, alkalinity, organic matter, N-NH4−, and metals from sample Up to Dw. Attention must be given to the pH, which is basic in upstream conditions and much more acidic in downstream; ORP, showing an oxidant environment upstream and reducing downstream; and the presence of Fe and Mn in Dw, where they can be possibly used as electron acceptors by anaerobic populations.
16S rRNA sequencing
Richness and diversity of microbial communities
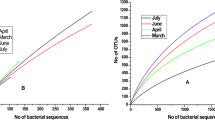
A total of 3113 OTUs were identified, considering all 8 samples. The coverage shows that the sequencing could represent the microbial communities well, varying from 97 to 99%. The richness and diversity of microbial communities in waste and surface water samples are shown in Fig. 3, in which different patterns can be observed for sites 1 and 2.
At the first site, there was a significant decrease in richness from the first (1_1) to the second depth (1_2), followed by an increase in the third one (1_3). This behavior shows the existence of a selective environment in 1_2, leading to the dominance of specific populations adapted to more reducing conditions and higher concentrations of organic matter and dissolved ions, possibly anaerobic hydrolytic or fermentative bacteria.
On the other hand, at the second site, there was an increase of richness with depth, indicating that there might be a higher availability of organic matter (increase in COD and TOC, see Table 1SM of the Supplementary Material), moisture, and nutrients with depth, associated with the inexistence of stressing conditions. The condition found at site 2—increase in richness with depth—was also observed by Gomez et al. (2011) and Wang et al. (2017).
Observing data from sites 1 and 2, it can be observed that solid waste samples with lower concentrations of the analyzed parameters (COD, alkalinity, EC, TKN, PO43−, SO42−, NH4−, NO3−, Na, Mg, and K, generally from site 2: 2_1, 2_2, and 2_3) had higher ecological indexes and number of OTUs. Zainun and Simarani (2018) and Song et al. (2015) found that more stabilized samples (i.e., from closed landfills or landfills in the stabilized methanogenic phase) had higher Chao and Shannon indexes than samples from active landfills.
Thus, samples with more stabilized content can supposedly harbor more microbial species, either because they have been more intensely colonized by different species or because more species can survive in environments with lower concentrations of contaminants. It is interesting to note, therefore, that the heterogeneities can be found inside the same waste deposit—and not necessarily in active and closed landfills—forming zones with specific content and its associated community, and adding complexity to the study of solid waste.
When compared with other studies in landfills, the observed ecological indexes and the number of OTUs were higher than those observed by Wang et al. (2017) in China, but lower than those observed by Xu et al. (2017), also in China, and Zainun and Simarani (2018), in Malaysia, showing that different patterns can be observed in landfills, depending on a variety of environmental conditions.
Regarding the sampling points Up and Dw, it can be observed that they had very similar richness and diversity indexes. However, the impact of the landfill leachate seems to have caused a mild increase in the communities’ richness (Chao and ACE indexes) and a decrease in diversity (Shannon index). Hence, more species probably grow in media with more organic matter and nutrients (Dw), but in this community, some species are dominant since they are better adapted to the higher levels of contaminants.
Taxonomic analysis at the level of phyla
Considering all the analyzed samples, 52 phyla were identified, in which 16 had a higher abundance than 1%, which surpasses the number identified by previous studies (Wang et al. 2017; Xu et al. 2017). The relative abundance of the different phyla for the distinct studied samples is presented in Fig. 4.
Representatives of the phyla Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, and Chloroflexi were identified in all samples, including in the Up and Dw sediment samples. Proteobacteria (37%), Firmicutes (28%), and Actinobacteria (13%) were the most abundant phyla in the landfill, whereas Proteobacteria (54%), Bacteroidetes (12%), and Acidobacteria (12%) were the most abundant in Up, and Proteobacteria (48%), Bacteroidetes (10%), and Firmicutes (9%) were the most abundant in Dw.
The phyla Proteobacteria were dominant in all samples, except for 1_2 (4%), where Firmicutes dominated (82%), and 1_3, where both named phyla had similar abundances (approximately 40%). These results agree well with other studies in landfills, in which Proteobacteria (Sawamura et al. 2010; Gomez et al. 2011; Zainun and Simarani 2018; Liu et al. 2019) and Firmicutes (Krishnamurthi and Chakrabarti 2013; Kochling et al. 2015; Wang et al. 2017; Xu et al. 2017) were the most abundant phyla.
Bacteria belonging to the phylum Firmicutes have been associated with a degradation of polysaccharides—such as cellulose, lignocellulose, and starch (Song et al. 2015; Xu et al. 2017; Zainun and Simarani 2018; Jiang et al. 2019)—playing an important role in the synthesis of fulvic-like substances and humification (Liu et al. 2019) and in the formation of dissolved organic matter (Jiang et al. 2019). This role seemed to be well associated with the physicochemical analysis performed in the present study, since the Up point had the lowest representation of Firmicutes, and site 1 (especially 1_2 and 1_3), which presented the highest concentrations of organic matter, showed significant communities of Firmicutes (see Figs. 2 and 4).
It is interesting to highlight that the occurrence of Firmicutes has been associated either with the initial methanogenic phase of landfills (Song et al., 2015) or with older landfill cells (Liu et al. 2019), which shows that this phylum does respond not only to the residue age but also to the decomposition stage and, consequently, to the contaminant concentrations. Thus, Fajardo et al. (2019) and Kasemodel et al. (2019) observed that Firmicutes were the most abundant phylum in samples with higher concentrations of Pb, Zn, and Cd, possibly because of their ability to form endospores under stressful conditions. Hence, in our study, the occurrence of such phylum has been more intensely verified in zones with a combination of higher organic matter (COD and TOC), nutrients, and metal concentrations (especially Ba, Cd, Pb, Ca, Mg, and Sr).
Representatives of the phylum Proteobacteria, on the other hand, even having been linked to organic matter degradation in anaerobic environments (Bareither et al. 2013) and wastewater treatment plants (Greay et al. 2019), were found in all analyzed samples with a similar significance (except for 1_2), showing that different roles were played by them, in cleaner (Up) or more contaminated (Dw and landfill samples) environments. Proteobacteria were also the dominant phylum in closed and active landfills studied by Zainun and Simarani (2018), and were dominant in control soil samples, playing important roles in soil ecosystems (Wang et al. 2017; Rajasekar et al. 2018; Fajardo et al. 2019).
The phyla Acidobacteria and Actinobacteria had low significance in 1_2 and 1_3, having been possibly displaced by Firmicutes, as described by Fajardo et al. (2019). Actinobacteria was associated with the degradation of organic matter (Wang et al. 2017) and cellulose, especially at intermediate and old stages of waste decomposition (Liu et al. 2019). Hence, samples from site 2, which seem to have more stabilized content, presented a higher significant number of this phylum. Additionally, representatives of this phylum have been recently associated with sites contaminated with metals (Salam and Varma 2019), showing that they can play an important role in the metabolism of Fe and Zn, found in high concentrations at site 2 (see Fig. 2).
Similarly, Nitrospirae was barely found at site 1 (1_1, 1_2, and 1_3) but was more significant in all other samples. This phylum was found in closed landfills, surrounded by trees and vegetative plants (Zainun and Simarani 2018), and was associated with nitrification (Song et al. 2015). Thus, it might have played an important role at site 2, where lower levels of ammoniacal nitrogen were found, linked to higher concentrations of nitrite (see Fig. 2).
The phylum Gemmatimonadetes was more significant in the landfill’s superficial samples (1_1 and 2_1) and in Up than in deeper samples or in Dw. Thus, its presence can be associated with environments with lower levels of contaminants, and possibly with a higher availability of oxygen. Wong et al. (2019) verified its presence only in landfill soil cover samples treated with biochar, which is supposed to enhance soil aeration and bacterial growth.
It is interesting to note that Zainun and Simarani (2018) associated Acidobacteria, Gemmatimonadetes, Verrucomicrobia, Actinobacteria, and Nitrospirae with samples from the closed landfill, which presented lower contaminant concentrations; the present study also made it possible to relate these phyla to more stabilized samples.
On the other hand, the phyla Deferribacteres, Nitrospinae, and Spirochaetes were characteristic of the sample Dw. Deferribacteres and Spirochaetes have been reported in cultures inoculated with oil, having metabolism associated with the fermentation of short-chain organic acids and the release of acetic acid and H2 (Silva et al. 2013). The phylum Deferribacteres includes chemoorganotrophic bacteria that respire preferentially anaerobically with various terminal electron acceptors, including Fe(III), Mn(IV), and nitrate (Alauzet and Jumas-Bilak 2014). It is supposed that such phyla play important roles in the degradation of organic matter from the leachate which stems from the landfill, possibly forming an iron-reducing environment, which agrees with the physicochemical characteristics presented in Fig. 2.
Finally, Nitrospinae has been associated with the oxidation of nitrite to nitrate (Pachiadaki et al. 2017; Jurczyk and Koc-Jurczyk 2017); as no nitrite was detected in Dw (see Fig. 2), but high concentrations of ammoniacal nitrogen, it seems that this phylum is part of a conjunction of nitrification reactions, rapidly consuming the nitrite produced by the oxidation of ammonia.
Figure 5 shows the weighted grouping of the studied samples, in which it is possible to note that their relation could not be explained by their in-depth position. The factor that seems to have mostly influenced the communities was the sample’s spatial localization (site 1 or site 2), rather than their depth. This conclusion has also been observed by Xu et al. (2017) in two sanitary landfills in China.
Additionally, it can be observed that surface water samples did not have any significant relation with solid waste samples, even the one contaminated by leachate (Dw). In other words, the microbiological populations depend more significantly on the kind of environmental matrix they come from (solid waste or surface water), than on physicochemical characteristics (contaminated or uncontaminated samples).
c analysis at the level of class, order, family, and genera
Site 1
The taxonomic analysis at the level of class (see Figure 1SM of the Supplementary Material) showed that Bacilli, which has been reported as being resistant to heavy metal contamination (Fajardo et al. 2019), was the most abundant one at site 1 (19%, 55%, and 32%, respectively, in 1_1, 1_2, and 1_3). Regarding other abundant classes, Alphaproteobacteria (16%) and Betaproteobacteria (11%) were also abundant in the first layer (1_1), whereas Clostridia (25%) was the second most abundant in 1_2 and Gammaproteobacteria (30%) in 1_3. Clostridia was also found in all the landfill samples and in Dw, but logically was not found in Up, since such sampling point is located in an upstream position, with oxidant conditions and low concentrations of organic matter.
Clostridia has been reported as being resistant to chemical stress, playing an important role in bioprocessing and biotransformations, acting in fermentation processes (Johnson 2019). It is capable of hydrolyzing a wide range of carbohydrates and producing hydrogen (Wong et al. 2018) and can be found in environments with low ORP and high biodegradable organic matter content (Dao et al. 2016). Therefore, its presence is probably associated with the degradation of organic matter in reducing conditions.
Regarding the analysis at the level of genera at site 1 (Fig. 6), it is interesting to note that the phylum dominance in samples 1_2 and 1_3 (Firmicutes) was kept until the genera level, showing that the genus Exiguobacterium represented 54% and 30% of relative abundance in those samples, respectively. The mentioned samples both had more reducing conditions and higher concentrations of organic matter, EC, alkalinity, TKN, N-NH4−, SO42−, and metals (see Fig. 2). Exiguobacterium has been reported as being gram-positive, able to grow in aerobic and anaerobic environments at a wide range of temperatures (5–40 °C), pH (6.5–12.0), and NaCl concentrations, having applications to alkaline wastewater treatment (Kulshreshtha et al. 2010). To the best of our knowledge, it is the first time Exiguobacterium has been reported in landfills.
The second most abundant genus in 1_2 was Gelria (7%), whereas Acinetobacter (13%) and Pseudomonas (12%) were the second most significant genera in 1_3. Gelria is an anaerobic, syntrophic, and endospore-forming genus (Plugge et al. 2002), known as end-stage fermenters involved in the metabolism of fatty acids (Fitzgerald et al.2019). Pseudomonas was associated with refuse decomposition (Song et al. 2015), denitrification, and pollutant degradation (Wang et al. 2017). Acinetobacter was regarded as sulfur-metabolizing bacteria (He et al. 2018; Ding et al. 2019), which is compatible with the concentrations of sulfate found in 1_3 (Fig. 2), and has also been associated with the thermophilic phase of composting (Jiang et al. 2019).
Site 2
Differently, at the second site, Alphaproteobacteria was the most abundant class (26%, 21%, and 19%, respectively, in 2_1, 2_2, and 2_3; see Figure 1SM). Alphaproteobacteria was the most dominant class in closed landfills (Zainun and Simarani 2018), showing that the physicochemical conditions at site 2 can be associated with more degraded content. The second most abundant classes were Betaproteobacteria (8.5%) and Gammaproteobacteria (8.5%) for 2_1 and Unindentified_actinobacteria in 2_2 and 2_3 (8.5% and 13% respectively).
Regarding the analysis at the genus level (Fig. 6), Sphingomonas was significant in site 2 and in 1_1, but was not identified in 1_2 or 1_3. This genus has been reported as abundant in areas contaminated by Pb, Cu, Zn, and Cd (Wang et al. 2018), and e-waste (Wu et al. 2019), which can be associated with the high Zn content in site 2.
Interestingly, Paenibacillus was more closely identified in sample 2_2, where extremely high concentrations of Zn were observed. This genus, together with Bacillus, was abundant in samples contaminated by heavy metals (Fajardo et al. 2019). Similarly, Arthrobacter was more abundant in sample 2_2 and has been reported as resistant to metal contamination and able to bioremediate sites contaminated by heavy metals (Hong et al. 2015; Salam and Varma 2019; Wu et al. 2019).
Sediment samples Up and Dw
Finally, regarding the sediment samples, Betaproteobacteria, Deltaproteobacteria, and Gammaproteobacteria were the most abundant classes both for Up and Dw (see Figure 1SM). Alphaproteobacteria was also abundant in Up, whereas Clostridia was profuse in Dw. It is interesting to note, regarding the order level (Figure 2SM), that many orders that were identified in Up were also identified in Dw, but at much smaller percentages (e.g., Sphingobacteriales, Rhizobiales, Myxoccocales, Burkholderiales, Xanthomonadales, Nitrosomonadales). Probably, bacteria belonging to those orders were sensitive to stressing conditions. An opposite behavior was observed for Rhodocyclales, Clostridiales, Unidentified_deferribacteres, Syntrophobacterales, and Methylophylales, which were much more significant in Dw, thus associated with the metabolization of pollutants.
At the genus level (Fig. 6), Caldithrix was only identified in Dw, and Gelria. It was barely identified in Up and was abundant in Dw. Caldithrix is obligately anaerobic, whose members are chemoorganoheterotrophs able to ferment polysaccharides (Alauzet and Jumas-Bilak 2014). Other genera which were plentiful in Dw (e.g., Crenotrix, Dechlorobacter, Anaerovorax, Desulfatiglans, Ignavibacterium, Desulfobulbus, Geothermobacter) can thus be associated with the degradation of compounds from the leachate originating from the deposit, especially under reducing conditions and using different electron acceptors. Therefore, these genera could be used as indicators of contamination by waste deposits.
Interestingly, Geobacter and Anaeromyxobacter were identified both in Up and Dw, but more significantly in the first one. Both genera are formed by exoelectrogenic bacteria, which are capable of transferring electrons outside the cells to extracellular acceptors, and thus play important biogeochemical roles in natural environments such as soils, sediments, and freshwater (Wang et al. 2019).
Particularly, Geobacter acts in the biogeochemical cycling of carbon and Fe (Lovley et al. 2011) and is capable of degrading a variety of organic contaminants, coupling oxidation of organic matter to the reduction of Fe (III) and Mn (IV) (Jiang et al. 2020). It is interesting to note that Geobacter could be identified in pristine (Up) and contaminated (Dw) samples, playing distinct roles, both in biogeochemical cycles and organic matter degradation. Consequently, the use of such genera as indicators of leachate contamination is not recommended as they fill different niches in a diversity of environments (Lovley et al. 2011).
Finally, regarding the analysis at the genera level in the study area, it is important to highlight that the vast majority of them could not be identified. Dominant genera in the landfill included Exiguobacterium, Pseudomonas, Streptococcus, Sphingomonas, Acinetobacter, and Gelria, in which occurrence was dependent on specific conditions at each site. On the other hand, Caldithrix, Crenothrix, Gelria, Methylonera, and Dechloromonas were abundant in a Dw sediment sample and were indicators of leachate contamination, whereas Geobacter and Anaeromyxobacter were profuse both in Up and Dw positions, which makes them inappropriate to be used as indicators.
Correlations between microbial communities and environmental factors
In order to visualize the influence of some environmental factors in microbial communities, canonical correlation analyses were performed, using 13 phyla with a relative abundance > 1%, 15 genera with relative abundance > 3%, and 19 factors (depth, moisture, pH, ORP, EC, alkalinity, COD, TKN, SO4, PO4, Pb, Fe, Zn, Mg, Ba, Ca, Sr, Na, and K).
The result of the CCA considering the communities at the phyla level is presented in Fig. 7, in which axes 1 and 2 were responsible for about 93% of the samples’ variability.
The CCA observed that the samples could be clearly separated into quadrants, according to their spatial location: samples from site 1 are in the first and forth quadrants (except for 1_1, which is in the middle of the first and second quadrants), presenting higher pollutant concentrations (COD, nutrients, Na, K, Mg, Ca, Sr); samples from site 2 are in the second quadrant and are represented by high Zn content (which is especially true for 2_2) and higher ORP values; and sediment samples obviously had higher moisture and were represented in the third quadrant.
It is interesting to note that studies conducted at deeper locations and longer intervals (Sawamura et al. 2010; Gomez et al. 2011; Xu et al. 2017), and at smaller intervals and shallower positions—such as the present study, Wang et al. (2017), and Zainun and Simarani (2018)—led to similar conclusions that the microbial heterogeneity is more strongly affected by the sample’s spatial position than by their in-depth location. Therefore, microbial heterogeneity can be explained mainly by buried content and the consequent physicochemical characteristics; water content and oxygen availability, which are more dependent on the samples’ in-depth location, did not play an important role in the distribution of microbiological communities.
It can also be observed that many phyla had their occurrence dependent on a conjunction, or a mixture, of parameters, which led to their location being in intermediate positions and far from the arrows indicating specific environmental factors. This was especially true for Proteobacteria, Bacteroidetes, and Chloroflexi.
Similarly, although they were found in all samples, the phylum Firmicutes was presented in the graph closer to sample 1_2 and on the same side of the arrows indicating COD, alkalinity, Na, K, Mg, and TKN, showing that these factors have significantly affected this phylum’s abundance. In the same way, Actinobacteria and Acidobacteria were more abundant in samples from site 2 and were thus represented in the second quadrant.
The longest arrows indicate the factors which most influenced the microbial communities: COD, Na, K, TKN, alkalinity, moisture, and ORP. It is important to highlight that moisture especially influenced the differentiation among landfill samples and surface water samples.
CCA analysis based on the most abundant genera is presented in Fig. 8; axes 1 and 2 were responsible for about 84% of the samples’ variability. It can be seen that Exiguobacterium and Gelria were strongly affected by COD and K concentrations, whereas Sphingomonas was influenced by Fe concentrations. Higher COD and K concentrations can be associated with younger leachates (Andrettola and Cannas 1992; Filho and Miguel 2017), indicating that Exiguobacterium and Gelria are possibly more profuse in environments with higher pollutant concentrations.
On the other hand, Arthrobacter was associated with high ORP (more oxidizing conditions), high EC, and deeper positions. The occurrence of other genera may not be strongly associated with specific conditions, as it is dependent on a mixture of parameters, as can be observed by their central position in the graph.
CCA analysis showed that landfills can harbor more and less contaminated zones, which influence the occurrence of distinct populations—typical of younger and older landfills—within the same deposit. Hence, even closed and abandoned landfills may have highly contaminated zones with communities and physicochemical characteristics typical of active landfills. Heterogeneity creating microenvironments inside landfills has been discussed by Cossu et al. (2019).
The association between the microorganisms and physicochemical parameters observed in the present study confirms the ability of communities from natural environments to degrade and remediate contaminants, and indicates their potential for biotechnological applications, enabling their use in treatment and remediation projects. For instance, Firmicutes, abundant in environments with higher metal content in the present study, has already been applied to remediate such contaminants (Fajardo et al. 2019); Exiguobacterium, abundant in samples with higher organic matter content and pH values, has already been reported in wastewater treatment plants, neutralizing highly alkaline effluents (Kulshreshta et al., 2010). The present study showed the relevance of such populations to landfills, indicating that they can be applied in biotechnological solutions for solid waste disposal.
Conclusions
Next-generation sequencing was conducted in order to assess the structure and diversity of bacterial communities in an unplanned closed landfill in Brazil. Proteobacteria and Firmicutes were the most abundant phyla, accounting for 65% and 59% of the sequences in landfill and surface water samples, respectively.
Several environmental factors influenced the occurrence of microbial communities, especially organic matter (COD and TOC), nitrogen (TKN), Na, K, Mg, and Zn. Microbial heterogeneity was more strongly affected by the sample’s spatial position than by their in-depth location. Therefore, specific populations were found in different analyzed samples, and their occurrence could be associated with local physicochemical conditions. Exiguobacterium, Pseudomonas, Streptococcus, Sphingomonas, Acinetobacter, and Gelria were abundant genera inside the landfill, but with locally dependent distribution. Caldithrix, Crenothrix, Gelria, Methylonera, and Dechloromonas were abundant in a Dw surface water sample affected by the leachate stemming from the deposit and thus functioned as indicators of contamination. On the other hand, Geobacter and Anaeromyxobacter were identified both in Up and Dw samples, and thus their presence cannot indicate leachate contamination.
This study provided up-to-date knowledge about microbial populations in landfills in Brazil and established important relationships with studies conducted in other countries and landfills. Therefore, it helped to better understand the factors affecting microbial communities and heterogeneity in waste deposits, which is fundamental for their appropriate management.
Notes
https://www.ncbi.nlm.nih.gov/sra/PRJNA566349
References
ABRELPE (2017). Overview of solid waste in Brazil in 2016. Organized by the Brazilian Association of Public Cleaning and Special Waste Companies (ABRELPE) (in Portuguese). São Paulo, Brazil. Available at: www.abrelpe.org.br/Panorama/panorama2016.pdf
Aharoni I, Siebner H, Dahan O (2017) Application of vadose-zone monitoring system for real-time characterization of leachate percolation in and under a municipal landfill. Waste Manage 67(2017):203–213
Alauzet C, Jumas-Bilak E (2014) The phylum Deferribacteres and the genus Caldithrix. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes. Springer, Berlin, Heidelberg
Andreottola G, Cannas P (1992) Chemical and biological characteristics of landfill leachate. In: Christensen TH, Cossu R, Stegmann R (eds) Landfilling of waste: leachate. Chapman and Hall
Álvares, C.M.B. (2000) Contribution to the knowledge of environment in the region of a dumpsite in São Carlos-SP, through geological, geophysical, topographic, and chemical studies. Master’s dissertation (in Portuguese) Escola de Engenharia de São Carlos, Universidade de São Paulo, São Carlos, 2000
APHA (2012) Standard methods for the examination of water and wastewater, 22nd edn. American Public Health Association; AWWA, WEF, Washington
Bareither A, Wolfe GL, Mcmahon KD, Benson CH (2013) Microbial diversity and dynamics during methane production from municipal solid waste. Waste Manage 33(2013):1982–1992
Belevi H, Baccini P (1989) Long-term behavior of municipal solid waste landfills. Waste Manag Res 7:43–56
Bichet, V. Grisey, E. Aleya, L. (2016) Spatial characterization of leachate plume using electrical resistivity tomography in a landfill composed of old and new cells (Belfort, France). Eng Geol. 211 (2016). Pags 61–73
Bossolan, N.R.S. (1993) Ecological aspects of the bacterial populations in groundwater under the influence of a controlled landfill. Master’s dissertation. Universidade Federal de São Carlos, São Carlos, 1993
CETESB (Environmental Agency of São Paulo State) List of contaminated areas, 2017. Available at http://areas.contaminadas.cetesb.sp.gov.br
Cetrulo TB, Marques RC, Cetrulo NM, Silva F, Moreira RM, Cortés DM, Malheiros TF (2018) Effectiveness of solid waste policies in developing countries: a case study in Brazil. Cleaner Prod 205:179–187
Cossu R, Morello L, Stegmann R (2019) Biochemical processes in landfill. In: Cossu R, Stegman R (eds) (2019) Solid Waste Landfilling: Concepts, Processes, Technologies, vol 2019. Elsevier, pp 773–796
Dao HTN, Kuroda K, Nakahara N, Danshita T, Hatamoto M, Yamaguchi T (2016) 16S rRNA gene-based comprehensive analysis of microbial community compositions in a full-scale leachate treatment system. J Biosci Bioeng 122(6):708–715. https://doi.org/10.1016/j.jbiosc.2016.06.003
Ding Y, Xiong J, Zhou B, Wei J, Qian A, Zhang H, Zhu W, Zhu J (2019) Odor removal by and microbial community in the enhanced landfill cover materials containing biochar-added sludge compost under different operating parameters. Waste Manag 87(2019):679–690
Edgar RC et al (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27.16:2194–2200
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10.10:996–998
Fajardo C, Costa G, Nande M, Botías P, Cantalejo JG, Martín M (2019) Pb, Cd, and Zn soil contamination: monitoring functional and structural impacts on the microbiome. Appl Soil Ecol 135(2019):56–64
Fetter CW (2018) Contaminant hydrogeology. Waveland Press, inc, p 2018
Filho JLP, Miguel MG (2017) Long-term characterization of landfill leachate: impacts of the tropical climate on its composition. Am J Environ Sci 13(2):116–127. https://doi.org/10.3844/ajessp.2017.116.127
Fitzgerald JA, Wall DM, Jackson SA, Murphy JD, Dobson ADW (2019) Trace element supplementation is associated with increases in fermenting bacteria in biogas mono-digestion of grass silage. Renew Energy 138:980–986
Gadotti, R. F. (1997) Evaluation of the groundwater and surface water contamination in an area close to a dumpsite in São Carlos city. Master’s dissertation (in Portuguese). Escola de Engenharia de São Carlos, Universidade de São Paulo, São Carlos
Gomez AM, Yannarell AC, Sims GK, Cadavid-Restrepo G, Herrera CXM (2011) Characterization of bacterial diversity at different depths in the Moravia Hill landfill site at Medellín, Colombia. Soil Biol Biochem 43:2011
Greay TL, Gofton AW, Zahedi A, Paparini A, Linge KL, Joll CA, Ryan UM (2019) Evaluation of 16S next-generation sequencing of hypervariable region 4 in wastewater samples: an unsuitable approach for bacterial enteric pathogen identification. Sci Total Environ 670(2019):1111–1124
Haas BJ et al (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21.3:494–504
He R, Yao X-Z, Chen M, Ma R-C, Li H-J, Wang C, Ding S-H (2018) Conversion of sulfur compounds and microbial community in anaerobic treatment of fish and pork waste. Waste Manag 76:383–393. https://doi.org/10.1016/j.wasman.2018.04.006
Hong C, Si Y, Xing Y, Li Y (2015) Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ Sci Pollut Res 22(14):10788–10799
Jayanthi B, Emenike CU, Agamuthu P, Simarani K, Mohamad S, Fauziah SH (2016) Selected microbial diversity of contaminated landfill soil of Peninsular Malaysia and the behavior towards heavy metal exposure. CATENA 147:25–31. https://doi.org/10.1016/j.catena.2016.06.033
Jiang Z, Shi M, Shi L (2020) Degradation of organic contaminants and steel corrosion by the dissimilatory metal-reducing microorganisms Shewanella and Geobacter spp. Int Biodeterior Biodegradation 147:104842. https://doi.org/10.1016/j.ibiod.2019.104842
Jiang Z, Lu Y, Xu J, Li M, Shan G, Li Q (2019) Exploring the characteristics of dissolved organic matter and succession of bacterial community during composting. Bioresour Technol 292:121942. https://doi.org/10.1016/j.biortech.2019.121942
Johnson EA (2019) Clostridia. In: Encyclopedia of microbiology, Fourth edn, pp 690–695
Jurczyk L, Koc-Jurczyk J (2017) Quantitative dynamics of ammonia-oxidizers during biological stabilization of municipal landfill leachate pretreated by Fenton’s reagent at neutral pH. Waste Manag 63:310–326
Kasemodel MC, Sakamoto IK, Varesche MBA, Rodrigues VGS (2019) Potentially toxic metal contamination and microbial community analysis in an abandoned Pb and Zn mining waste deposit. Sci Total Environ 675:367–379
Kochling T, Sanz JL, Gavazza S, Florencio L (2015) Analysis of microbial community structure and composition in leachates from a young landfill by 454 pyrosequencing. Appl Microbiol Biotechnol 99:5657–5668
Krishnamurthi S, Chakrabarti T (2013) Diversity of bacteria and archaea from a landfill in Chandigarh, India, as revealed by culture-dependent and culture-independent molecular approaches. Systematic and Applied Microbiol 36(2013):56–68
Kulshreshtha NM, Kumar A, Dhall P, Gupta S, Bisht G, Pasha S, Singh VP, Kumar R (2010) Neutralization of alkaline industrial wastewaters using Exiguobacterium sp. Int Biodeterior Biodegradation 64(3):191–196
Kumar S (2016) Municipal solid waste management in developing countries. CRC Press
Idowu IA, Atherton W, Hashim K, Kot P, Alkhaddar R, Alo BI, Shaw A (2019) An analyses of the status of landfill classification systems in developing countries: Sub Saharan Africa landfill experiences. Waste Manag 87:761–771
Lavagnolo MC (2018) Landfilling in Developing Countries. In: Cossu R, Stegman R (eds) Solid waste landfilling: concepts, processes, technologies, vol 2019. Elsevier, pp 773–796
Liu S, Xi B, Qiu Z, He X, Zhang H, Dang Q, Zhao X, Li D (2019) Succession and diversity of microbial communities in landfills with depths and ages and its association with dissolved organic matter and heavy metals. Sci Total Environ 651(2019):909–916
Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA, Nevin KP (2011) Geobacter: the microbe electric’s physiology, ecology, and practical applications. Adv Microb Physiol:1–100. https://doi.org/10.1016/b978-0-12-387661-4.00004-5
Mangimbulude, J. C. (2013) Hydrochemistry and natural attenuation in leachate from tropical landfills. Thesis (doctorate) of the Department of Ecological Science, VU University Amsterdam, the Netherlands
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J\ 2011
Menezes, D.B. (1995) Diagnosis of the impacts of a dumpsite in São Carlos in the environment. Master’s dissertation (in Portuguese). Escola de Engenharia de São Carlos, Universidade de São Paulo. São Carlos, 1995
Moretto, R.L.; Siqueira Neto, A.C.; Elis, V.R.; Miguel, M.G. Detection of leachate pockets in experimental cell of municipal solid waste with aid of geophysics. Sardinia, Sixteenth International Waste Management and Landfill Symposium, 2017
Moreira, J.V.F. (2019) DNA metabarcoding of the microbiota present in the leachate of the sanitary landfill of the city of Foz do Iguacu-PR, aiming at bioremediation processes. (in Portuguese). Undergraduate thesis (Biotechnology). Instituto Latino-Americano de Ciências da Vida e da Natureza, Universidade Federal da Integração Latino-Americana
Morita AKM, Wendland E (2019a) Hydrogeochemical characterization of an area impacted by an old deposit in a Guarani Aquifer recharge zone. Geociências (UNESP) 38(4):1017–1028
Morita AKM, Wendland E (2019b) Characterization of the solid waste and leachate produced by an old waste deposit. In: Annals of the 6th Symposium of Solid Waste. São Carlos, EESC-USP
Morita AKM, Pelinson NS, Elis VR, Wendland E (2020a) Long-term geophysical monitoring of an abandoned dumpsite area in a Guarani Aquifer recharge zone. J Contam Hydrol 230:103623. https://doi.org/10.1016/j.jconhyd.2020.103623
Morita AKM, Pelinson NS, Wendland E (2020b) Persistent impacts of an abandoned non-sanitary landfill in its surroundings. Environ Monit Assess 192(7):463. https://doi.org/10.1007/s10661-020-08451-7
Pachiadaki MG et al (2017) Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science 358(1046–1051):2017
Pan J, Wang X, Cao A, Zhao G, Zhou C (2019) Screening methane-oxidizing bacteria from municipal solid waste landfills and simulating their effects on methane and ammonia reduction. Environ Sci Pollut Res 26:37082–37091(2019). https://doi.org/10.1007/s11356-019-06545-5
Plugge CM, Balk M, Zoetendal EG, Stams AJ (2002) Gelria glutamica gen. nov., sp. nov., a thermophilic, obligately syntrophic, glutamate-degrading anaerobe. Int J Syst Evol Microbiol 52:401–407
PMSC. (2011) Detailed environmental investigation (final report) (in Portuguese). São Carlos local council, São Carlos, 2011
Quast C, Pruesse E et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013:D590–D596
Quintaes BR, Campos JC, Miguel MAL, Leite AMO, Hinojosa MAG (2014) Use of molecular tools in studies about microbial communities in leachates from urban solid waste deposits (in Portuguese). Revista Brasileira de Ciências Ambientais\ 31(2014)
Rajasekar A, Sekar R, Medina-Roldan E, Bridge J, Moy CKS, Wikinson S (2018) Next-generation sequencing showing potential leachate influence on bacterial communities around a landfill in China. Can J Microbiol 64(8):537–549. https://doi.org/10.1139/cjm-2017-0543
Salam M, Varma A (2019) Bacterial community structure in soils contaminated with electronic waste pollutants from Delhi NCR, India. Electronic J Biotechnol 41(2019):72–80
Santos, A.L.; Peixoto, R.; Rosado, A.S. (2009) New approaches to understanding microbial diversity in wastewater, landfills and leachate treatment. \ 13(4): 631-648, doi:https://doi.org/10.4257/oeco.2009.1304.07
Sawamura H, Yamada M, Endo K, Soda S, Ishigaki T, Ike M (2010) Characterization of microorganisms at different landfill depths using carbon-utilization patterns and 16S rRNA gene based T-RFLP. J Biosci Bioeng 109:2
Shinzato, MPB. (2014) Mobilization of pollutants in a deactivated waste deposit. Doctoral thesis (in Portuguese). Escola de Engenharia de São Carlos, Universidade de São Paulo, 2014
Silva TR, Verde LCL, Neto EVS, Oliveira VM (2013) Diversity analyses of microbial communities in petroleum samples from Brazilian oil fields. Int Biodeterior Biodegrad 81:57–70
Song L, Wang Y, Zhao H, Long D (2015) Composition of bacterial and archaeal communities during landfill refuse decomposition processes. Microbiol Res 181:105–111
Souto, G. D. A. B.; Povinelli, J. (2011) Tables of typical characteristics of the leachate from landfills in Brazil: acidic and methanogenic phases In: Congresso Brasileiro de Engenharia Sanitária e Ambiental (Brazilian Conference), 26
Stamps BW, Lules CN, Suflita JM, Masoner JR, Cozzarelli IM, Kolpin DW, Stevenson BS (2016) Municipal solid waste landfills harbor distinct microbiomes. Front Microbiol 7:2016
Wang J, Deng H, Wu S-S, Deng Y-C, Liu L, Han C, Jiang YB, Zhong W-H (2019) Assessment of abundance and diversity of exoelectrogenic bacteria in soil under different land use types. CATENA 172:572–580. https://doi.org/10.1016/j.catena.2018.09.028
Wang T, Yuan Z, Yao J (2018) A combined approach to evaluate activity and structure of soil microbial community in long-term heavy metals contaminated soils. Environ Eng Res 23(2018):62–69
Wang X, Cao A, Zhao G, Zhou C, Xu R (2017) Microbial community structure and diversity in a municipal solid waste landfill. Waste manage 66:2017
Wendland E, Barreto CEAG, Gomes LH (2007) Water balance in the Guarani Aquifer outcrop zone based on hydrogeologic monitoring. J Hydrol (Amsterdam) 342(2007):261–269
Wong JTF, Chenb X, Deng W, Chai Y, Ng CWW, Wong MH (2019) Effects of biochar on bacterial communities in a newly established landfill cover topsoil. J\ Environ Manag 236(2019):667–673
Wong YM, Show PL, Wu TY, Leong HY, Ibrahim S, Juan JC (2018) Production of bio-hydrogen from dairy wastewater using pretreated landfill leachate sludge as an inoculum. JBioscience Bioeng 127(2019):150–159
Wu Z, Gao G, Wang Y (2019) Effects of soil properties, heavy metals, and PBDEs on microbial community of e-waste contaminated soil. Ecotoxicol Environ Safety 180(2019):705–714
Xu S, Lu W, Liu Y, Ming Z, Liu Y, Meng R, Wang H (2017) Structure and diversity of bacterial communities in two large sanitary landfills in China as revealed by high-throughput sequencing (MiSeq). Waste Manage 63(2017):41–48
Zainun MY, Simarani K (2018) Metagenomics profiling for assessing microbial diversity in both active and closed landfills. Sci Total Environ 616–617(2018):269–278
Zolnikov TR, Silva RC, Tuesta AA, Marques CP, Cruvinel VRN (2018) Ineffective waste site closures in Brazil: a systematic review on continuing health conditions and occupational hazards of waste collectors. Waste Manag 80:26–39
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brazil (CAPES) - Finance Code 001, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, grant number 168734/2018-4) and by the São Paulo Research Foundation (FAPESP, grant numbers 2015/03806-1 and 2018/24615-8).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Diane Purchase
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• Proteobacteria and Firmicutes were the most abundant phyla in the landfill.
• Lower COD, alkalinity, PO43−, SO42−, N-NH4−, N-NO3−, TKN, Na, Mg, and K values were associated with higher ecological indexes.
• COD, Na, K, TKN, and ORP strongly influenced the populations.
• Exiguobacterium was identified in samples with higher organic matter concentrations.
• Zones representative of active and closed landfills co-exist inside the deposit.
Electronic supplementary material
ESM 1
(DOCX 38 kb)
Rights and permissions
About this article
Cite this article
Morita, A.K.M., Sakamoto, I.K., Varesche, M.B.A. et al. Microbial structure and diversity in non-sanitary landfills and association with physicochemical parameters. Environ Sci Pollut Res 27, 40690–40705 (2020). https://doi.org/10.1007/s11356-020-10097-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-020-10097-4